-
PDF
- Split View
-
Views
-
Cite
Cite
Ágnes Ősz, Lőrinc Sándor Pongor, Danuta Szirmai, Balázs Győrffy, A snapshot of 3649 Web-based services published between 1994 and 2017 shows a decrease in availability after 2 years, Briefings in Bioinformatics, Volume 20, Issue 3, May 2019, Pages 1004–1010, https://doi.org/10.1093/bib/bbx159
Close - Share Icon Share
Abstract
The long-term availability of online Web services is of utmost importance to ensure reproducibility of analytical results. However, because of lack of maintenance following acceptance, many servers become unavailable after a short period of time. Our aim was to monitor the accessibility and the decay rate of published Web services as well as to determine the factors underlying trends changes.
We searched PubMed to identify publications containing Web server-related terms published between 1994 and 2017. Automatic and manual screening was used to check the status of each Web service. Kruskall–Wallis, Mann–Whitney and Chi-square tests were used to evaluate various parameters, including availability, accessibility, platform, origin of authors, citation, journal impact factor and publication year.
We identified 3649 publications in 375 journals of which 2522 (69%) were currently active. Over 95% of sites were running in the first 2 years, but this rate dropped to 84% in the third year and gradually sank afterwards (P < 1e-16). The mean half-life of Web services is 10.39 years. Working Web services were published in journals with higher impact factors (P = 4.8e-04). Services published before the year 2000 received minimal attention. The citation of offline services was less than for those online (P = 0.022). The majority of Web services provide analytical tools, and the proportion of databases is slowly decreasing. Conclusions. Almost one-third of Web services published to date went out of service. We recommend continued support of Web-based services to increase the reproducibility of published results.
Introduction
The advent of the omics techniques, including genomics, transcriptomics, proteomics and metabolomics enabled the entire spectrum of the data for a particular cellular component to be obtained in a single experiment. Omics experiments almost always generate big data. It is becoming an increasingly challenging task to analyse such data—to find a needle in a multidimensional haystack. New, Web-based analytical tools arose to help, and many of these enable the storage, integrated analysis and biological interpretation of such databases. The Omictools search engine (https://omictools.com/) provides a collection of more than a thousand Web-based resources available for omics data analysis [1].
Today, many journals offer platforms for publishing Web-based applications or databases. For example, Nucleic Acids Research (Oxford Academic Journals) has published an annual Web server [2] and a database issue [3] for >10 years in a row. Catalogues of these Web servers and databases are also available (http://bioinformatics.ca/links_directory/ and http://www.oxfordjournals.org/nar/database/c/). Another major player in this field is Bioinformatics (Oxford Academic Journals), a journal dedicated to genome bioinformatics and computational biology research. Both Nucleic Acids Research and Bioinformatics request a 2-year maintenance of the published services from the authors after publication (Table 1).
Features and maintenance requirements of the top five journals publishing Web-based tools and resources
| Journal . | Publisher . | Published Web services up to 1 January 2017 . | IF (2016) . | H-index (2016) . | SJR ranking (2016) . | Requirements for acceptance . | Suggestions for acceptance . | Required maintenance period . | Suggested maintenance . |
|---|---|---|---|---|---|---|---|---|---|
| Nucleic Acid Research (including Web server and Database issue) | Oxford | 1159 | 10.2 | 414 | D1(7.4) | Must be freely available | Available source code through an open-source license | Full 2 years following publication | Using of stable URLs |
| Bioinformatics | Oxford | 686 | 7.3 | 300 | D1(4.9) | Must be freely available to non-commercial users. Test data should be made available. At the minimum, authors must provide one of: Web server, source code or binary | Available source code through an open-source license | Full 2 years following publication | Using of stable URLs |
| BMC Bioinformatics | BMC | 260 | 2.5 | 163 | Q1(1.5) | Must be freely available to any researcher wishing to use them for non-commercial purposes without restrictions | Available source code through an open-source license | No maintenance period required | Using of stable URLs.An archive of the source code of the current version of the software should be included as a supplementary file |
| PLoS One | PLoS | 176 | 2.8 | 218 | Q1(1.2) | Software and databases should be open source, deposited in an appropriate archive and conform to the open-source definition | Dependency on commercial software does not preclude a paper from consideration, although complete open-source solutions are preferred | No maintenance period required | NA |
| Proteins: Structure, Function, and Bioinformatics | Wiley | 86 | 2.3 | 169 | Q2(1.3) | No special requirements | NA | No maintenance period required | NA |
| Journal . | Publisher . | Published Web services up to 1 January 2017 . | IF (2016) . | H-index (2016) . | SJR ranking (2016) . | Requirements for acceptance . | Suggestions for acceptance . | Required maintenance period . | Suggested maintenance . |
|---|---|---|---|---|---|---|---|---|---|
| Nucleic Acid Research (including Web server and Database issue) | Oxford | 1159 | 10.2 | 414 | D1(7.4) | Must be freely available | Available source code through an open-source license | Full 2 years following publication | Using of stable URLs |
| Bioinformatics | Oxford | 686 | 7.3 | 300 | D1(4.9) | Must be freely available to non-commercial users. Test data should be made available. At the minimum, authors must provide one of: Web server, source code or binary | Available source code through an open-source license | Full 2 years following publication | Using of stable URLs |
| BMC Bioinformatics | BMC | 260 | 2.5 | 163 | Q1(1.5) | Must be freely available to any researcher wishing to use them for non-commercial purposes without restrictions | Available source code through an open-source license | No maintenance period required | Using of stable URLs.An archive of the source code of the current version of the software should be included as a supplementary file |
| PLoS One | PLoS | 176 | 2.8 | 218 | Q1(1.2) | Software and databases should be open source, deposited in an appropriate archive and conform to the open-source definition | Dependency on commercial software does not preclude a paper from consideration, although complete open-source solutions are preferred | No maintenance period required | NA |
| Proteins: Structure, Function, and Bioinformatics | Wiley | 86 | 2.3 | 169 | Q2(1.3) | No special requirements | NA | No maintenance period required | NA |
Features and maintenance requirements of the top five journals publishing Web-based tools and resources
| Journal . | Publisher . | Published Web services up to 1 January 2017 . | IF (2016) . | H-index (2016) . | SJR ranking (2016) . | Requirements for acceptance . | Suggestions for acceptance . | Required maintenance period . | Suggested maintenance . |
|---|---|---|---|---|---|---|---|---|---|
| Nucleic Acid Research (including Web server and Database issue) | Oxford | 1159 | 10.2 | 414 | D1(7.4) | Must be freely available | Available source code through an open-source license | Full 2 years following publication | Using of stable URLs |
| Bioinformatics | Oxford | 686 | 7.3 | 300 | D1(4.9) | Must be freely available to non-commercial users. Test data should be made available. At the minimum, authors must provide one of: Web server, source code or binary | Available source code through an open-source license | Full 2 years following publication | Using of stable URLs |
| BMC Bioinformatics | BMC | 260 | 2.5 | 163 | Q1(1.5) | Must be freely available to any researcher wishing to use them for non-commercial purposes without restrictions | Available source code through an open-source license | No maintenance period required | Using of stable URLs.An archive of the source code of the current version of the software should be included as a supplementary file |
| PLoS One | PLoS | 176 | 2.8 | 218 | Q1(1.2) | Software and databases should be open source, deposited in an appropriate archive and conform to the open-source definition | Dependency on commercial software does not preclude a paper from consideration, although complete open-source solutions are preferred | No maintenance period required | NA |
| Proteins: Structure, Function, and Bioinformatics | Wiley | 86 | 2.3 | 169 | Q2(1.3) | No special requirements | NA | No maintenance period required | NA |
| Journal . | Publisher . | Published Web services up to 1 January 2017 . | IF (2016) . | H-index (2016) . | SJR ranking (2016) . | Requirements for acceptance . | Suggestions for acceptance . | Required maintenance period . | Suggested maintenance . |
|---|---|---|---|---|---|---|---|---|---|
| Nucleic Acid Research (including Web server and Database issue) | Oxford | 1159 | 10.2 | 414 | D1(7.4) | Must be freely available | Available source code through an open-source license | Full 2 years following publication | Using of stable URLs |
| Bioinformatics | Oxford | 686 | 7.3 | 300 | D1(4.9) | Must be freely available to non-commercial users. Test data should be made available. At the minimum, authors must provide one of: Web server, source code or binary | Available source code through an open-source license | Full 2 years following publication | Using of stable URLs |
| BMC Bioinformatics | BMC | 260 | 2.5 | 163 | Q1(1.5) | Must be freely available to any researcher wishing to use them for non-commercial purposes without restrictions | Available source code through an open-source license | No maintenance period required | Using of stable URLs.An archive of the source code of the current version of the software should be included as a supplementary file |
| PLoS One | PLoS | 176 | 2.8 | 218 | Q1(1.2) | Software and databases should be open source, deposited in an appropriate archive and conform to the open-source definition | Dependency on commercial software does not preclude a paper from consideration, although complete open-source solutions are preferred | No maintenance period required | NA |
| Proteins: Structure, Function, and Bioinformatics | Wiley | 86 | 2.3 | 169 | Q2(1.3) | No special requirements | NA | No maintenance period required | NA |
Reproducibility is a primary requirement for scientific publications [4], but it depends on many factors. Accessibility and continuous maintenance are the most fundamental factors that enable the reproducibility of study results of in silico studies and databases. An important cornerstone in this process is the maintenance of a constant Uniform Record Locator (URL) — a Web link that directs researchers to references, data sets and other informational resources [5]. Multiple studies investigated the accessibility of Web references in PubMed abstracts, and the reported lifespan of the websites was relatively short—up to 23% of the URL references became inactive after 1 year and up to 52% after 5 years [6–8].
How can a Web service persist longer? Three major bioinformatics centres, the US National Centre for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov/), the European Bioinformatics Institute (EMBL-EBI, http://www.ebi.ac.uk/) and the Swiss Institute for Bioinformatics (SIB, http://www.sib.swiss/>) [9] maintain a few databases and tools mainly by governmental support. However, these institutions do not have a mechanism to ‘import’ services developed elsewhere. In general, no funding agencies or grant schemes enable independent academic authors to maintain published Web services or keep them accessible for longer periods of time.
What is the half-life of a published Web service — more specifically, what is the probability that a service remains online after 1, 2 or 5 years? With an increasing volume of Web services and publication options, life scientists and other researchers expect long-lasting maintenance from the provider. Our aims were to evaluate the decay rate of Web-based services and to provide a comprehensive status report of factors that affect their endurance. We evaluated available online databases and tools published via PubMed in the past 22 years for this purpose.
Methods
Data set
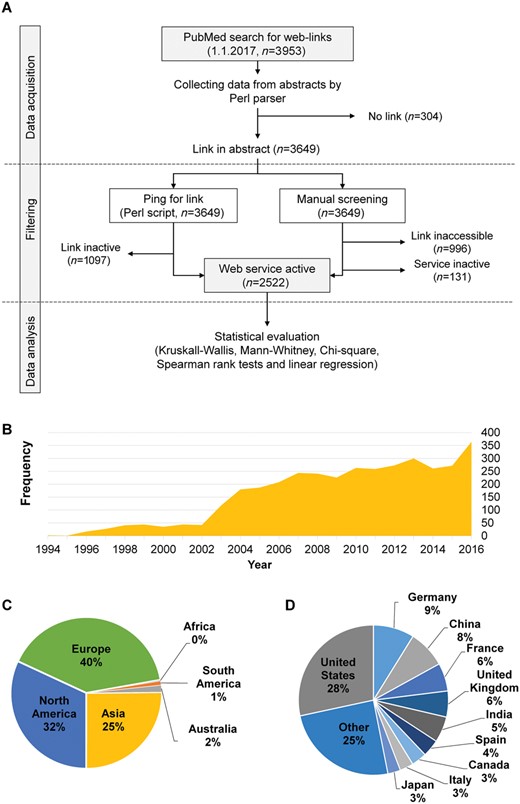
Figure 1A summarizes the study workflow. A literature search was performed in PubMed (http://www.pubmed.com/) to collect Web services published before 1 January 2017, and used following search terms: ‘(www[Title/Abstract] OR http[Title/Abstract]) AND (online tool[Title/Abstract] OR web server[Title/Abstract] OR server[Title/Abstract])’. We downloaded all abstracts in a .txt file (Supplementary File 1) and parsed this with a Perl script (Supplementary File 2). Journal name, publication year, title, PMID and URL were collected into a ‘.tsv’ file (Supplementary Table S1).
Evaluation of Web services available in PubMed. Overview of the study protocol (A). Annual publications of new Web services from 1994 to 2016 show an average yearly increase of 30.8% (B). The proportion of published Web services based on the affiliation of the last author by continents (C), and by country (D).
Screening and filtering
Each published URL status was automatically pinged by using a Perl script (Supplementary File 3) at least three different times based on a Parser output file. The status information was inserted into the same file (Supplementary Table S1).
To validate the results of the automatic screening and to collect additional data, an additional manual screening was executed between January and May of 2017 separately for each study. In addition to website status (active/inactive), classification (database/tool/both), platform (online/desktop/both) and registration request (yes/no) information were collected. Impact factor as of 2016, overall citations and the affiliations of the last author (country and continent) were downloaded from the SCOPUS database and linked to the individual publications.
Statistical analysis
Results
The number of Web servers keeps increasing
A total of 3953 Web services published in 476 journals from 1994 to 2016 were found for the search term in PubMed. The Parser script identified 3649 articles containing website links in 375 journals. The remainder (n = 304) contained either supplementary data links or other non-relevant information. For those selected, author affiliation data (n = 2996), citations data (n = 2932) and impact factor data (n = 3223) were acquired from SCOPUS (Supplementary Table S1).
The data show an ongoing increasing trend in the number of Web service publications (Figure 1B). The average annual increase in the total number of tools is 30.8%. Although the majority of publications originated from Europe (40%) (Figure 1C), with respect to individual countries, the largest proportion of the tools stemmed from the United States (28%) followed by Germany (9%) and China (8%) (Figure 1D). The bulk of the publications (65%) was published in only five journals, with Nucleic Acids Research and Bioinformatics alone publishing more than half of all Web services.
Availability of Web services
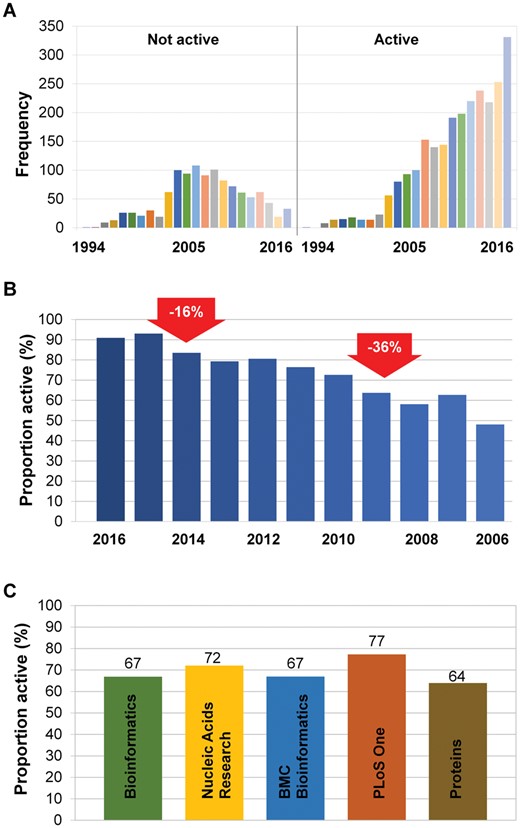
A total of 73% of the Web services were accessible during the automatic or manual screening on at least one occasion. For the accessible websites, a further 4% (n = 131) were inactive, so altogether, 69% of websites were functioning at the time of our study (n = 2522). The proportion of accessible sites shows a significant inverse correlation with the age of the publication (P < 1e-16). The total number of active and inactive services for each year is shown in Figure 2A.Interestingly, the highest proportion of accessible websites was found in the second year (which was 95% in 2015, in our 2017 survey). In the first 2 years after publication, 8% of the websites were offline, then the proportion of active sites indicated two noteworthy declines, one after the second year (a further 11% decay) and another after the sixth year (an additional 7% decay in 2010). Thus, after the seventh year, more than a third of all Web services were inactive (red arrows in Figure 2B). The mean half-life of Web services published in the past 5 years was 10.39 years.
Dynamic change through time. A remarkably high proportion (31%) of published Web services are already inactive. The distribution of Web services from 1994 until 2016 based on accessibility (A). The relative frequency of active Web services between 2008 and 2016. The arrows indicate the rates of the decrease in the proportion of active Web services (B). The relative proportion of active tools by journal (C).
When comparing the proportion of active sites among the top five journals, PLoS One had the highest rate of active sites (total = 77%) (Figure 2C). However, when we performed a regression analysis to compute significance and included age as an additional parameter, the difference between PLoS One and the other journals was not significant. PLoS One is a relatively new journal in this field, and the mean age of the papers it contains is therefore low. In this model, Nucleic Acids Research was the only journal that showed statistical significance compared with the other journals (P = 9.6e-09), and the Web services published in Nucleic Acids Research had a 10% higher chance of accessibility compared with services published elsewhere during the same period. The regression showed that age was linked to a 3.5% yearly decay of working sites (P = 2e-16).
We assigned the missing tools to eight major categories. These encompass genome analysis (this includes genome annotation, genome assembly, genome analysis, genome database, phylogenomics, comparative genomics, genome variant analysis, genome editing, DNA structure analysis, epigenomics, sequence alignment, genome bowser), transcriptome analysis (including gene expression analysis, RNA modification analysis, RNA interference, non-coding RNA analysis, RNA structure analysis, gene expression regulation), proteome analysis (including protein sequence analysis, protein comparison analysis, protein structure analysis, metabolomics, drug discovery, protein annotation), network analysis tools (including biological network analysis, biological network data, mathematical modelling, synthetic biology), phenotype (GWAS analysis, linkage analysis, QTL mapping, eQTL mapping, phenomics), text mining, visualization tools and others (education, metagenome, climate change, etc.). Of these, proteome analysis (35.5% of all discontinued tools), genome analysis (20.0%), network analysis tools (14.2%) and transcriptome analysis (13.8%) represent the major categories, while all others combined reach about 16% of discontinued tools. The top 100 (in terms of citations) unavailable Web services as well as their respective categories is presented in Supplementary Table S2.
Many Web links vanished over time or included a mistyping error in the URL. We recovered 329 revised links (9% of all links) by a manual internet search or automatic redirection, and 93% of these were online. The earliest published Web service still active was the ‘SBase protein domain library’ published in Nucleic Acids Research in 1994 [10], which is currently running in a novel Web domain (http://pongor.itk.ppke.hu/? q=bioinfoservices).
Features of active tools
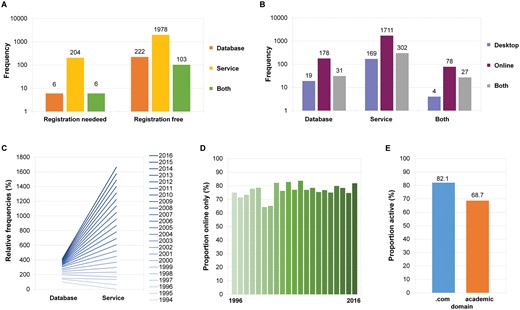
Most of published Web tools with a required registration offer an analysis service (P = 0.001, Figure 3A). No significant correlation is present between the environment type (online/desktop) and the tool type (Figure 3B). Historically, most of the first Web tools offered access to databases. While the increase in the number of new databases is slowing down, the growth in services remains linear, which shifts the proportion in favour of services (Figure 3C). Most tools are online only (64–84%), and this seems to be constant with no significant trend in either direction (Figure 3D).
Features of active Web tools. Most that require a registration offer a service (A). Distribution of the various type of Web tools (service, database, both types) by environment (online only, desktop only, both environments) on a logarithmic scale (B). The yearly distribution of active Web services by type shows an increased proportion of services (C). The yearly proportion of active Web services available online only does not show a significant trend (D). Sites with .com domain were more likely to work (E).
The proportion of active tools also shows differences among the various continents. However, because of the low number of publications originating in Africa (n = 6) and South America (n = 27), no meaningful comparison is possible. Considering publishing in the top 10 countries, the greatest proportion of active tools belongs to Canada (82%) and the least to Japan (44%) (Table 2).
Distribution of Web services in the top 10 most publishing countries by active status
| Continent . | Country . | N active . | N inactive . | Percentage active . |
|---|---|---|---|---|
| North America | Canada | 81 | 18 | 81.8 |
| Asia | India | 128 | 39 | 76.6 |
| Other countries | 536 | 205 | 72.3 | |
| Europe | Spain | 76 | 35 | 68.5 |
| Europe | Italy | 57 | 31 | 64.8 |
| North America | The United States | 550 | 300 | 64.7 |
| Europe | Germany | 171 | 94 | 64.5 |
| Europe | France | 115 | 66 | 63.5 |
| Europe | UK | 102 | 66 | 60.7 |
| Asia | China | 132 | 112 | 54.1 |
| Asia | Japan | 36 | 46 | 43.9 |
| Continent . | Country . | N active . | N inactive . | Percentage active . |
|---|---|---|---|---|
| North America | Canada | 81 | 18 | 81.8 |
| Asia | India | 128 | 39 | 76.6 |
| Other countries | 536 | 205 | 72.3 | |
| Europe | Spain | 76 | 35 | 68.5 |
| Europe | Italy | 57 | 31 | 64.8 |
| North America | The United States | 550 | 300 | 64.7 |
| Europe | Germany | 171 | 94 | 64.5 |
| Europe | France | 115 | 66 | 63.5 |
| Europe | UK | 102 | 66 | 60.7 |
| Asia | China | 132 | 112 | 54.1 |
| Asia | Japan | 36 | 46 | 43.9 |
Distribution of Web services in the top 10 most publishing countries by active status
| Continent . | Country . | N active . | N inactive . | Percentage active . |
|---|---|---|---|---|
| North America | Canada | 81 | 18 | 81.8 |
| Asia | India | 128 | 39 | 76.6 |
| Other countries | 536 | 205 | 72.3 | |
| Europe | Spain | 76 | 35 | 68.5 |
| Europe | Italy | 57 | 31 | 64.8 |
| North America | The United States | 550 | 300 | 64.7 |
| Europe | Germany | 171 | 94 | 64.5 |
| Europe | France | 115 | 66 | 63.5 |
| Europe | UK | 102 | 66 | 60.7 |
| Asia | China | 132 | 112 | 54.1 |
| Asia | Japan | 36 | 46 | 43.9 |
| Continent . | Country . | N active . | N inactive . | Percentage active . |
|---|---|---|---|---|
| North America | Canada | 81 | 18 | 81.8 |
| Asia | India | 128 | 39 | 76.6 |
| Other countries | 536 | 205 | 72.3 | |
| Europe | Spain | 76 | 35 | 68.5 |
| Europe | Italy | 57 | 31 | 64.8 |
| North America | The United States | 550 | 300 | 64.7 |
| Europe | Germany | 171 | 94 | 64.5 |
| Europe | France | 115 | 66 | 63.5 |
| Europe | UK | 102 | 66 | 60.7 |
| Asia | China | 132 | 112 | 54.1 |
| Asia | Japan | 36 | 46 | 43.9 |
Most tools were developed by universities or academic institutes (.org, .edu, .gov and .univ), but 117 services had a .com domain—however, the .com domains were more likely to be active (82.1% compared with 68.7% in non-profit sector, Figure 3E). NCBI, EMBL-EBI and SIB maintain 121 tools, 85 of which are active (70.3%, compared with 69.1% of services published elsewhere).
The citation and impact factors of Web tools
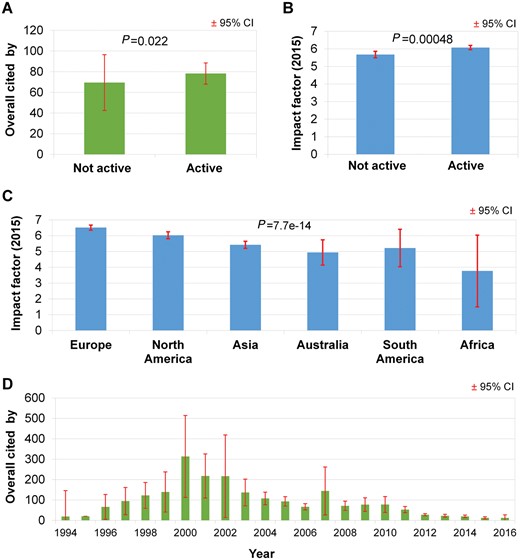
Active tools received more citations than inactive tools (P = 0.022); however, the numerical difference is surprisingly small (Figure 4A). At the same time, journals with a higher impact factor are more likely to retain an active tool — again, with an almost negligible numerical difference (P = 0.00048, Figure 4B). Distribution of impact factor between continents revealed highly significant differences (P = 7.7e-14). The mean impact factor was highest for studies published from Europe and lowest for studies from Africa (Figure 4C).
Impact of Web services shows significant differences based on accessibility. The mean citation rate of active Web tools is higher than for inactive tools (A). The mean impact factor of Web services by active status (B). The distribution of the mean impact factor by continent shows that Europe is first and Africa is last (C). The mean citation of Web services per year shows minimal attention to papers published before the year 2000 (D). The red lines show the 95% confidence interval in all graphs.
After the year 2000, citations show a strong correlation with the age of the publication. Interestingly, the number of citations is lower for pre-2000 tools (Figure 4D). The most cited publication (ClustalW2) [11] has already received over 18 000 citations.
Discussion
In the present study, we validated the time-related decay of Web-based services and databases separately by an automated and by a manual analysis. To our knowledge, no similar study has evaluated the activity of Internet-based tools to date. However, the presence and availability of URLs have already been appraised in multiple publications. For example, a previous study verified the presence and availability of URLs in Medline abstracts [8]. They identified 1630 unique links, and only 63% of these were available according to an automatic screening. Thus, they revealed similar proportion of availability of published URLs as in our study.
A more recent study found that 35% of URL references were offline within 18 months after the original publication in Annals of Emergency Medicine [12]. URL references in publications between 1999 and 2004 were analysed in five biomedical informatics journals [13], and again had a similar accessibility rate (69% online). Dellavalle et al. [7] revealed that 13% of Internet references in three of the top 1% cited US scientific journals were inactive at 27 months after publication. We can confirm these observations with 16% loss of active Web services within the first 2 years after acceptance.
Another study that investigated the New England Journal of Medicine and The Lancet revealed 15 and 18% of inaccessible Internet references [14]. However, it was possible to identify a correct address for 2–4% of these lost links. Wren [8] reported formatting or spelling errors in 12% of published URLs in Medline. Later, they found that citations of published Web services implicitly foresee permanent online availability of these services [15]. Nevertheless, many highly cited tools were also discontinued. It is highly probable that there is also a certain level of redundancy and more advanced services have replaced those outdated—however, it was not possible to perform such a detailed literature search, which would enable to identify other cutting-edge services providing the same functionality. However, we have to note that not each unavailable service was discontinued. We were able to identify a new, updated link for 329 of the broken addresses (23% of offline Web services) by a manual Internet search.
The correct identification of the published Web services is necessary for reproducibility [16]. Resources, including software, seem to be not uniquely identifiable in 46% of publications [17]. The Resource Identification Initiative is designed to help researchers to identify and correctly cite resources from the biomedical literature [18]. Although usage of these initiatives is increasing, misidentification and a lack of accessibility still limit reproducibility [18]. The implementation of a permanent and common archival system and a unified URL citation has already been suggested a few times [13, 14, 19, 20]. Here, we still found numerous studies with the problem of lost references and Web links—a permanent solution for this issue is not the implementation of a new service. Rather, PubMed itself could add a ‘web reference’ category to each study and check the activity of the sites regularly in an automated manner.
Multiple studies raised awareness that the utility of many services is restricted because of unpublished or failing source codes [21, 22]. The Open Source Initiative targets this issue by recommending openly available source codes for software [23]. In our analysis, the source code was available for only 22% of the active sites.
Containers, virtual machines and source code repositories represent possible solutions to ensure the continued availability of Web-based bioinformatics tools and databases. Containers enclose a runtime environment, including the application, dependencies, libraries, configuration files, etc., as one package. A container can help to eliminate differences related to the operating system and/or to the underlying infrastructure. Containers are faster and smaller than virtual machines—the later imitate a dedicated hardware and enclose therefore the entire operating system as well. A physical server can also host multiple virtual machines simultaneously. Available containers contain Docker (https://www.docker.com/), Solaris Containers (http://www.oracle.com/technetwork/server-storage/solaris/containers-169727.html), LXC (https://linuxcontainers.org/) and FreeBSD jails (https://www.freebsd.org). Virtual machines include Virtualbox (https://www.virtualbox.org/), Parallels (https://www.parallels.com/eu/), VMware products (https://www.vmware.com/) and QEMU (https://www.qemu.org/). Finally, a minimal solution would be the utilization of a source code repository. These can not only host the code but can also enable review and management by other developers. GitHub (https://github.com/), Google Code (https://code.google.com/) and SourceForge.net (https://sourceforge.net/) are among the most commonly used source code repositories.
In summary, we evaluated Web services published over a time span of 22 years. Over 95% of sites were running in the first 2 years, but this rate declined to 84% in the third year and became gradually lower afterwards. Tools published before the year 2000 received minimal attention. The majority of Web tools provide an analysis tool, while the proportion of databases is also growing. Based on our results, we suggest that large-scale funding initiatives, such as ELIXIR (https://www.elixir-europe.org/), should create a mechanism for the maintenance of important services.
We validated the time-related decay of Web-based bioinformatics tools and databases by an automated and by a manual analysis separately.
Over 95% of Web services are running in the first 2 years, but this rate becomes gradually lower afterwards.
The impact and citation of Web services show significant differences based on accessibility.
Funding
The study was supported by the NVKP_16-1-2016-0037 grant of the National Research, Development and Innovation Office (NKFIH), Hungary.
Ágnes Ősz is an assistant research fellow at the MTA TTK EI Lendület Cancer Biomarker Research Group and at the Semmelweis University 2nd Department of Pediatrics, Budapest, Hungary. Her main interests include big data and cross-sectional analysis.
Lőrinc Sándor Pongor is a PhD fellow at the MTA TTK EI Lendület Cancer Biomarker Research Group and at the Semmelweis University, 2nd Department of Pediatrics Budapest, Hungary. His main research interests include algorithm development for next generation data analysis.
Danuta Szirmai is a final-year student engaged at the MTA TTK EI Lendület Cancer Biomarker Research Group.
Balázs Győrffy is the head of MTA TTK EI Lendület Cancer Biomarker Research Group, the head of node of ELIXIR Hungary, and a scientific advisor at the Semmelweis University 2nd Department of Pediatrics, Budapest, Hungary. He published over 150 scientific papers in bioinformatics and oncology.