-
PDF
- Split View
-
Views
-
Cite
Cite
Adriano Barbosa-Silva, Dorina Bratfalean, Wei Gu, Venkata Satagopam, Paul Houston, Lauren B Becnel, Serge Eifes, Fabien Richard, Andreas Tielmann, Sascha Herzinger, Kavita Rege, Rudi Balling, Paul Peeters, Reinhard Schneider, on behalf of the European Translational Information and Knowledge Management Services (eTRIKS) Consortium, Presenting and sharing clinical data using the eTRIKS Standards Master Tree for tranSMART, Bioinformatics, Volume 35, Issue 9, May 2019, Pages 1562–1565, https://doi.org/10.1093/bioinformatics/bty809
Close - Share Icon Share
Abstract
Standardization and semantic alignment have been considered one of the major challenges for data integration in clinical research. The inclusion of the CDISC SDTM clinical data standard into the tranSMART i2b2 via a guiding master ontology tree positively impacts and supports the efficacy of data sharing, visualization and exploration across datasets.
We present here a schema for the organization of SDTM variables into the tranSMART i2b2 tree along with a script and test dataset to exemplify the mapping strategy. The eTRIKS master tree concept is demonstrated by making use of fictitious data generated for four patients, including 16 SDTM clinical domains. We describe how the usage of correct visit names and data labels can help to integrate multiple readouts per patient and avoid ETL crashes when running a tranSMART loading routine.
The eTRIKS Master Tree package and test datasets are publicly available at https://doi.org/10.5281/zenodo.1009098 and a functional demo installation at https://public.etriks.org/transmart/datasetExplorer/ under eTRIKS—Master Tree branch, where the discussed examples can be visualized.
1 Introduction
The European Translational Information and Knowledge Management Services (eTRIKS, https://www.etriks.org/, 2017) is tasked with providing tools and services to support data management and analysis for >60 diverse biomedical research projects which have been funded by the Innovative Medicines Initiative (IMI). As Europe’s largest public-private partnership, IMI funds projects ranging from molecular and systems biology to clinical trials and full translational research projects. The community translational research system under use is tranSMART (Athey et al., 2013; Dunn et al., 2017), first developed by the pharma industry, and then gifted to a global translational research development community. The tranSMART system has undergone extensive development extended by its own community and the eTRIKS project, which has focused on an implementation that serves IMI projects users based in the European Union (EU). The flexibility and capability of tranSMART is well presented in a recent paper showing the availability of workflows within a sandbox environment (Satagopam et al., 2016). tranSMART serves as the central knowledge management system for eTRIKS, while other tools and complimentary services applicable to the data value chain, such as data harmonization, sharing, analysis, visualization and preservation, have been developed.
To expedite medical breakthroughs the sharing of clinical research data is vital owing to legislative incentives and increased public pressure, many clinical trial registries are expanding their remit to share not only basic summary trial registration data but also results. Wider data sharing is one way of tackling reporting bias by increasing visibility of successful studies as well as failed ones. Additionally, data standards play a pivotal role in tackling the omnipresent problem of reproducibility. Begley et al. reproduced 53 experiments from landmark publications to find 47 out of 53 could not be replicated; a very worrying trend for preclinical studies that are used as the scientific basis for target identification for new drug development (Begley and Ellis, 2012).
The Data FAIRport initiative in 2014 prescribed a set of guiding principles known as FAIR: Findable, Accessible, Interoperable, Reusable which should be applied where data is deemed scientifically valuable (Wilkinson et al., 2016). Those principles have gained official recognition from G20, NIH and the Directorate General for Research and Innovation of the European Commission. The consistent application of common semantics and data structures, as outlined within data standards, is a key factor to ensure interoperability and reusability of data. The eTRIKS Data Standards Work Package created a Standards Starter Pack (https://doi.org/10.5281/zenodo.50398/, 2016), which outlines the FAIR principles and recommendations for the main clinical and genomic standards as well as supporting vocabularies and minimum information guidelines that should be applied in the entire translational research landscape. eTRIKS has also produced the IMI Data catalogue which centralizes metadata of ongoing and past IMI projects. It is part of the service that eTRIKS provides in its key knowledge management performance with a focus on the findability of project level study description metadata. Furthermore, this well received initiative facilitates broader sharing and accessibility of data (http://datacatalog.elixir-luxembourg.org/ckan/, 2017).
For clinical research data, The Clinical Data Interchange Standards Consortium (CDISC, https://www.cdisc.org/, 2018) data standards have been implemented in over 90 countries, and are now mandated by Food and Drug Administration of the United States (FDA, 2014) and Pharmaceuticals and Medical Devices Agenda (PMDA) in Japan (https://www.pmda.go.jp/files/000206449.pdf, 2018) in order to increase the uptake of data standards, which, when applied, contribute to higher data quality. The lack of implementing standards will render datasets from different cohorts inadequate when integrating with complementary research data for meta-analyses (Elefsinioti et al., 2016). A recent paper by the American College of Medical Genetics and Genomics (Acmg, 2017) discussed the importance of using the information from one patient cohort to benefit other patients. The ACMG’s framework for data sharing will work best if standards are implemented within the framework, as within tranSMART, and datasets are gathered by utilizing those standards from the beginning of the research, as is also recommended by CDISC.
2 Implementation
The eTRIKS Standard Master Tree is based on the standards for clinical data representation developed by CDISC, mainly the Study Data Tabulation Model (SDTM) standard. The proposal of eTRIKS was to create a hierarchical navigation tree in which the raw data, collected at the multiple cohorts, should be promptly mapped to the elements of this tree so that data are loaded automatically with the correct topology into tranSMART i2b2 (Informatics for Integrating Biology and the Bedside) framework. The requirement for this is that all the data collected from a patient will be organized and formatted using the SDTM model. SDTM modeling increases the ability to compare information among systems and/or organizations, whilst also decreasing the time to initiate a new research study. The use of these data standards improves the data quality, their interoperability and their management, which allows easier, faster and more reliable data aggregation.
The eTRIKS Standard Master Tree presents the clinical data within tranSMART i2b2. eTRIKS has radically updated the original tranSMART engine that sorts and presents the clinical data within the system. Users can choose to map their clinical data content to a favorite terminology prior to the SDTM modelling using global standards such as OMICS, NCI (https://www.cancer.gov/digital-standards, 2017) or LOINC (https://loinc.org/, 2017), as long as the SDTM variable names as maintained. Further, the clinical data is mapped to a ‘clinical mapping file’, which requires a good working knowledge of the CDISC foundational standards, in order to represent the SDTM structure of the clinical data correctly in the hierarchy of the tranSMART i2b2 repository (Abend et al., 2009).
In practice if one thinks about the outcome of a ‘glucose test’, this test may be named ‘sugar test’ or ‘glucose test’ in different differing cohorts, which may be well understood by experts but not a machine as the same concept. The use of standard name ‘Glucose Tolerance Test’ (NCBI’s, MeSH Unique ID: D005951) would avoid any confusion or wrong interpretation and enable data query across cohorts. Further to this, considering that the metabolite ‘glucose’ could be measured in different samples (e.g. blood, urine), the test results could be reported in different units (mg/dl or mmol/l) and/or the test could be performed at different periods of the time (screening, visit 1, visit 2, etc.), error prone aspects during the data analysis. If the problem is proposed, ‘How to standardize the manner by which this information should be organized and formatted for effective and precise cohorts comparisons?’ One answer should be: ‘Use a Standard Master Ontology Tree’ or in this case, the eTRIKS Standard Master Tree. The application of this tool coupled with a good application of controlled vocabularies will increase greatly the Reusability and Interoperability Principles mentioned above.
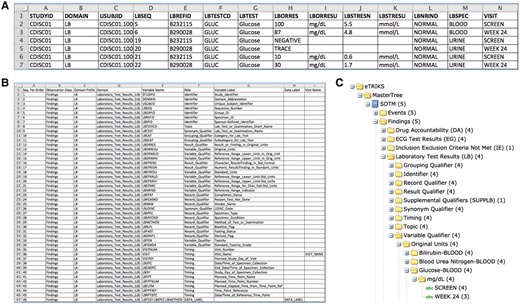
In the ‘Glucose Tolerance Test’ example, upon mapping to the tranSMART Standard Master Tree, the outcome of this test would already be represented as displayed in Figure 1A below. The test result is reported in this example by means of 14 variables (columns A-N) for the subject CDISC01.100008 (column C). Note that column G collects one variable called LBTEST (Lab Test Examination Name), which is filled with the standard value ‘Glucose’ and another variable LBSPEC (Specimen Type) is used to distinguish ‘BLOOD’ from ‘URINE’ samples. In terms of readout values, the variable LBORRES (Result or Finding in Original Units) records the original values as collected reported units in LBORRESU (Original Units) the unit itself (e.g. mg/dl). The example shows results converted to numeric type and this reported value to a standard unit, which is achieved by using the pair variables LBSTRESN (Numeric Result/Finding in Standard Units) and LBSTRESU (Standard Units), for values and units (e.g. mmol/l), respectively. The SDTM Implementation Guide provides a comprehensive description including four sessions: 1—Overview of topics for specific general observation class associated with specific domains, 2—Specification for table of variables, 3—Rules for correct implementation of standards and 4—Examples.
Content representation of the eTRIKS Master Tree Package. (A) SDTM data file for one patient (USUBJID) for the LB domain. (B) SMOT_Lite definition session for the LB domain. (C) Hierarchical (i2b2) tree created for the LB domain and displayed in the tranSMART web app
To avoid the all too common pitfalls of redundant data eTRIKS developed the Standard Master Tree, using the comprehensive SDTM domain structure, to support and give structure and context to the data so it can be easily identified. The Standard Master Tree follows a basic and easy-to-understand logic, which was built upon the premise of tranSMART rules for data loading. This means that multiple data collected for one patient for the same domain (e.g. Laboratory Test Results—LB) should be distinguished based on Data Labels. This way, for the LB domain, results of ‘Glucose’ and ‘Creatinine’ tests for example, could be loaded in the same run. Moreover, multiple results for the same test should also be distinguished based of the Visit Names. Respecting these two basic rules, any results from any sort of laboratory tests and even results for any other domains, can be easily represented in tranSMART via the Standard Master Tree.
3 Features
The eTRIKS Standard Master Tree model consists of a package of three main components: (i) a CDISC clinical dataset reported as define.xml metadata and converted in .txt tabulate files composing of 16 SDTM domains represented as review data as collected for 4 fictitious subjects (Fig. 1A); (ii) the tranSMART standard master ontology tree as TM SMOT-SDTM_lite.txt definition file, where all the information concerned about the correct positioning of the SDTM variables can be found (Fig. 1B); and the (iii) Mapper script where users can map their data files to the TM SMOT-SDTM_Lite definition, avoiding manual work. The script reads a target directory containing the input SDTM files and maps all the collected variables against the SMOT-SDMT_Lite.txt master tree file mentioned above. This is achieved with a single command line: ‘php mapper.php SMOT_Lite.txt CDISC01_ClinicalData’ (further details are explained on the package’s README file). Figure 1A depicts an example for the Glucose test of one such subject. Figure 1B depicts part of the TM SMOT-SDTM_Lite file where definitions for the domain LB is displayed (Note the seven columns required for the annotation of each of the SDTM variables used in this domain). This information can be found easily on the SDTM implementation guide as should be adopted by the data curators. Finally, Figure 1C displays the graphical hierarchy tree, known as tranSMART i2b2 tree, where the loaded data can be further queried and used to create comparison subsets on the tranSMART i2b2 web app, these can be visualized in a sandbox implementation available at http://public.etriks.org/transmart/datasetExplorer under the eTRIKS—Master Tree branch.
The strategy for clinical research data standards representation proposed above offers a readily available method to integrate multiple translational research datasets while meeting the Interoperability and Reusability aspects of the FAIR principles. Once the data is within the eTRIKS Standard Master Tree, it can then take advantage of the tranSMART environment, where it will receive a unique study and server specific identifier and the metadata can be given greater and essential specificity. With an effective tranSMART search tool where multiple datasets and/or studies can be pooled and queried, coupled with an entry within the eTRIKS data catalogue the data has undergone FAIR-ification to a satisfactory degree. Now the data is Findable and also Accessible, and it can begin its hopefully long life adding scientific value to any number of future studies or aggregated data comparisons.
4 Conclusion
The tranSMART Standard Master Tree presented here adds to other efforts to make other software data interoperable with tranSMART. Projects such as ‘ODM to i2b2’ converts data stored in XML/ODM based systems such as OpenClinica and REDCap into i2b2 format (https://github.com/CTMM-TraIT/trait_odm_to_i2b2, 2018); and ‘REDCap2SDTM’ converts electronic data capture system data to SDTM (Yamamoto et al., 2017). Taken together, this software could benefit from the Master Tree concept in order to standardize the manner that SDTM studies should appear within a tranSMART navigation tree to users.
If the tools and processes above are adopted in the scope of the NIH funded projects, it will contribute greatly to creating an overseas bridge for data sharing initiatives with the EU/EFPIA-funded (IMI) translational medicine research projects, of which over 60 are being supported by the eTRIKS project. While not all of the eTRIKS supported projects have implemented the tranSMART Standards Master Tree they have all received the appropriate guidance and advice from eTRIKS experts or as laid out in the eTRIKS standards starter pack. Tremendous curation efforts were necessary to guarantee that IMI data was collected in good quality, once that, frequently, the big challenge for translational research projects lies on the quality of the data itself, not only its metadata. The adoption of the technologies and standards developed and presented in this paper will support a significant step towards a position where IMI data can be shared, and the findings reproduced to benefit the health care research community, allowing a standardized representation of SDTM data across multiple tranSMART servers. The eTRIKS Standard Master Tree package can be downloaded at https://doi.org/10.5281/zenodo.1009098.
Acknowledgements
Experiments presented in this paper were carried out using the HPC facilities of University of Luxembourg (http://hpc.uni.lu).
Funding
This work has received support from the EU/EFPIA Innovative Medicines Initiative Joint Undertaking eTRIKS grant no. 115446.
Conflict of Interest: none declared.
References
FDA. (