-
PDF
- Split View
-
Views
-
Cite
Cite
Yu Jiang, Hongmei Zhang, Shan V Andrews, Hasan Arshad, Susan Ewart, John W Holloway, M Daniele Fallin, Kelly M Bakulski, Wilfried Karmaus, Estimation of eosinophil cells in cord blood with references based on blood in adults via Bayesian measurement error modeling, Bioinformatics, Volume 36, Issue 6, March 2020, Pages 1923–1924, https://doi.org/10.1093/bioinformatics/btz839
Close - Share Icon Share
Abstract
Eosinophils are phagocytic white blood cells with a variety of roles in the immune system. In situations where actual counts are not available, high quality approximations of their cell proportions using indirect markers are critical.
We develop a Bayesian measurement error model to estimate proportions of eosinophils in cord blood, using the cord blood DNA methylation profiles, based on markers of eosinophil cell heterogeneity in blood of adults. The proposed method can be directly extended to other cells across different reference panels. We demonstrate the method’s estimation accuracy using B cells and show that the findings support the proposed approach. The method has been incorporated into the estimateCellCounts function in the minfi package to estimate eosinophil cells proportions in cord blood.
estimateCellCounts function is implemented and available in Bioconductor package minfi.
Supplementary data are available at Bioinformatics online.
1 Introduction
Eosinophils are white blood cells that perform a variety of functions, e.g. helping combat multicellular parasites and certain infections in vertebrates. Eosinophils are implicated in various diseases including asthma and allergy. The proportion of eosinophils relative to other cells in umbilical cord blood has the ability to predict respiratory illnesses in high risk infants (Berek, 2016; Junge et al., 2014). It is important to correctly identify eosinophils and estimate their proportions in cord blood.
To estimate eosinophil counts in biological samples, cell counting is often needed. However, this is not feasible in many settings due to the low proportion of eosinophils in the blood, limitations of fresh cord blood samples and difficulty in discriminating eosinophils from other granulocytes (Ethier et al., 2014). An indirect method for estimating cell compositions, which uses DNA methylation (DNAm) profiles to infer cell proportions, has been developed (Houseman et al., 2015) and incorporated into an R package minfi. This approach estimates eosinophil proportions in peripheral blood in adults. However, estimation bias is noted when the method is applied to samples collected from cord blood or other tissues due to the lack of eosinophil-specific reference data (Aryee et al., 2014). We develop a Bayesian measurement error model aiming to correct the bias in the estimation of eosinophil cell proportions in cord blood when using a reference database based on blood in adults.
2 Materials and Methods
As noted earlier, and determine the size of measurement errors. Thus, the choice of prior parameters a, b, c and d is critical and should reasonably represent the range of errors. We select these parameters utilizing cell types that have both cord and adult blood references available. Here, we briefly discuss the approach used to specify the four parameters and the details are in Supplementary Section S2. In the selection of parameters a and b, using the existing method available in the minfi package (in the function estimateCellCounts), we are able to infer the cell type proportions for each sample using the reference profiles constructed based on cord blood (this gives for a cell) as well as the proportions via reference profiles based on blood in adults (this gives for that cell). Consequently, we are able to estimate the magnitude of measurement errors of each sample for each of the six cell types (by calculating the differences between and ). These measurement errors are then used to determine the values of a and b in the prior distribution of . To specify the prior parameters c and d objectively and informatively, we implement two strategies. When inferring a and b, we use information on cells in the same or a similar category to eosinophils, which potentially increases the accuracy of inferred systematic error. Granulocytes are selected for this purpose since eosinophilic cells are a subset of this cell type. In addition, taking into account that systematic errors may vary between studies, we summarize measurement errors inferred from six independent studies.
To demonstrate the effectiveness of the proposed method, we use B cells and assess the accuracy of the estimated B cell proportions in six studies (Supplementary Section S3). We also examine the impact of different prior distributions of on the inference of cell proportions (Supplementary Section S4). The results show that using the proposed informative prior distributions improves the estimated cell proportions; it gives smaller mean squared errors and estimation bias compared to alternative priors, informative or none-informative.
3 Illustration of the R function for cell proportion estimation with correction
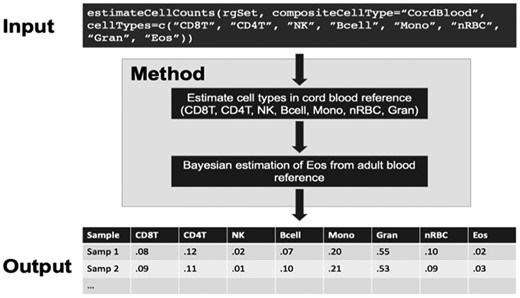
We have incorporated the Bayesian method for estimating cell proportions into the estimateCellCounts function in the minfi package. As shown in Fig. 1, the entire Bayesian estimation model and computation is in the background. The format of input data and output of the updated estimateCellCounts function are similar to the format of its previous version. It takes DNAm data in the format of a RGChannelSet as the input data and returns cell counts for all samples. To estimate the proportion of eosinophils, users should ensure that the cellTypes option includes ‘Eos’, and the compositeCellType is defined as ‘CordBlood’. The arguments in the estimateCellCounts function are specified as follows:
estimateCellCounts (rgSet, compositeCellType = ‘CordBlood’, cellTypes = c (‘CD8T’, ‘CD4T’, ‘NK’, ‘Bcell’, ‘Mono’, ‘nRBC’, ‘Gran’, ‘Eos’).
Flowchart of Eos cell proportion estimation in cord blood using estimateCellCounts function in minfi packages
4 Summary
The proposed method has been built into the estimateCellCounts function in the minfi package. The use of the function is the same as that for the default cord blood estimateCellCounts function except that ‘Eos’ needs to be specified in the cellTypes vector in order to infer its cell proportions. The added computational time is only 1.6 s (0.8% of total running time of estimateCellCounts) with the inclusion of the Bayesian measurement error modeling for an analysis of 70 samples. The current measurement error model is developed for the estimation of eosinophil cells. However, it is not restricted to this cell and can be directly applied to estimate proportions of other cell types as long as the cell has information in one reference database. In addition, the proposed method is not restricted to cells in cord blood. It can be extended to whole blood or other tissues when reference data are not available.
Acknowledgements
We are thankful to the computation support from the High Performance Computing facility at the University of Memphis.
Funding
National Institutes of Health (NIH) R01AI121226 (PIs: Zhang, Holloway), R01HL132321 and R03HD092776 (PI: Karmaus), the start-up funds for Dr. Jiang from the School of Public Health at the University of Memphis. Dr. Bakulski is supported by the NIH R01AG055406 (PIs: Bakulski, Ware), P30ES017885 (PI: Loch-Caruso), R01ES025531 (PI: Fallin) and R01ES025574 (PI: Schmidt).
Conflict of Interest: none declared.
References