-
PDF
- Split View
-
Views
-
Cite
Cite
Ivan Yu Chernyshov, Philip V Toukach, REStLESS: automated translation of glycan sequences from residue-based notation to SMILES and atomic coordinates, Bioinformatics, Volume 34, Issue 15, August 2018, Pages 2679–2681, https://doi.org/10.1093/bioinformatics/bty168
Close - Share Icon Share
Abstract
Glycans and glycoconjugates are usually recorded in dedicated databases in residue-based notations. Only a few of them can be converted into chemical (atom-based) formats highly demanded in conformational and biochemical studies. In this work, we present a tool for translation from a residue-based glycan notation to SMILES.
The REStLESS algorithm for translation from the CSDB Linear notation to SMILES was developed. REStLESS stands for ResiduEs as Smiles and LinkagEs as SmartS, where SMARTS reaction expressions are used to merge pre-encoded residues into a molecule. The implementation supports virtually all structural features reported in natural carbohydrates and glycoconjugates. The translator is equipped with a mechanism for conversion of SMILES strings into optimized atomic coordinates which can be used as starting geometries for various computational tasks.
REStLESS is integrated in the Carbohydrate Structure Database (CSDB) and is freely available on the web (http://csdb.glycoscience.ru/csdb2atoms.html).
Supplementary data are available at Bioinformatics online.
1 Introduction
Carbohydrates are of great interest for modern biomedical sciences (Ohtsubo and Marth, 2006; Varki, 2017). Molecular modeling is among the most popular tools for drug design and molecular docking, including those for carbohydrates (Jo et al., 2017; Sliwoski et al., 2014; Yuriev and Ramsland, 2015), and it always starts from a primary structure. In glycomics, data on primary structures are usually recorded using specialized semantic encoding schemes (notation languages). There is a number of notation languages for glycans (Lütteke, 2015) used by glycoinformatic projects such as databases (Hayes, 2011; Lütteke et al., 2006; Tiemeyer et al., 2017; Toukach and Egorova, 2016), ontologies (Ranzinger et al., 2015) and visualization tools (Cheng et al., 2017; Tsuchiya et al., 2017). However, only a few of them support the full variety of structural features of natural carbohydrates and derivatives, and to this day none of them has been supported in popular molecular modeling and visualization software. This gap has been partially closed by Sweet-II (Bohne et al., 1999) and GLYCAM (http://www.glycam.org/cb) tools allowing generation of 3D structures from encoded glycan sequences using the LINUCS or GLYCAM notation, respectively. However, these tools work only for fully defined structures built of monosaccharides typical for mammalian glycans. Recently, the Self-Contained Sequence Representation (SCSR; Chen et al., 2011) and the Hierarchical Editing Language for Macromolecules (HELM; Zhang et al., 2012) notations have been developed to bridge the gap between bio- and cheminformatics. However, carbohydrates are almost completely unsupported in these formats. An alternative to residue-based notations is Web3 Unique Representation of Carbohydrate Structures (WURCS; Matsubara et al., 2017) combining both residue and atom-based approaches. It is convenient for representation of glycans in databases, but, due to its dual nature, it has a number of problems, such as poor human readability and lack of support of some structural features of glycoconjugates. Unlike SMILES, WURCS is not chemically complete and is not supported by general cheminformatic software. Without automated interpretation of dedicated carbohydrate notations, input of such complex molecules in computational tools is a tedious task hampering the usage of glycoinformatics in everyday research.
In this paper, we report REStLESS (ResiduEs as Smiles and LinkagEs as SmartS)—the algorithm and the tool filling the gap between the residue-based notation used to store structural information in databases and the atom-based notation applicable for structure, conformation and energy calculations. Particularly, we present a translator from the CSDB Linear (Toukach, 2011) notation, used in the Carbohydrate Structure Database (CSDB) (Toukach and Egorova, 2016), into SMILES strings, widely used as a standard descriptor of primary structures in general organic chemistry. CSDB Linear is human-readable and is intuitively comprehended by carbohydrate researchers; it can be obtained by automated translation from another popular carbohydrate notation, namely GlycoCT (Herget et al., 2008), or by retrieval of ca. 19 000 natural carbohydrate structures from CSDB. SMILES (Weininger, 1988) is the gold standard for molecular data representation and is supported by a multitude of other chemoinformatic tools. A web tool and an automated interface built upon our translation algorithm are freely available on the Internet.
2 Methods and implementation
In the CSDB Linear code molecules are described as assemblies of residue and linkages. Each residue contains information on anomeric and absolute configurations, residue base name (stereochemistry descriptor, e.g. Glc for glucose), ring size and modifiers (e.g. N for amino group at position 2). Briefly, the linkage between residues is described as a pair of atom indexes in the linked residues. It is assumed that the linkage is formed with elimination of water or ammonia. This fact encouraged us to use SMARTS reaction expressions (http://www.daylight.com/dayhtml/doc/theory/theory.smarts.html) to aggregate monomeric residues into a molecule. SMILES strings were pre-generated for each combination of the base name, ring size and modifiers, which fully define the molecular connectivity, giving a cache of 941 SMILES-encoded monomers, which can be easily expanded. According to the CSDB content analysis, this list covers virtually all carbohydrate and non-carbohydrate constituents present in natural glycans, glycopolymers and glycoconjugates (Toukach and Egorova, 2016). Carbon atoms within the residues are enumerated using isotopic specification in order to link specific positions of residues. The SMILES strings of residues are concatenated into the SMILES code of a target molecule using the RDKit (http://www.rdkit.org) implementation of SMARTS reaction expressions, which are prepared depending on the type of the linked atoms. In a few cases, bonding leads to formation of a new stereocenter (e.g. in glycopyruvates). If the configuration of such center is specified in the CSDB Linear code, it is configured during post-processing. Not every structure can be translated into a single SMILES string. This may occur due to unspecified absolute configurations, ring sizes or bond positions. In this case, our algorithm produces all possible structures, for each of which a SMILES string is generated. If a residue contains only one stereo center and its configuration is undefined, the corresponding atom is set as non-configured in SMILES. The same is done for an anomeric atom if the anomeric configuration is not known. For more details about the SMILES generation algorithm, see Supplementary Material S1.
Supplementary Table S1 contains structural features of natural glycans with indication of support by CSDB Linear notation and by the REStLESS translator. If an input CSDB Linear code describes a repeating unit of a regular polymer, the start and the end of the repeating fragment are represented by dummy atoms with zero atomic numbers (Supplementary Fig. S3). The CSDB Linear notation allows superclasses and aliases if certain residues in a structure are underdetermined or unsupported by a monomer subdatabase. Such residues are displayed as dummy atoms with an assigned isotopic number in a SMILES code.
Generation of the atomic coordinates and subsequent 3D models is implemented in MOL format and is visualized using the JSmol library for browsers (Hanson, 2013). The SMILES strings obtained from a carbohydrate or derivative structure can have undefined configurations of stereocenters due to unknown configurations of some atoms in some residues. For such structures, a set of fully-defined SMILES strings is derived. Atomic coordinates for each fully-defined SMILES are generated by RDKit. However, we found out that residue conformation in molecules containing multiple pyranoses is often simulated erroneously (twist, boat, inverted chair). To overcome this problem, the torsion angles of each pyranose ring were adjusted to model either the 1C4 or 4C1 conformation. The high temperature molecular dynamics simulations (Frank et al., 2007) were used to identify the preferred conformation for each of 381 pyranoses. The MM3 force field, reported as appropriate for carbohydrates (Toukach and Ananikov, 2013), was used to calculate 1 ns trajectories at 1000 K in the TINKER suite (https://dasher.wustl.edu/tinker/). The choice of the preferred conformation followed counting the number of steps during which a pyranose adopted a 1C4 or 4C1 conformation. For more details about the 3D modeling algorithm, see Supplementary Material S2.
The atomic coordinate generation algorithm worked well for molecules containing up to 200–250 non-hydrogen atoms. Generation of bigger structures might exceed a timeout introduced to save server resources during bulk operations. We overcame this problem by caching of the atomic coordinates at the first user request and by pre-generation of 37 571 MOL-files of 19 946 structures of carbohydrates and derivatives stored in CSDB.
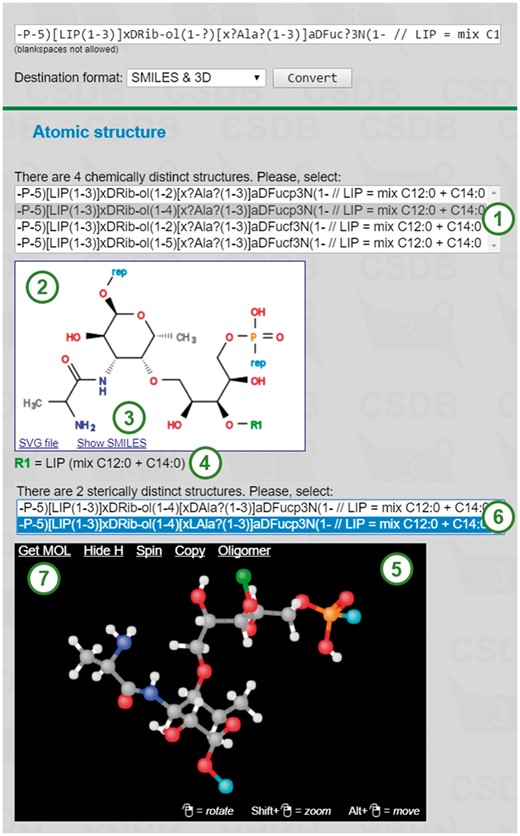
The REStLESS translator is equipped with a web interface and is additionally incorporated in the export modules of CSDB. An example of its input (above the bold line) and output (below the bold line) is shown in Figure 1. The input CSDB Linear code can be obtained from CSDB, entered manually, or translated from GlycoCT by built-in routines. If multiple SMILES strings were constructed for a single CSDB Linear code (e.g. if the latter has uncertainties, such as ribitol linkage position in this example), the list of corresponding structures is displayed as a selector (1) above the image. The white panel contains a structural formula (2) corresponding to a SMILES string selected in the selector. The bounds of polymer repeating units are depicted as ‘rep’. Superclasses and aliases are denoted as ‘R1’, ‘R2’, etc. and are explained below the image (4). The structural formula can be downloaded in the SVG format, and the SMILES string can be shown by clicking on ‘Show SMILES’ (3). The lower panel contains a rendered 3D structure (5) visualized by JSmol. You can move, zoom and rotate the structure. If several stereoisomers are possible for a single SMILES string, their list is displayed above the applet (6). In this example they differ by the absolute configuration of alanine. There are several links for 3D model processing at the top of the JSmol applet (7): 3D models can be downloaded in MOL format by clicking on ‘Get MOL’, hydrogen atoms can be hidden by ‘Hide H’, and CSDB Linear code of the model can be copied to clipboard by ‘Copy’. If a structure is polymeric, SMILES and atomic coordinates of its oligomeric repeating unit can be obtained by clicking on ‘Oligomer’.
REStLESS tool user interface exemplified on (1–4)-linked 5-phospho-d-ribitol 3-N-alanyl-3-deoxy-α-d-fucopyranose polymer
To save server resources during bulk operations, timeouts of 5 s and 60 s were introduced for the structural formula and atomic coordinate generation, respectively. To start calculation with longer timeouts manually, click on the image with an error message.
The automated programming interface (API) of the REStLESS translator was designed for unmanned processing by other glycoinfomatic projects. It is documented in the Supplementary Material S4.
3 Conclusion
The translator from CSDB Linear to SMILES is implemented as a part of CSDB (http://csdb.glycoscience.ru) and verified on all CSDB content. The underlying algorithm is a proof of concept for the generation of SMILES from any language that describes a molecule as a set of covalently-linked residues and can be used for the development of translators from residue-based notations into SMILES. In addition to the translator, a generator of atomic coordinates suitable for molecular modeling was created. The major feature of the translator is a possibility to translate uncertainties in CSDB Linear code into all possible SMILES strings and corresponding structures, which opens up an opportunity to use popular atomistic approaches to molecular modeling on a wide variety of natural glycans in a bulk mode.
Funding
Research in carbohydrate geometry modeling was funded by Russian foundation for Basic Research, grant 18-04-00094. Programing of web-services was funded by Russian Science Foundation, grant 14-50-00126.
Conflict of Interest: none declared.
References