-
PDF
- Split View
-
Views
-
Cite
Cite
Heng Li, Minimap2: pairwise alignment for nucleotide sequences, Bioinformatics, Volume 34, Issue 18, September 2018, Pages 3094–3100, https://doi.org/10.1093/bioinformatics/bty191
Close - Share Icon Share
Abstract
Recent advances in sequencing technologies promise ultra-long reads of ∼100 kb in average, full-length mRNA or cDNA reads in high throughput and genomic contigs over 100 Mb in length. Existing alignment programs are unable or inefficient to process such data at scale, which presses for the development of new alignment algorithms.
Minimap2 is a general-purpose alignment program to map DNA or long mRNA sequences against a large reference database. It works with accurate short reads of ≥100 bp in length, ≥1 kb genomic reads at error rate ∼15%, full-length noisy Direct RNA or cDNA reads and assembly contigs or closely related full chromosomes of hundreds of megabases in length. Minimap2 does split-read alignment, employs concave gap cost for long insertions and deletions and introduces new heuristics to reduce spurious alignments. It is 3–4 times as fast as mainstream short-read mappers at comparable accuracy, and is ≥30 times faster than long-read genomic or cDNA mappers at higher accuracy, surpassing most aligners specialized in one type of alignment.
Supplementary data are available at Bioinformatics online.
1 Introduction
Single Molecule Real-Time (SMRT) sequencing technology and Oxford Nanopore technologies (ONT) produce reads over 10 kbp in length at an error rate ∼15%. Several aligners have been developed for such data (Chaisson and Tesler, 2012; Li, 2013; Lin and Hsu, 2017; Liu et al., 2016, 2017; Sedlazeck et al., 2018; Sović et al., 2016). Most of them were five times as slow as mainstream short-read aligners (Langmead and Salzberg, 2012; Li, 2013) in terms of the number of bases mapped per second. We speculated there could be substantial room for speedup on the thought that 10 kb long sequences should be easier to map than 100 bp reads because we can more effectively skip repetitive regions, which are often the bottleneck of short-read alignment. We confirmed our speculation by achieving approximate mapping 50 times faster than BWA-MEM (Li, 2016). Suzuki and Kasahara (2018) extended our work with a fast and novel algorithm on generating base-level alignment, which in turn inspired us to develop minimap2 with added functionality.
Both SMRT and ONT have been applied to the sequencing of spliced mRNAs (RNA-seq). While traditional mRNA aligners work (Iwata and Gotoh, 2012; Wu and Watanabe, 2005), they are not optimized for long noisy sequence reads and are tens of times slower than dedicated long-read aligners. When developing minimap2 initially for aligning genomic DNA only, we realized minor modifications could enable the base algorithm to map mRNAs as well. Minimap2 becomes a first RNA-seq aligner specifically designed for long noisy reads. We have also extended the original algorithm to map short reads at a speed faster than several mainstream short-read mappers.
In this article, we will describe the minimap2 algorithm and its applications to different types of input sequences. We will evaluate the performance and accuracy of minimap2 on several simulated and real datasets and demonstrate the versatility of minimap2.
2 Materials and methods
Minimap2 follows a typical seed-chain-align procedure as is used by most full-genome aligners. It collects minimizers (Roberts et al., 2004) of the reference sequences and indexes them in a hash table, with the key being the hash of a minimizer and the value being a list of locations of the minimizer copies. Then for each query sequence, minimap2 takes query minimizers as seeds, finds exact matches (i.e. anchors) to the reference, and identifies sets of colinear anchors as chains. If base-level alignment is requested, minimap2 applies dynamic programming (DP) to extend from the ends of chains and to close regions between adjacent anchors in chains.
Minimap2 uses indexing and seeding algorithms similar to minimap (Li, 2016), and furthers the predecessor with more accurate chaining, the ability to produce base-level alignment and the support of spliced alignment.
2.1 Chaining
2.1.1 Finding the optimal chaining scores
We note that if anchor i is chained to j, chaining i to a predecessor of j is likely to yield a lower score. When evaluating Equation (1), we start from anchor i − 1 and stop the process if we cannot find a better score after up to h iterations. This approach reduces the average time to O(hN). In practice, we can almost always find the optimal chain with h = 50; even if the heuristic fails, the optimal chain is often close.
2.1.2 Backtracking
Let P(i) be the index of the best predecessor of anchor i. It equals 0 if or otherwise. For each anchor i in the descending order of f(i), we apply repeatedly to find its predecessor and mark each visited i as ‘used’, until P(i) = 0 or we reach an already ‘used’ i. This way we find all chains with no anchors used in more than one chains.
2.1.3 Identifying primary chains
In the absence of copy number changes, each query segment should not be mapped to two places in the reference. However, chains found at the previous step may have significant or complete overlaps due to repeats in the reference (Li and Durbin, 2010). Minimap2 used the following procedure to identify primary chains that do not greatly overlap on the query.
Let Q be an empty set initially. For each chain from the best to the worst according to their chaining scores: if on the query, the chain overlaps with a chain in Q by 50% or higher percentage of the shorter chain, mark the chain as secondary to the chain in Q; otherwise, add the chain to Q. In the end, Q contains all the primary chains. We did not choose a more sophisticated data structure (e.g. range tree or k-d tree) because this step is not the performance bottleneck.
2.1.4 Estimating per-base sequence divergence
Suppose a query sequence harbors n seeds of length k, m of which are present in a chain. We want to estimate the sequence divergence ϵ between the query and the reference sequences in the chain. This is useful when base-level alignment is too expensive to perform.
2.1.5 Indexing with homopolymer compressed k-mers
SmartDenovo (https://github.com/ruanjue/smartdenovo; J. Ruan, personal communication) indexes reads with homopolymer-compressed (HPC) k-mers and finds the strategy improves overlap sensitivity for SMRT reads. Minimap2 adopts the same heuristic.
The HPC string of a string s, denoted by , is constructed by contracting homopolymers in s to a single base. An HPC k-mer of s is a k-long substring of . For example, suppose , and the first HPC 4-mer is .
To demonstrate the effectiveness of HPC k-mers, we performed read overlapping for the example E. coli SMRT reads from PBcR (Berlin et al., 2015), using different types of k-mers. With normal 15 bp minimizers per 5 bp window, minimap2 finds 90.9% of ≥2 kb overlaps inferred from the read-to-reference alignment. With HPC 19-mers per 5 bp window, minimap2 finds 97.4% of overlaps. It achieves this higher sensitivity by indexing 1/3 fewer minimizers, which further helps performance. HPC-based indexing reduces the sensitivity for current ONT reads, though.
2.2 Aligning genomic DNA
2.2.1 Alignment with 2-piece affine gap cost
2.2.2 The Suzuki–Kasahara formulation
When performing global alignment, we do not need to compute Hrt in each cell. We use 16-way vectorization throughout the alignment process. When extending alignments from ends of chains, we need to find the cell (r, t) where Hrt reaches the maximum. We resort to 4-way vectorization to compute . Because this computation is simple, Equation (5) is still the dominant performance bottleneck.
In practice, our 16-way vectorized implementation of global alignment is three times as fast as Parasail’s 4-way vectorization (Daily, 2016). Without banding, our implementation is slower than Edlib (Šošić and Šikic, 2017), but with a 1000 bp band, it is considerably faster. When performing global alignment between anchors, we expect the alignment to stay close to the diagonal of the DP matrix. Banding is applicable most of the time.
2.2.3 The Z-drop heuristic
When minimap2 breaks a global alignment between two anchors, it performs local alignment between the two subsequences involved in the global alignment, but this time with the one subsequence reverse complemented. This additional alignment step may identify short inversions that are missed during chaining.
2.2.4 Filtering out misplaced anchors
Due to sequencing errors and local homology, some anchors in a chain may be wrong. If we blindly align regions between two misplaced anchors, we will produce a suboptimal alignment. To reduce this artifact, we filter out anchors that lead to a >10 bp insertion and a >10 bp deletion at the same time, and filter out terminal anchors that lead to a long gap towards the ends of a chain. These heuristics greatly alleviate the issues with misplaced anchors, but they are unable to fix all such errors. Local misalignment is a limitation of minimap2 which we hope to address in future.
2.3 Aligning spliced sequences
In alignment, a deletion no shorter than is regarded as an intron, which pays no cost to gap extensions.
If RNA-seq reads are not sequenced from stranded libraries, the read strand relative to the underlying transcript is unknown. By default, minimap2 aligns each chain twice, first assuming – as the splicing signal and then assuming –, the reverse complement of –, as the splicing signal. The alignment with a higher score is taken as the final alignment. This procedure also infers the relative strand of reads that span canonical splicing sites.
In the spliced alignment mode, minimap2 further increases the density of minimizers and disables banded alignment. Together with the two-round DP-based alignment, spliced alignment is several times slower than genomic DNA alignment.
2.4 Aligning short paired-end reads
3 Results
Minimap2 is implemented in the C programming language and comes with APIs in both C and Python. It is distributed under the MIT license, free to both commercial and academic uses. Minimap2 uses the same base algorithm for all applications, but it has to apply different sets of parameters depending on input data types. Similar to BWA-MEM, minimap2 introduces ‘presets’ that modify multiple parameters with a simple invocation. Detailed settings and command-line options can be found in the minimap2 manpage (supplementary information). In addition to the applications evaluated in the following sections, minimap2 also retains minimap’s functionality to find overlaps between long reads and to search against large multi-species databases such as nt from NCBI.
3.1 Aligning long genomic reads
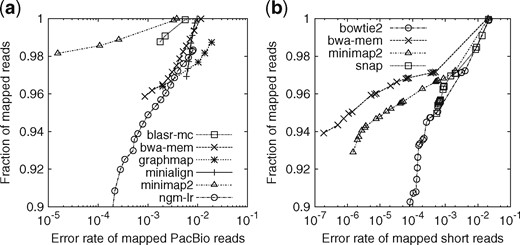
As a sanity check, we evaluated minimap2 on simulated human reads along with BLASR (v1.MC.rc64; Chaisson and Tesler, 2012), BWA-MEM (v0.7.15; Li, 2013), GraphMap (v0.5.2; Sović et al., 2016), Kart (v2.2.5; Lin and Hsu, 2017), minialign (v0.5.3; https://github.com/ocxtal/minialign) and NGMLR (v0.2.5; Sedlazeck et al., 2018). We excluded rHAT (Liu et al., 2016) and LAMSA (Liu et al., 2017) because they either crashed or produced malformatted output. In this evaluation, minimap2 has higher power to distinguish unique and repetitive hits, and achieves overall higher mapping accuracy (Fig. 1a). Minimap2 and NGMLR provide better mapping quality estimate: they rarely give repetitive hits high mapping quality. Apparently, other aligners may occasionally miss close suboptimal hits and be overconfident in wrong mappings. On run time, minimap2 took 200 CPU seconds, comparable to minialign and Kart, and is over 30 times faster than the rest. Minimap2 consumed 6.8GB memory at the peak, more than BWA-MEM (5.4GB), similar to NGMLR and less than others.
Evaluation on aligning simulated reads. Simulated reads were mapped to the primary assembly of human genome GRCh38. A read is considered correctly mapped if its longest alignment overlaps with the true interval, and the overlap length is ≥10% of the true interval length. Read alignments are sorted by mapping quality in the descending order. For each mapping quality threshold, the fraction of alignments (out of the number of input reads) with mapping quality above the threshold and their error rate are plotted along the curve. (a) Long-read alignment evaluation. 33 088 ≥1000 bp reads were simulated using pbsim (Ono et al., 2013) with error profile sampled from file ‘m131017_060208_42213_*.1.*’ downloaded at http://bit.ly/chm1p5c3. The N50 read length is 11 628. Aligners were run under the default setting for SMRT reads. Kart outputted all alignments at mapping quality 60, so is not shown in the figure. It mapped nearly all reads with 4.1% of alignments being wrong, less accurate than others. (b) Short-read alignment evaluation. 10 million pairs of 150 bp reads were simulated using mason2 (Holtgrewe, 2010) with option ‘–illumina-prob-mismatch-scale 2.5’. Short-read aligners were run under the default setting except for changing the maximum fragment length to 800 bp
On real human SMRT reads, the relative performance and fraction of mapped reads reported by these aligners are broadly similar to the metrics on simulated data. We are unable to provide a good estimate of mapping error rate due to the lack of the truth. On ONT ∼100 kb human reads (Jain et al., 2018), BWA-MEM failed. Kart, minialign and minimap2 are over 70 times faster than others. We have also examined tens of ≥100 bp INDELs in IGV (Robinson et al., 2011) and can confirm the observation by Sedlazeck et al. (2018) that BWA-MEM often breaks them into shorter gaps. The issue is much alleviated with minimap2, thanks to the 2-piece affine gap cost.
3.2 Aligning long spliced reads
We evaluated minimap2 on SIRV control data (AC: SRR5286959; Byrne et al., 2017) where the truth is known. Minimap2 predicted 59 918 introns from 11 018 reads. 93.8% of splice juctions are precise. We examined wrongly predicted junctions and found the majority were caused by clustered splicing signals (e.g. two adjacent sites). When INDEL sequencing errors are frequent, it is difficult to find precise splicing sites in this case. If we allow up to 10 bp distance from true splicing sites, 98.4% of aligned introns are approximately correct. It is worth noting that for SIRV, we asked minimap2 to model the splicing signal only without extra bases. This is because SIRV does not honor the evolutionarily prevalent signal (Irimia and Roy, 2008).
We next aligned real mouse reads (Byrne et al., 2017) with GMAP (v2017-06-20; Wu and Watanabe, 2005), minimap2, SpAln (v2.3.1; Iwata and Gotoh, 2012) and STAR (v2.5.3a; Dobin et al., 2013). In general, minimap2 is more consistent with existing annotations (Table 1): it finds more junctions with a higher percentage being exactly or approximately correct. Minimap2 is over 40 times faster than GMAP and SpAln. While STAR is close to minimap2 in speed, it does not work well with noisy reads.
Evaluation of junction accuracy on 2D ONT reads
| . | GMAP . | minimap2 . | SpAln . | STAR . |
|---|---|---|---|---|
| Run time (CPU min) | 631 | 15.9 | 2076 | 33.9 |
| Peak RAM (GByte) | 8.9 | 14.5 | 3.2 | 29.2 |
| # aligned reads | 103 669 | 104 199 | 103 711 | 26 479 |
| # chimeric alignments | 1904 | 1488 | 0 | 0 |
| # non-spliced alignments | 15 854 | 14 798 | 17 033 | 10 545 |
| # aligned introns | 692 275 | 693 553 | 692 945 | 78 603 |
| # novel introns | 11 239 | 3113 | 8550 | 1214 |
| % exact introns | 83.8 | 94.0 | 87.9 | 55.2 |
| % approx. introns | 91.8 | 96.9 | 92.5 | 82.4 |
| . | GMAP . | minimap2 . | SpAln . | STAR . |
|---|---|---|---|---|
| Run time (CPU min) | 631 | 15.9 | 2076 | 33.9 |
| Peak RAM (GByte) | 8.9 | 14.5 | 3.2 | 29.2 |
| # aligned reads | 103 669 | 104 199 | 103 711 | 26 479 |
| # chimeric alignments | 1904 | 1488 | 0 | 0 |
| # non-spliced alignments | 15 854 | 14 798 | 17 033 | 10 545 |
| # aligned introns | 692 275 | 693 553 | 692 945 | 78 603 |
| # novel introns | 11 239 | 3113 | 8550 | 1214 |
| % exact introns | 83.8 | 94.0 | 87.9 | 55.2 |
| % approx. introns | 91.8 | 96.9 | 92.5 | 82.4 |
Notes: Mouse cDNA reads (AC: SRR5286960; R9.4 chemistry) were mapped to the primary assembly of mouse genome GRCm38 with the following tools and command options: minimap2 (‘-ax splice’); GMAP (‘-n 0 –min-intronlength 30 –cross-species’); SpAln (‘-Q7 -LS -S3’); STARlong (according to http://bit.ly/star-pb). The alignments were compared to the EnsEMBL gene annotation, release 89. A predicted intron is novel if it has no overlaps with any annotated introns. An intron is exact if it is identical to an annotated intron. An intron is approximate if both its 5′- and 3′-end are within 10 bp around the ends of an annotated intron. Chimeric alignments are defined in the SAM spec (Li et al., 2009).
Evaluation of junction accuracy on 2D ONT reads
| . | GMAP . | minimap2 . | SpAln . | STAR . |
|---|---|---|---|---|
| Run time (CPU min) | 631 | 15.9 | 2076 | 33.9 |
| Peak RAM (GByte) | 8.9 | 14.5 | 3.2 | 29.2 |
| # aligned reads | 103 669 | 104 199 | 103 711 | 26 479 |
| # chimeric alignments | 1904 | 1488 | 0 | 0 |
| # non-spliced alignments | 15 854 | 14 798 | 17 033 | 10 545 |
| # aligned introns | 692 275 | 693 553 | 692 945 | 78 603 |
| # novel introns | 11 239 | 3113 | 8550 | 1214 |
| % exact introns | 83.8 | 94.0 | 87.9 | 55.2 |
| % approx. introns | 91.8 | 96.9 | 92.5 | 82.4 |
| . | GMAP . | minimap2 . | SpAln . | STAR . |
|---|---|---|---|---|
| Run time (CPU min) | 631 | 15.9 | 2076 | 33.9 |
| Peak RAM (GByte) | 8.9 | 14.5 | 3.2 | 29.2 |
| # aligned reads | 103 669 | 104 199 | 103 711 | 26 479 |
| # chimeric alignments | 1904 | 1488 | 0 | 0 |
| # non-spliced alignments | 15 854 | 14 798 | 17 033 | 10 545 |
| # aligned introns | 692 275 | 693 553 | 692 945 | 78 603 |
| # novel introns | 11 239 | 3113 | 8550 | 1214 |
| % exact introns | 83.8 | 94.0 | 87.9 | 55.2 |
| % approx. introns | 91.8 | 96.9 | 92.5 | 82.4 |
Notes: Mouse cDNA reads (AC: SRR5286960; R9.4 chemistry) were mapped to the primary assembly of mouse genome GRCm38 with the following tools and command options: minimap2 (‘-ax splice’); GMAP (‘-n 0 –min-intronlength 30 –cross-species’); SpAln (‘-Q7 -LS -S3’); STARlong (according to http://bit.ly/star-pb). The alignments were compared to the EnsEMBL gene annotation, release 89. A predicted intron is novel if it has no overlaps with any annotated introns. An intron is exact if it is identical to an annotated intron. An intron is approximate if both its 5′- and 3′-end are within 10 bp around the ends of an annotated intron. Chimeric alignments are defined in the SAM spec (Li et al., 2009).
We have also evaluated spliced aligners on a human Nanopore Direct RNA-seq dataset (http://bit.ly/na12878ont). Minimap2 aligned 10 million reads in <1 wall-clock hour using 16 CPU cores. 94.2% of aligned splice junctions consistent with gene annotations. In comparison, GMAP under option ‘-k 14 -n 0 -min-intronlength 30 –cross-species’ is 160 times slower; 68.7% of GMAP junctions are found in known gene annotations. The percentage increases to 84.1% if an aligned junction within 10 bp from an annotated junction is considered to be correct. On a public Iso-Seq dataset (human Alzheimer brain from http://bit.ly/isoseqpub), minimap2 is also faster at higher junction accuracy in comparison to other aligners in Table 1.
We noted that GMAP and SpAln have not been optimized for noisy reads. We are showing the best setting we have experimented, but their developers should be able to improve their accuracy further.
3.3 Aligning short genomic reads
We evaluated minimap2 along with Bowtie2 [v2.3.3; (Langmead and Salzberg, 2012)], BWA-MEM and SNAP [v1.0beta23; (Zaharia et al., 2011)]. Minimap2 is 3–4 times as fast as Bowtie2 and BWA-MEM, but is 1.3 times slower than SNAP. Minimap2 is more accurate on this simulated dataset than Bowtie2 and SNAP but less accurate than BWA-MEM (Fig. 1b). Closer investigation reveals that BWA-MEM achieves a higher accuracy partly because it tries to locally align a read in a small region close to its mate. If we disable this feature, BWA-MEM becomes slightly less accurate than minimap2. We might implement a similar heuristic in minimap2 in future.
To evaluate the accuracy of minimap2 on real data, we aligned human reads (AC: ERR1341796) with BWA-MEM and minimap2, and called SNPs and small INDELs with GATK HaplotypeCaller v3.5 (Depristo et al., 2011). This run was sequenced from experimentally mixed CHM1 and CHM13 cell lines. Both of them are homozygous across the whole genome and have been de novo assembled with SMRT reads to high quality. This allowed us to construct an independent truth variant dataset (Li et al., 2017) for ERR1341796. In this evaluation, minimap2 has higher SNP false negative rate (FNR; 2.6% of minimap2 vs 2.3% of BWA-MEM), but fewer false positive SNPs per million bases (FPPM; 7.0 versus 8.8), similar INDEL FNR (11.2% versus 11.3%) and similar INDEL FPPM (6.4 versus 6.5). Minimap2 is broadly comparable to BWA-MEM in the context of small variant calling.
3.4 Aligning long-read assemblies
Minimap2 can align a SMRT assembly (AC: GCA_001297185.1) against GRCh38 in 7 min using eight CPU cores, over 20 times faster than nucmer from MUMmer4 (Marçais et al., 2018). With the paftools.js script from the minimap2 package, we called 2.67 million single-base substitutions out of 2.78Gbp genomic regions. The transition-to-transversion ratio (ts/tv) is 2.01. In comparison, using MUMmer4’s dnadiff pipeline, we called 2.86 million substitutions in 2.83Gbp at ts/tv = 1.87. Given that ts/tv averaged across the human genome is about 2 but ts/tv averaged over random errors is 0.5, the minimap2 callset arguably has higher precision at lower sensitivity.
The sample being assembled is a female. Minimap2 still called 201 substitutions on the Y chromosome. These substitutions all come from one contig aligned at 96.8% sequence identity. The contig could be a segmental duplication absent from GRCh38. In constrast, dnadiff called 9070 substitutions on the Y chromosome across 73 SMRT contigs. This again implies our minimap2-based pipeline has higher precision.
4 Discussions
Minimap2 is a versatile mapper and pairwise aligner for nucleotide sequences. It works with short reads, assembly contigs and long noisy genomic and RNA-seq reads, and can be used as a read mapper, long-read overlapper or a full-genome aligner. Minimap2 is also accurate and efficient, often outperforming other domain-specific alignment tools in terms of both speed and accuracy.
The capability of minimap2 comes from a fast base-level alignment algorithm and an accurate chaining algorithm. When aligning long query sequences, base-level alignment is often the performance bottleneck. The Suzuki–Kasahara algorithm greatly alleviates the bottleneck and enables DP-based splice alignment involving >100 kb introns, which was impractically slow 10 years ago. The minimap2 chaining algorithm is fast and highly accurate by itself. In fact, chaining alone is more accurate than all the other long-read mappers in Figure 1a (data not shown). This accuracy helps to reduce downstream base-level alignment of candidate chains, which is still several times slower than chaining even with the Suzuki–Kasahara improvement. In addition, taking a general form, minimap2 chaining can be adapted to non-typical data types such as spliced reads and multiple reads per fragment. This gives us the opportunity to extend the same base algorithm to a variety of use cases.
Modern mainstream aligners often use a full-text index, such as suffix array or FM-index, to index reference sequences. An advantage of this approach is that we can use exact seeds of arbitrary lengths, which helps to increase seed uniqueness and reduce unsuccessful extensions. Minimap2 indexes reference k-mers with a hash table instead. Such fixed-length seeds are inferior to variable-length seeds in theory, but can be computed much more efficiently in practice. When a query sequence has multiple seed hits, we can afford to skip highly repetitive seeds without affecting the final accuracy. This further alleviates the concern with the seeding uniqueness. At the same time, at low sequence identity, it is rare to see long seeds anyway. Hash table is the ideal data structure for mapping long noisy sequences.
Acknowledgements
We owe a debt of gratitude to H. Suzuki and M. Kasahara for releasing their masterpiece and insightful notes before formal publication. We thank M. Schatz, P. Rescheneder and F. Sedlazeck for pointing out the limitation of BWA-MEM. We are also grateful to minimap2 users who have greatly helped to suggest features and to fix various issues.
Funding
This work was supported by the National Human Genome Research Institute [1R01HG010040-01].
References