-
PDF
- Split View
-
Views
-
Cite
Cite
Johannes Rainer, Laurent Gatto, Christian X Weichenberger, ensembldb: an R package to create and use Ensembl-based annotation resources, Bioinformatics, Volume 35, Issue 17, September 2019, Pages 3151–3153, https://doi.org/10.1093/bioinformatics/btz031
Close - Share Icon Share
Abstract
Bioinformatics research frequently involves handling gene-centric data such as exons, transcripts, proteins and their positions relative to a reference coordinate system. The ensembldb Bioconductor package retrieves and stores Ensembl-based genetic annotations and positional information, and furthermore offers identifier conversion and coordinates mappings for gene-associated data. In support of reproducible research, data are tied to Ensembl releases and are kept separately from the software. Premade data packages are available for a variety of genomes and Ensembl releases. Three examples demonstrate typical use cases of this software.
ensembldb is part of Bioconductor (https://bioconductor.org/packages/ensembldb).
Supplementary data are available at Bioinformatics online.
1 Introduction
When the human genome was released as a first draft (Lander et al., 2001), researchers started to manage these kinds of large, fragmented and rapidly evolving complex datasets by creating genome browsing and database systems such as EMBL-EBI Ensembl (Birney et al., 2004). The availability of a reference genome allows definition of a coordinate system, in which genomic data, such as collaboratively defined gene models, are described unambiguously by chromosomal positions. Ensembl publishes several data releases per year, rendering it a valuable resource for consistent and tightly integrated data. These data are used in high-throughput genomic data analyses, which are frequently carried out in the R statistical programing language using tools provided by Bioconductor (Huber et al., 2015).
Here we present ensembldb, a Bioconductor package enabling the creation and usage of comprehensive, locally stored Ensembl-based offline annotation databases. In addition to gene model annotations we include protein annotations in the pre-built databases, offer a fast and powerful filter mechanism and provide functions for the mapping of arbitrary positions between the genome, exome, transcriptome and proteome.
2 Implementation and available data
Ensembl is one of the main annotation resources for genomic data with a web service for online data access and APIs enabling programmatic data access. The ensembldb package provides functions to retrieve annotations for any of the >300 species available through Ensembl and EnsemblGenomes using their Perl API and to store information in small custom databases, which can be distributed as self-contained SQLite files or MySQL databases. The annotations included in our EnsDb databases comprise (i) genomic coordinates for all genes, transcripts and exons of a species and their relation to each other; (ii) general metadata information such as gene and transcript biotypes, NCBI Entrez gene IDs; and (iii) protein annotations including amino acid sequences, positions of protein domains within these (from e.g. Pfam; Finn et al., 2016) and mappings of Ensembl protein identifiers to UniProt accession numbers. Some of these annotations are also available in other Bioconductor annotation resources, in particular TxDb databases from the GenomicFeatures package (Lawrence et al., 2013) providing genomic coordinates, or org*db packages that contain gene-related annotations. With ensembldb, all this information is bundled conveniently into a single database. We distribute pre-built EnsDb databases covering all Ensembl core species for a range of Ensembl releases using Bioconductor’s AnnotationHub resource, which can be thought of as a queryable repository for annotation data. These locally stored databases enable offline access to Ensembl annotations in Bioconductor, in contrast to the biomaRt package (Durinck et al., 2009) that, while also providing Ensembl annotations, requires active internet connectivity.
In Bioconductor, the AnnotationDbi package provides a common interface for retrieving annotation data. Furthermore, the GenomicFeatures package defines means for representation, organization and structured retrieval of transcript models and genomic positions of genes and their exons. The ensembldb package is compliant with both interfaces, such that data retrieval and data access is handled in a standardized way.
In addition, we developed a powerful filtering framework in ensembldb, which directly translates to SQL queries for performance increase (benchmarks provided in the supplement). It is based on our AnnotationFilter classes, available as a separate Bioconductor package to encourage usage beyond ensembldb. This filtering framework can be classified into two main groups: one to query arbitrary textual information, such as gene symbols or UniProt accession numbers, and the other to handle positional information of genes, exons, transcripts and protein domains. Filters can be combined with logical expressions to create tailored queries and retrieve only specific data from the databases. This is particularly useful for visualizing transcript models from certain genomic regions: ensembldb facilitates plotting with Bioconductor packages ggbio (Yin et al., 2012) and Gviz (Hahne and Ivanek, 2016).
Generally, results returned by ensembldb are compatible with the standards defined by Bioconductor, such that data can be easily exchanged with other packages for further analysis.
3 Usage and examples
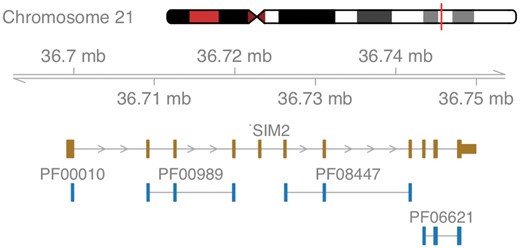
The first example illustrates filtered data retrieval using ensembldb in the context of Down syndrome, a genetic disorder characterized by the presence of all or parts of a third copy of chromosome 21. In our example, we are interested in transcription factors encoded on Chromosome 21 with a basic helix-loop-helix DNA-binding domain, as described by Pfam ID PF00010: given a variable edb of type EnsDb, the simple command gene(edb, filter = ∼ protein_domain_id == “PF00010” & seq_name == “21”) returns the genomic annotations for three genes: SIM2, a master regulator of neurogenesis thought to contribute to some phenotypes of Down syndrome (Gardiner and Costa, 2006), and the two genes OLIG1 and OLIG2, triplication of which was shown to cause developmental brain defects (Chakrabarti et al., 2010). Visualization of the genomic neighborhood is accomplished by passing the filter to an ensembldb function extracting data for plotting using Gviz, as shown in Figure 1.
Schematic overview of the gene SIM2. Shown are the exons of the gene in brown and all co-locating Pfam protein domains in blue
Another hallmark of ensembldb is its capability to convert any position within a protein, transcript or the genome to any other of these three entities, extending the genome to transcript mapping functionality of GenomicFeatures. For example, one of the known variants responsible for human red hair color is located at position 16:89920138 (dbSNP ID rs1805009) on the human genome (version GRCh38) and is readily converted by ensembldb to position 294 on the respective protein given by Ensembl ID ENSP00000451605 using the command genomeToProtein(GRanges(“16”, IRanges(89920138, width = 1)), edb) with edb as defined in the first example. We furthermore find that this protein is encoded by the MC1R gene issuing the AnnotationDbi-compatible query command select(edb, keys = “ENSP00000451605”, keytype = “PROTEINID”, columns = “SYMBOL”).
Our annotation packages also contain protein sequence information. Thus, with the call proteins(edb, filter = ∼ protein_id == “ENSP00000451605”)$protein_sequence, we get the protein sequence for the selected ID to find on position 294 an aspartic acid (‘D’), which is in agreement with the reference amino acid of variant Asp294His (Valverde et al., 1995) described by the dbSNP ID cited above.
Expanded code with descriptions and results for these two examples is provided as Supplementary Material and as a Bioconductor package vignette.
Finally, by providing gene annotations and positional information of exons on the genome and supporting the standard Bioconductor interfaces for data retrieval, ensembldb can be easily integrated into genome analysis pipelines. An extended example is given in the Supplementary Material, where we present a modified version of the standard Bioconductor RNA-seq workflow (Love et al., 2015).
4 Conclusion
Here we have described the Bioconductor package ensembldb, which utilizes annotation resources from Ensembl and integrates them into Bioconductor. The separation of source code and annotation data facilitates reproducible research by allowing ensembldb to access any set of annotations published in the past. With an extensive filtering system, searches can be customized to meet very specific requirements and powerful coordinate mapping functions enable conversion of coordinates between proteins, transcripts, and the genome. Providing protein and protein domain annotations along with genome-centered annotations makes ensembldb also an asset for any post-genome data analysis that aims to combine data from these various domains.
For each new Ensembl release, we create EnsDb annotation databases for all Ensembl vertebrates and plan to provide future continuous support for them via AnnotationHub.
Conflict of Interest: none declared.
Acknowledgements
The authors thank the Department of Innovation, Research and University of the Autonomous Province of Bozen/Bolzano for covering the Open Access publication costs and for funding this work. The authors thank Dr Sebastian Gibb for suggestions and feedback and are grateful to Peter P. Pramstaller for enabling this work and for all the support.
References