-
PDF
- Split View
-
Views
-
Cite
Cite
Allan J R Ferrari, Milan A Clasen, Louise Kurt, Paulo C Carvalho, Fabio C Gozzo, Leandro Martínez, TopoLink: evaluation of structural models using chemical crosslinking distance constraints, Bioinformatics, Volume 35, Issue 17, September 2019, Pages 3169–3170, https://doi.org/10.1093/bioinformatics/btz014
Close - Share Icon Share
Abstract
A software was developed to evaluate structural models using chemical crosslinking experiments. The user provides the types of linkers used and their reactivity, and the observed crosslinks and dead-ends. The software computes the minimum length of a physically inspired linker that connects the reactive atoms of interest, and reports the consistency of each distance with the experimental observation. Statistics on model consistency with the links are provided. Tools to evaluate the correlation of crosslinks in ensembles of models were developed. TopoLink was used to evaluate the potential crosslinks of all structures of the CATH database. The number of crosslinks expected as a function of protein size and linker length can be used as guide for experimental design.
TopoLink is available as free software at http://m3g.iqm.unicamp.br/topolink, and distributed as source code with a user-friendly graphical interface for Windows. A web server is also provided.
Supplementary data are available at Bioinformatics online.
1 Introduction
Chemical crosslinking mass spectrometry (XLMS) is a technique for the evaluation of interatomic distances in biomolecular structures (Schneider et al., 2018). It provides upper bounds for the distances between amino acid residues, by the identification of pairs of atoms in the surface of proteins, which are within the linker reach. The distance information can be used for modeling protein complexes and tertiary structures (Dos Santos et al., 2018; Sinz, 2006).
The evaluation of structural models using chemical crosslinking depends on the determination of reactive conformations of the linker on the surface of the structure (Matthew Allen Bullock et al., 2016). Usually, the shortest path along the surface has been associated to the reactivity between a pair of residues (Degiacomi et al., 2017; Kahraman et al., 2011; Matthew Allen Bullock et al., 2016).
TopoLink is a package to compute these distances using a physical representation of the linker and evaluate structural models. With the experimental setup in mind, the user provides a description of the experiments performed, the types of potential crosslinks and the experimental observations. TopoLink determines the consistency of the model with the experimental data.
2 Approach
The user provides the reactivity and reach of the linkers and an account of observed crosslinks and dead-ends (products of the reaction of one residue with the linker but for which the other reactive group of the linker did not bind another residue, either because there is no other reactive residue within the linker reach or due to inactivation reactions).
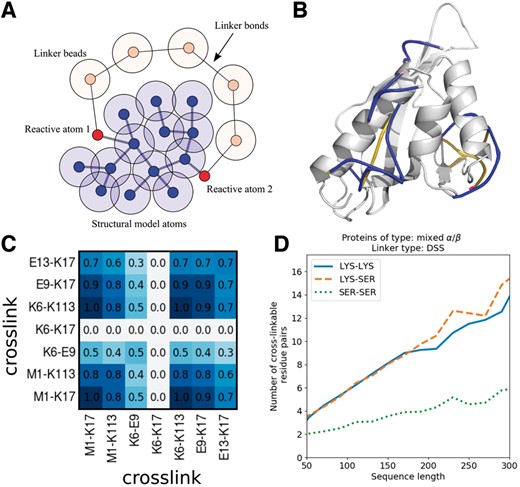
The topological distances are obtained by defining a linker as a sequence of beads with atomic dimensions and bound by harmonic potentials. The length of this linker is minimized while avoiding the overlap of the beads with the protein atoms (see the Supplementary Material for details). A sketch of the model used for optimization is shown in Figure 1A, and a typical result illustrated in Figure 1B.
(A) Representation of the model used to compute the topological distances. (B) Links obtained for an example structure. (C) Matrix of joint-observation of crosslinks in an ensemble of models. (D) Statistics of XL in general protein structures: number of cross-linkable pairs of residues as function of protein sequence length
The linker is flexible and, if Cα or Cβ atoms are used as references, the side-chain of the reactive residues is ignored and the flexibility of the linker will incorporate the flexibility of the side-chains. By default, we suggest using Cβ atoms, such that the linker will preserve the directionality of the side-chain.
2.1 Setup of the calculation
A minimal input file is shown below:
structure protein.pdb
experiment DSS
# Res Chain ReNum Atom Res Chain ResNum Atom Dist
linktype LYS all all CB LYS all all CB 17.8
# Res Chain ResNum Res Chain Resnum
observed LYS A 37 LYS A 53
end experiment DSS
compute observed
Here, the linker DSS can potentially bind any pair of Lys residues (through the ‘all’ keywords) if the distance between their CB atoms is <17.8 Å. A crosslink between Lys 37 and Lys 53, both from chain A, was observed. The last line indicates that we are interested in observed links, but we could compute the topological distance of ‘all’ possible crosslinks, among other options described in the ‘Usage’ guide.
Additional input and output parameters are available for more comprehensive experimental setups, including multiple experiments, linker types, reactivities. The setup of this calculations can be performed from a graphical user interface or using our online server.
2.2 Output
TopoLink will report the topological distances between the Cβ atoms and the consistency of the observations with the model. For each link a PDB file can be generated to observe the linker on the protein structure, as shown in Figure 1B. Finally, TopoLink outputs general statistics on the consistency of the experimental observations with the model: The number of crosslinks that are observed and are consistent with the model, the number of observations that are missing according to the structure, and the number of observations that are inconsistent with the model. The results are reported for each experiment independently, and for the complete set of experiments.
If many models are analyzed with TopoLink, a set of output files is obtained. Tools to study the consistency of the links with the set of structures are provided, as illustrated in Figure 1C. Dark elements of the matrix indicate pairs of crosslinks which are satisfied simultaneously in most models. The link-correlation package allows the computation of pair-exclusion and correlation matrices as well. We also provide a tool, called linkensemble, to compute the minimum set of models required to account for all experimental observations.
2.3 XL statistics
Using TopoLink, we have computed the potential crosslinkable distances of the CATH database of 21 091 non-homologous proteins (S40 v4.1) (Sillitoe et al., 2015). Results were organized in terms of protein size, linker length, and residue type, and can be accessed in our web server (at the ‘XL Statistics’ link). An example is shown in Figure 1D: The number of crosslinkable distances of a DSS linker increases with protein sequence length essentially linearly, and about 8 XLs are expected to be observed linking pairs of Lysine residues for proteins of ∼150 residues.
3 Conclusion
A package to study the consistency between XLMS experiments and structural models was developed. The software uses a physically inspired distance calculation algorithm, and its input is thought in terms of the experimental design. A graphical user interface and a web server are available. Statistical analysis tools are provided for multiple-model runs.
Funding
We thank the funding agency Fapesp [Grants 2010/16947-9, 2013/05475-7, 2013/08293-7, 2016/13195-2 and 2018/14274-9] for financial support.
Conflict of Interest: none declared.
References