-
PDF
- Split View
-
Views
-
Cite
Cite
Mark N Puttick, MCMCtreeR: functions to prepare MCMCtree analyses and visualize posterior ages on trees, Bioinformatics, Volume 35, Issue 24, December 2019, Pages 5321–5322, https://doi.org/10.1093/bioinformatics/btz554
Close - Share Icon Share
Abstract
The fossil record is incomplete, so molecular divergence time analysis is a crucial tool in estimating evolutionary timescales. MCMCtree contained in the PAML software provides Bayesian methods to estimate divergence times of genomic-sized sequences. Here, I present MCMCtreeR, a flexible R package to prepare time priors for MCMCtree analysis and plot time-scaled phylogenies. The package provides functions to refine parameters and visualize time-calibrated node prior distributions so that these priors accurately reflect confidence in known, usually fossil, time information. After the parameters have been chosen, the package produces output files ready for MCMCtree analysis. Following analysis, the package has tools to compare prior and posterior calibrated node age distributions and produce plots of the time-scaled phylogenies. The plotting functions allow for the inclusion of age uncertainty on time-scaled phylogenies, including the display of full posterior distributions on nodes. Options also allow for the inclusion of the geological timescale, and these plotting functions are applicable with posterior age estimates from any Bayesian divergence time estimation software.
MCMCtreeR is an R package available on CRAN (https://CRAN.R-project.org/package=MCMCtreeR). MCMCtreeR depends on the R packages ape, sn and stats4.
1 Introduction
The fossil record is incomplete, so it is not possible to build a reliable timescale of life with fossils alone. The molecular clock has been vital in understanding species’ divergence times, as it allows for the incorporation of fossil information, typically as node priors, to calibrate estimated divergence times from modelled changes in DNA or amino acid changes (Yang and Donoghue, 2016).
Bayesian estimation of molecular clock parameters has become an effective method of divergence time estimation (Rannala and Yang, 1996; dos Reis et al., 2016), mainly as it allows for the integration across uncertainties in node ages and other model parameters. These methods use Bayes’ theorem to estimate the posterior distribution of parameters, including node age estimates, that are informed by the model and prior information. For calibrated nodes, the prior probability is a probabilistic distribution that reflects a user’s uncertainty in age values. When calibrating these internal nodes of a phylogeny, the user will generally want to use available fossil data to define a prior distribution that accounts for all the uncertainties surrounding this source of information, i.e. specifying sensible maximum and minimum bounds. This Bayesian divergence time estimation is implemented in several programmes, including BEAST2 (Bouckaert et al., 2014), MrBayes (Ronquist et al., 2012) and RevBayes (Hohna et al., 2016). However, it is generally only feasible to run analyses of genomic-sized datasets (Morris et al., 2018) in the software MCMCtree (Yang, 2007), mainly due to its use of an approximate likelihood calculation (dos Reis and Yang, 2011). The MCMCtreeR R package presented here facilitates these analyses, particularly by allowing users to choose the most appropriate calibrated prior node distributions for divergence time analyses.
For MCMCtree divergence time analyses, users need to specify calibrated node priors and their associated parameters. The R package MCMCtreeR provides functions to estimate parameters for time-calibrated node distributions to reflect the uncertainty in the fossil record, by allowing the visualization of distributions and automatically calculating appropriate parameter values given user-supplied temporal information. Furthermore, the package can write output files with the parameters that describe these distributions in the correct MCMCtree format, so they can be immediately read into the programme and analysed. MCMCtreeR also provides functions to plot time-scaled trees and summarize the full posterior age uncertainty on each node, with labels of geologic and absolute time. This inclusion of a posteriori age uncertainty is a vital component of Bayesian divergence time analysis, as exact values such as median estimates are not sufficient to summarize across the full age posterior (Warnock et al., 2017).
2 Materials and methods
2.1 Estimation of parameters for calibrated node priors
The functions here help users choose distribution parameters to reflect age information for prior age distributions, visualize time priors and produce output files ready to be used in MCMCtree.
MCMCtree allows users to choose from different distributions to place prior ages on internal nodes: fixed estimates, upper age, uniform (bound), Skew t, Skew normal, Cauchy and Gamma. For all of these distributions, MCMCtreeR allows users to refine parameter values that reflect confidence in prior time information, visualize distributions and write node information to files ready to input into MCMCtree. As input, the package assumes the user has a bifurcating tree topology, age information for internal nodes and taxon names that descend from calibrated nodes. The functions refine parameter values so that the resulting distribution spans user-supplied minimum bounds (lower age) and maximum bounds (upper age). By default, MCMCtreeR treats minimum ages as ‘hard’ (i.e. no probability of a younger age) constraints, and maximum ages are ‘soft’ (i.e. non-zero probability of older ages); the treatment of probabilities for these ages is fully customizable, and it is worth noting this differs from the MCMCtree defaults. The functions ensure that 97.5% of the distribution falls between these minima and maxima. For distributions with multiple parameters, typically one parameter is fixed while the other parameters are used to produce the desired age range.
2.2 Visualizing of trees and associated uncertainty
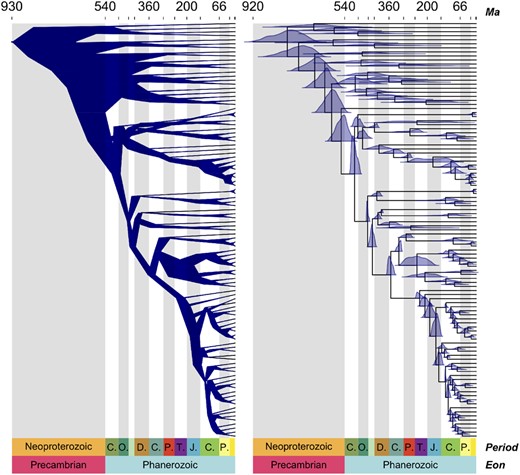
The function can take any time-scaled tree from any software that can be imported into R in the ape format (Paradis and Schliep, 2019). The package also contains methods to read and summarize posterior age estimates from MrBayes and RevBayes. Node uncertainty can be summarized using simple bars showing the 95% highest posterior density (HPD), the full posterior density plotted on the nodes of the tree and branch widths displayed as the range of uncertainty for their upper and lower ages (Fig. 1).
Examples of plotting options from the function MCMC.tree.plot() showing the ‘cladogram’ option (left), and the ‘distributions’ option with full posterior distributions displayed on nodes (right) with data from Morris et al. (2018)
3 Conclusions
MCMCtreeR provides a range of functions to prepare MCMCtree divergence time analysis. The package also allows for visualization of trees and age uncertainty from analysis using any software. Full documentation is available on CRAN with two vignettes also available at https://github.com/PuttickMacroevolution/MCMCtreeR/tree/master/vignettes.
Acknowledgements
Thank you to Rachel Warnock, Sandra Álvarez-Carretero, and an anonymous reviewer for comments that made this article much better.
Funding
This work was supported by a Royal Commission for the Exhibition of 1851 fellowship.
Conflict of Interest: none declared.
References