-
PDF
- Split View
-
Views
-
Cite
Cite
Jordi Bartolome, Rui Alves, Francesc Solsona, Ivan Teixido, EasyModel: user-friendly tool for building and analysis of simple mathematical models in systems biology, Bioinformatics, Volume 36, Issue 3, February 2020, Pages 976–977, https://doi.org/10.1093/bioinformatics/btz659
Close - Share Icon Share
Abstract
EasyModel is a new user-friendly web application that contains ready-for-simulation versions of the BioModels Database, and allows for the intuitive creation of new models. Its main target audience is the experimental biologist and students of bioinformatics or systems biology without programming skills. Expert users can also benefit from it by implementing basic models quickly and downloading the code for further tailoring.
Freely available on the web at https://easymodel.udl.cat. Implementation is described in its own section.
1 Introduction
Mathematical modeling in molecular system biology still requires a considerable amount of technical skill and has a time consuming learning curve. As molecular biology becomes increasingly quantitative, mathematical modeling becomes a required skill in the molecular biologist’s tool set. Many tools for modeling and simulation of biological systems are available (Alves et al., 2006). Most of them are standalone. Yet, powerful platforms for mathematical computation (PMC) exist, such as MatLab, Maple or Mathematica. These platforms offer a wide range of mathematical solutions, with nuanced and flexible graphical user interfaces (GUI). The many advantages of using these PMC for mathematical modeling of biological systems comes at the cost of having to become an expert in the platform. That cost can be partially defrayed by a user friendly application that uses a PMC as the motor for calculations, as other modeling tools have already demonstrated (Benque et al., 2012; Helikar et al., 2012; Peters et al., 2017). Here, we present EasyModel, a web-based application for mathematical modeling in systems biology. It emphasizes user-friendliness for beginner users.
2 EasyModel
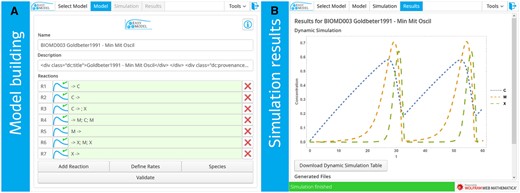
EasyModel simulates molecular biology networks using ordinary differential equations. The networks are conceptualized in terms of reactions that consume substrates, generate products and whose flux can be modulated by activators or inhibitors. EasyModel enables kinetic modeling and analysis for novel users, using the powerful Wolfram webMathematica calculus environment and its wide scope of implemented numerical and analytic methods. This is done in two ways. First, the application contains hundreds of ready-to-use models from the BioModels Database (Le Novère et al., 2006). It also provides a few additional models that illustrate how different kinetic functions, such as power laws, can be implemented. Second, an user-friendly GUI (Fig. 1) allows the user to handle those models and simulate or modify them. Users can also implement, simulate and analyze their own models in a simple way. On the background, EasyModel also caters for more advanced users by allowing them to download the code for more advanced tailoring and/or local use.
Minimal Cell Cycle model, as implemented in BioModel 003: A minimal cascade model for the mitotic oscillator involving cyclin and cdc2 kinase (Goldbeter, 1991). (A) How to input the model into EasyModel. (B) The results of the simulation, using default display options
2.1 Usage and features
When guest users enter the application, they are presented with a brief tutorial on how to use it. EasyModel complies with SBML (Hucka et al., 2003) Level 3 Version 2 specification, enabling the interchange of models with other modeling tools.
Users can use the full functionality of the tool as guests. Initially, they must select a model repository (public or private), followed by either choosing a preexisting model, importing a model from a SBML file, or creating a new model. They can also create an account if they want to store their own models privately.
Models are implemented using a simple notation with substrates separated from products by arrows. Users can either select a predefined rate law or define a new mathematical function for the kinetics of each process. Kinetic parameters may be also be linked to model species. EasyModel validates the model before simulation and analysis, warning the user if additional information or modifications are needed.
After model validation, users define how long the system is to be simulated and what type of analyses are to be performed. Both, time course and steady state simulations are available, as is sensitivity analysis with respect to model parameters and independent variables for both types of simulation. A linear stability analysis of steady states can also be performed.
After simulation, the results are represented in plots and tables. Both are downloadable. Users may also download the generated Mathematica notebook, and the model in SBML format.
2.2 Implementation
EasyModel web application was developed on the Java 8 EE programming platform, using the Vaadin 8 UI framework. It is deployed on a Tomcat 9 server. Model simulation is performed using webMathematica 3.4.3. Data are stored in a MySQL 5.7 server. SBML support and compliance was implemented using the JSBML Java library (Dräger et al., 2011). Source code is available at https://github.com/jordibart/easymodel licensed under the GNU GPL. All the dependencies of our project are open-source except for webMathematica, which requires a commercial license.
3 Conclusion
EasyModel bridges a gap that exists for most simulation software tools in systems biology: the trade-off between having a usable GUI that makes it an easy entry point for newcomers and the ability to fine-tune and expand the simulation code for more advanced uses. Future plans for the application include: to allow combination of individual models, scanning of parametric values and independent variables, stochastic simulations, with ‘delay’ and special structure, among others.
Funding
This work was partially supported by Ministerio de Economia, Industria y Competitividad [TIN2017-84553-C2-2-R]; Ministerio de Educacion [PRX18/00142] and by Bridge Grants from Universitat de Lleida and INSPIRES.
Conflict of Interest: none declared.
References