-
PDF
- Split View
-
Views
-
Cite
Cite
Abdullah Çağlar Öksüz, Erman Ayday, Uğur Güdükbay, Privacy-preserving and robust watermarking on sequential genome data using belief propagation and local differential privacy, Bioinformatics, Volume 37, Issue 17, September 2021, Pages 2668–2674, https://doi.org/10.1093/bioinformatics/btab128
Close - Share Icon Share
Abstract
Genome data is a subject of study for both biology and computer science since the start of the Human Genome Project in 1990. Since then, genome sequencing for medical and social purposes becomes more and more available and affordable. Genome data can be shared on public websites or with service providers (SPs). However, this sharing compromises the privacy of donors even under partial sharing conditions. We mainly focus on the liability aspect ensued by the unauthorized sharing of these genome data. One of the techniques to address the liability issues in data sharing is the watermarking mechanism.
To detect malicious correspondents and SPs—whose aim is to share genome data without individuals’ consent and undetected—, we propose a novel watermarking method on sequential genome data using belief propagation algorithm. In our method, we have two criteria to satisfy. (i) Embedding robust watermarks so that the malicious adversaries cannot temper the watermark by modification and are identified with high probability. (ii) Achieving ϵ-local differential privacy in all data sharings with SPs. For the preservation of system robustness against single SP and collusion attacks, we consider publicly available genomic information like Minor Allele Frequency, Linkage Disequilibrium, Phenotype Information and Familial Information. Our proposed scheme achieves 100% detection rate against the single SP attacks with only 3% watermark length. For the worst case scenario of collusion attacks (50% of SPs are malicious), 80% detection is achieved with 5% watermark length and 90% detection is achieved with 10% watermark length. For all cases, the impact of ϵ on precision remained negligible and high privacy is ensured.
Supplementary data are available at Bioinformatics online.
1 Introduction
Digital watermarking is one of the most important technological milestones in digital data hiding. It is used as a technique to hide a message or pattern within the data itself for various reasons like copyright protection or source tracking of digitally shared data. Watermarks may contain information about the legal owner of data, distribution versions and access rights (Lee and Jung, 2001). Although watermarking has a wide range of applications, implementation schemes require different configurations for each use case and data type. For embedding copyright information in data and source tracking, robustness (Barni and Bartolini, 2004) against modifications is the crucial factor to preserve. The factors influencing such configurations alter depending on the characteristics of data, as well. Non-existent redundancy, the existence of correlations and prior knowledge for inference and the utility’s impact on attacker inference in sequential data are such factors that prevent the explicit implementation of digital data watermarking methods on sequential data.
We propose a novel watermarking scheme for sharing sequential genomic data consisting of single-nucleotide polymorphism (SNPs) with three states to be used by medical service providers (SPs). Each SP has access to the uniquely watermarked version of some individuals’ genomic data. The preliminaries expected from this watermarking scheme are robustness against watermark tampering attacks such as modification and removal, imperceptibility for not revealing watermark locations, preserving utility of original data to a degree through a minimum number of changes and satisfy local differential privacy (LDP) in watermarks so that the watermarked versions of the data are indistinguishable from actual human genomic data and provide plausible deniability to data owners. By doing so, the watermarked data will not be shared in an unauthorized way and the source(s) of a leak will be easily identified by the data owner. To solve this multi-objective optimization problem, we use the belief propagation (BP) algorithm, which helps us to determine optimal watermarking indices on data that may preserve robustness with the highest probabilities. In BP, we consider the public knowledge about the human genome like Minor Allele Frequencies (MAFs) of SNPs, point-wise correlations between SNPs, called Linkage Disequilibrium (LD) and the prior knowledge of genotype and phenotype information that may potentially leak probabilities about watermarked points. Through conversion of prior information (MAF, LD and so on) into the marginal probability distribution of the three SNP states, we manage to infer the state probabilities of each SNP. Our contributions are as follows:
We introduce a novel method for watermarking sequential data concerning the privacy of data and the robustness of watermark at the same time. We present the method’s strengths and weaknesses in various attack scenarios and provide insight into the weaknesses.
Our method uses prior information (MAFs, phenotype information and so on) and inherent correlations to infer the state probabilities of SNPs. Using these inferred probabilities, we select SNPs that satisfy the following two criteria in a non-deterministic setup: a low probability of robustness decrease (change resistance) when attacked and a low utility loss (efficient index selection) when changed. By giving priority to these SNP points for watermarking, we guarantee the preservation of robustness and utility in data against various attacks. Besides, the identification probabilities of single SNPs using prior information are decreased with this method.
We test the robustness and limitations of our method using collusion (i.e. a comparison using multiple watermarked copies of the same data) and modification attacks and demonstrate how to reach a high probability of detection with various parameters, such as the watermark length, the number of SPs, the number of malicious SPs and the ϵ coefficient of LDP.
We introduce randomly distributed non-genome-conflicting noise generated for the data to act naturally as watermarks and create imperceptible watermark patterns from the normal human genome if not attacked with collusion. Hence, rather than creating a fixed number of point-wise changes and tracking these changes for source tracking, we evaluate the whole data and reach a high probability of detection with a minimum number of changes.
We introduce watermarking schemes that satisfy ϵ-LDP and plausible deniability in data along with it for data owners who value additional manners of enhanced privacy.
We provide a summary of related background on genomics, specifically Minor Allele Frequency (MAF) and LD, and LDP, in Supplementary Appendix Section 1.
2 Related works
Recent advances in molecular biology and genetics and next-generation sequencing increased the amount of genomics data significantly (Carter, 2019). While achieving a breakthrough in the genomics field, genomics data posses an important privacy risk for individuals by carrying sensitive information, i.e. kinship, disease, disorder or pathological conditions (Grishin et al., 2019). Thus, collecting, sharing and conducting research on genomic data became difficult due to privacy regulations (CMS, 1996). Furthermore, Humbert et al. (2013) show that sharing genomic data also threatens the relatives due to kin relation of genomic data. To this end, several works have been conducted to find emerging ways of privacy-preserving collection and analysis of the genomic and medical data in the last decade. Some of the privacy-preserving techniques used for medical data collection are k-anonymity, l-diversity, de-identification, perturbation, anonymization, or t-closeness (Kargupta et al., 2003; Li and Li, 2006; Machanavajjhala et al., 2007; Samurai and Sweeney, 1998; Wylie and Mineau, 2003). These methods, however, provide limited privacy protection and are prone to inference attacks. Ayday et al. (2013) proposed obfuscation methods in which the output domain is divided into several sections and one section is reserved for genomic data protection.
Digital watermarking is a technique usually used for copy protection by inserting a pattern to the digital signal such as a song, image, or video (Cox et al., 2008). It is an attack counter-measure for the case of leakage or sharing without consent. Watermarking does not prevent leakage; it is used as a detection technique for malicious parties. Watermarking schemes for sequential data, especially for genomic data, are very rare. Iftikhar et al. (2015) proposed a robust and distortion-free watermarking scheme, GenInfoGuard, for genomic data. Similar to ours, Liss et al. (2012) proposed a permanent watermarking scheme in synthetic genes that embeds binary string messages on open-frame synonymous amino-acid codon regions. Heider and Barnekow (2008) proposed the use of artificial dummy strands to act like watermarks on DNA.
Ayday et al. (2019) proposed a robust watermarking scheme for sharing sequential data against potential collusion attacks by using non-linear optimization. Our objective model is similar to theirs. Different from their study, we consider prior information in which besides correlations, all sequential genomic data related information like familial genomes, phenotype states can be included by using factor nodes in the BP algorithm. Besides, we designed single SP, collusion and removal attacks that incorporate all information so that the worst-case scenarios are assumed and the attack model becomes more inclusive. Another difference between our method and theirs is the incorporation of ϵ-LDP as an extra measure of privacy without impacting security. Andrés et al. (2012) proposed a method of embedding noise in sequential location data for geo-indistinguishability without violating the differential privacy. Inspired by their study and their new differential privacy criteria, we implemented a local setup that prevents the LDP violations in every data index.
3 Problem definition
We present the data, system and threat models and the objective of our system. Frequently used symbols and notations are presented in Supplementary Appendix Section 2.1, Table 3.
3.1 Data model
Sequential data contain ordered data points , where dl is the length of the data. The values of xi can be in different states from the set {} depending on the data type. For example, xi can be an hour, minute, or second triplets ranging from 0 to 23, 59, 59, respectively, for timestamp data. For our system, we will use 0, 1 and 2 for the SNP states of homozygous major, heterozygous and homozygous minor, respectively. The data length is dl and the number of points that will be watermarked at the end of the algorithm is wl.
3.2 System model
We consider a system between the data owner (Alice) and multiple SPs with whom Alice shares sequential genome data. SPs can be medical researchers, medical institutions, or bio-technical companies. Alice may decide to share the whole data or parts of it to receive different services. Besides, the parts shared with each SP may differ.
For all cases listed above, Alice wants to ensure solely that her data will not be shared unauthorized by SPs. If the data is shared, she wants to preserve a degree of differential privacy and detect the malicious SP(s) who shared the data. She uses watermarking and shares a watermarked version of the data, which satisfies the degree of privacy she desires. These versions are produced by removing certain parts or modifying the data. Data indices most optimal for the satisfaction of the mentioned criteria and the purposes of SPs who may use the data for including but not limited to research should be calculated beforehand by considering the structure, distribution and vulnerabilities of the data. Therefore, the proposed solution to the watermarking problem should have the element of flexibility for differing requirements of Alice and SPs, and the different pieces of information that might be used for the inference of data. To calculate the complex probability distributions of multi-variable sequential data among many things we use BP. Please see Supplementary Appendix Section 2 for the details of BP and our proposed solution. Other graph inference methods could be used but BP is adapted because of its approximation efficiency in non-loopy graph networks.
Watermarking is mostly done by changing the status of data indices. Adding dummy variables is an example of methods that do not change the actual values but common methods used for watermarking are usually removal or modification. Since a slight addition in sequential data causes a shift in other indices, it impacts the rest of the retrieval and embedding processes like the butterfly effect. We stick with the watermarking method by removal or modification. In a broader sense, non-sharing can be considered as modifying the status of a certain index into ‘non-available’. Normally, the security of a watermarking scheme increases along with the length of the watermark against attacks. However, a robust watermark should be short and as efficient as possible to maximize the detection probability of malicious SPs without reducing utility significantly.
3.3 Threat model
The objective of our proposed system and watermarking, in general, is identifying the source(s) of leakage when the data is shared in an unauthorized way. In the threat model, contrary to our objective, the only goal of malicious SPs is to share the data undetected. SPs can achieve this goal by decreasing the robustness of the watermark, which prevents the identification of the leakage source(s). Malicious SPs can identify high probability watermark points and tamper with the watermark pattern by removal or modification. For such scenarios, we presume that malicious SPs will not do blind attacks without the prior information of watermarked indices. These types of attacks will decrease the utility of data more than the robustness of the watermarking scheme and render the data useless. In favor of SPs though, we assume that they have all the prior information, such as parental genome data, observable traits for truly reflecting a worst-case scenario. Hence, we introduce three attack models that incorporate the additional one within both based on probabilistic identification that tests the robustness of the watermarks that our proposed method generates.
Single SP attack:
In this attack, a single malicious SP is expected to use the prior information available to infer the actual states of the data and identify the watermarked indices without collaborating with other SPs. Examples of prior information include MAFs, genotype and phenotype information of parents. For each data point, malicious SP finds the posterior probability of each state given the prior information prior information and compares it with the expected probability of given state . If the difference between the posterior and expected probabilities for the given state is high, it may indicate a watermarked index. We assume that the malicious SP knows the watermark length wl. Hence, SPs select the top wl indices with the highest differences in probability as watermarked and implement an attack.
Another vulnerability that malicious SPs can exploit is the inherent correlations and their values in the data to infer the actual states of correlated indices. For genomic data, linkage disequilibrium (LD); non-random association of certain alleles is an example of such correlations. LD is a property of certain alleles; not their loci. The correlation of alleles {A, B} in loci {IA, IB} will not hold if either A or B changes. The asymmetric correlation observed in LD is a valid method of representation for other sequential data types. We treat the correlations in the data as pairwise and asymmetric in the proposed system.
Collusion attack:
In addition to the knowledge obtained via a single SP attack, multiple SPs that receive the same proportion of data can vertically align their data to identify watermarked points. When SPs align their data, there will be indices with different states that can be considered as definitely watermarked. The proportion of data shared with SPs may differ, which will decrease the efficiency of alignment. However, for the construction of a strong model against worst-case scenarios, the system considers the same data is shared among all SPs. Potentially watermarked indices received from collusion attack can be used along with prior information obtained from running a single SP attack. This type of attack detects further more watermarked indices than the single SP attack.
Removal attack:
Using the knowledge from collusion attacks, malicious SPs may remove the watermarked indices or more data points from the received data. In collusion attacks, these indices are changed to other states of SNPs rather than being removed, which reduces the robustness of the detection algorithms. We design this attack to simulate a scenario of partial-sharing and its main focus is to remove the index sets that are longer than wl. Removal or change of indices longer than wl is not a preferred strategy in other attack types due to the increased reduction of utility per unit data, which should be desired less by the malicious SPs.
4 Proposed solution
When Alice wants to share her data with , they employ the following protocol. sends a request to Alice providing the indices required from her data, denoted as Ii. Alice then generates a list of available indices most suitable for watermarking Ji that satisfies and . The BP-based watermarking algorithm generates Ji, which we discuss in the sequel. Finally, Alice inserts a watermark into the indices of Ji. If the data is in binary form, it is as simple as changing 0 to 1 or vice versa. Otherwise, for the given state xi, a different state yi from the set and is chosen to be a part of the watermark pattern. In non-binary selection, if the given index contains correlation with other indices, the selection is determined by the probabilities and statistics of the correlated indices so that the watermark would not be vulnerable to correlation attacks. Otherwise, it is a random selection with uniform distribution.
Our method relies on the BP algorithm that uses prior information and previous shared versions of the data to identify the indices with maximized detection probabilities of malicious SPs, ensured privacy and minimized utility loss when modified for watermarking. BP is an iterative message-passing algorithm used for the inference of unobserved random variables from the observed ones. We use this algorithm to infer the probability distributions of indices given the multi-variable prior information, attack scenarios and privacy criteria. Normally, the factorization of prior information marginal probabilities could be used for a part of the inference of state probabilities. However, probability calculation gets exponentially complex as the dimensions of the data and the variety of prior information increases. Because BP approximates the actual state probabilities in a finite number of iterations, it is much more efficient than factorized calculation. The main idea is to represent the probability distribution of variable nodes by factorization into products of local functions in factor nodes. Therefore, given new prior information (e.g. complex correlations, phenotype indicators and disease history), factor nodes can be extended or re-formulated.
4.1 Nodes and messages
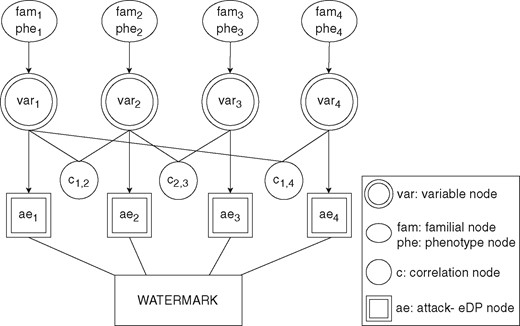
We describe the general setup and the details of the proposed BP-based algorithm for genomic data. BP consists of factor nodes, variable nodes and messages between them. Connections between variable nodes and factor nodes we use are given in the factor graph (see Fig. 1).
Factor graph representation of variable nodes and attack-eLDP interactions with other factor nodes: familial nodes, phenotype nodes and correlation nodes
The notations for the messages at the vth iteration are as follows:
: Message from variable node vari to factor or attack and ϵ-LDP (attack-eLDP) node k.
: Message from familial node to variable node k.
: Message from phenotype node to variable node k.
: Message from correlation node to variable node k.
: Message from attack-eLDP node to be used as parameters for the watermarking algorithm.
4.1.1 Variable nodes
Variable (decision) nodes represent unknown variables. Each variable node sends and receives messages to/from factor nodes to learn and update its beliefs. Its purpose is to infer the marginal state probabilities of all indices that can be obtained from prior information. For genomic data, this information is the publicly known statistics, such as LD correlations, familial genomic traits and phenotype features.
For each node, we have a marginal probability distribution of states . Each variable node, vari, represents the marginal probability distributions of the ith unknown variable in the format of so that each P corresponds to the probability of one y and all sums up to 1. The probability distributions in variable nodes are calculated by multiplying the probability distributions coming from the neighboring factor nodes, such as correlation, familial and phenotype nodes. The message from variable node i to factor node k indicates that at vth iteration where . Equation 1 provides the function for the representation of a message from variable node i to correlation factor node k:
where Z is a normalization constant and for all y must be equal to 1.
4.1.2 Factor nodes
Factor nodes represent the functions of factorized joint probability distributions of variable nodes. The messages received or sent by them might be dependent on multiple variable nodes as well as a single variable node. Factor nodes might also be independent and fixed from the start. For genomic data, the correlation between SNPs (LD) can be given as the example of the first case. In such scenario, variable node is connected to a correlation factor node along with the correlated variable node . For the second case of dependency on a single variable, a message passed into the AE-node is determined by the current state of any variable node can be given as an example. For the third case of dependency, family genomic information predetermined from the start can be given as an example. Let us assume for an SNP x, genomic information obtained from the family (father and mother) of certain individual L is (homozygous major) and (heterozygous). Then, we can safely predict the marginal probability distribution of that individual’s SNP as P(L, x)=[0.5, 0.5, 0] using the Mendelian Law of Segregation. This probability distribution is constant and not dependent on any value that the variable node might get. Therefore, throughout the algorithm, this probability distribution is propagated unchanged for any SNP xi and receives no message from its corresponding variable node.
Correlation factor nodes: We use LD to enhance the robustness of the system against correlation attacks. Hence, malicious SPs will not be able to use the SNPs, which are correlated with other SNPs with high probability, for watermark detection. For every SNP pair, correlation coefficients are calculated before the iteration and the pairs with coefficients σsj higher than the ρ threshold are marked as correlated and sensitive. Correlation coefficients may differ dependent on the states of each data point and their impact on estimating the probability distributions are typically asymmetric. For each sensitive SNP pair, there is one correlation node that keeps track of the correlations inside the data.
The intuition for calculating the message to be sent by correlation node is derived from the definition of r-squared, or the coefficient of determination, that explains how good the proportion of variance in the dependent variable predicts the proportion of variance in the independent variable (Glantz et al., 2016). Since our system uses and infers marginal probability distributions in BP, we use as a metric of how well we can predict the probability distribution of one state using the probability distributions of other correlated states (Miller, 1994). The messages from correlation node to jth variable node are calculated as
In these equations,
where s is the neighbor variable node, σsj denotes the correlation coefficient, and denotes the coefficient of determination. For further insight, please see the example in Supplementary Appendix Section 2.2.1.
Familial factor nodes: Familial factor node, calculates message using the Mendelian Inheritance Law of Segregation and sends it to variable node k. Please see Supplementary Appendix Section 1, Table 1 for Mendelian inheritance probabilities using the Law of Segregation. In the message, fi and mi corresponds to the ith SNP values of father and mother, respectively.
For example, if the father has and the mother has for the ith SNP, the message from familial node is as follows:
Phenotype factor nodes: Phenotype factor node, , is designed for representing probabilistic observable traits (e.g. certain hereditary diseases, tongue-curling, or hair color) that can be used for the inference of SNP states. In our experiments, we simulated this node with Mendelian phenotype traits only mainly due to the challenges of finding an exact causative gene and determining its exact predication value in non-Mendelian inheritance (van Heyningen and Yeyati, 2004). However, other observable traits are also viable with re-formulation. calculates message using the Mendelian Inheritance Law of Dominance and sends it to variable node k. Please see Supplementary Appendix Section 1, Table 2, for Mendelian inheritance probabilities using the Law of Dominance. In the message, corresponds to the dominance trait of the observed phenotype in ith SNP. can be either dominant or recessive. For example, if the data owner is known to have blue eyes (recessive gene), which we encode in the ith SNP, the message from phenotype node is as follows:
Attack-eLDP nodes: This bold definition is not used like a title, but as a definition similar to Correlation, Familial and Phenotype Factor Nodes.
The attack and ϵ-LDP (attack-eLDP) node is designed to simulate the inference power of the attackers on data and calculates the inverse probabilities that will keep the attacker uncertainty at maximum against single SP and collusion attacks while keeping the LDP criteria intact by updating the watermarked state options which violate ϵ-LDP. This node receives a message from the variable node. Although acting as another factor node, it does not send the message to the variable node. Instead, the attack-eLDP node sends its message along with a variable node message to the watermarking algorithm as parameters.
Inside the attack-eLDP node, the attack part re-calculates the watermarking probabilities of all indices based on the variable node probability distributions and previously shared versions of states to simulate single SP and collusion attacks. In every ’s watermarking, a set of previous sharings for each index i or set of indices I are used as a prior condition. Then the probabilities of the potential next states are calculated using binomial distribution given S. Finally, updated probability distributions are sent to the watermarking algorithm as the watermarking probability of each state. The calculation procedure followed by the node’s attack part is described in the sequel:
is the number of states equal to y in set
and are calculated from the variable node’s message.
, where A is the updated marginal watermarking probability distribution of ith index for the .
The eLDP part checks whether any states violate the LDP (Kairouz et al., 2014) condition for Alice who wants to have a plausible deniability factor for the versions of data she shares. The condition is satisfied if no state violates Equation 3 in Supplementary Appendix Section 1.2. Probability distributions of the violating states converge by continuous averaging with variable nodes until non-violation. This incorporation creates watermarks for all the SNPs of the data owner and acts as a lower bound of privacy ensured along with lower and upper bounds on confidence degree by its very definition. For further insight, please see the example in Supplementary Appendix Section 2.2.2.
4.2 Watermarking algorithm
SNP state inferences are assumed to be conducted by malicious SPs as well, given their prior information on the data for SP and Correlation Attacks (cf. Section 3.3). We consider attacker inference strength and privacy criteria at the same time during watermarking in which modifying the actual state of the data is mandatory. Modifying more indices than necessary results in losing utility on data. These modifications increase the detection probabilities of changed indices by malicious SPs and decrease efficiency. These modifications must be interpreted as actual data, not to give further means to malicious SPs for detecting watermarked indices. For example, watermarking a with is meaningless. Because no state of the other than homozygous major is observed, any change will be artificial and interpreted as watermarked.
Our watermarking scheme uses a probabilistic watermarking pattern rather than a deterministic one. To this end, we use a different set of indices and states to be watermarked for each SP. If we use fixed watermarked indices, it presents a risk of compromising watermark robustness against modifications and removals in single SP attacks and collusion attacks. If we use fixed watermarked states for each index, the data do not reflect the population distribution, and using the probabilistic inference, attackers can identify the indices that show discrepancies with the population.
Given these criteria, we calculate a watermark score that helps us to list indices better to watermark in descending order. This score is calculated by comparing the attack-eLDP marginal probability distributions with the original states of data. Firstly, the probability of the actual state in attack-eLDP distribution is subtracted from one. This gives us the probability of that index being watermarked. Then these indices are sorted in descending order to give priority to indices most likely to be watermarked. For further insight, see the watermarking algorithm given in Supplementary Appendix Section 3.
5 Evaluation
We evaluated the proposed scheme in various aspects like security against detection (robustness), the length of the watermark, utility loss and privacy guarantees. These aspects and their correspondence to the dependent variables are also provided. We give the details of the data model, the experimental setup and the results of the experiments in the sequel.
5.1 Data model and experimental setup
For the evaluation, we used the SNP data of 1000 Genomes Project (IGSR, 2013). The data set contains the 7690 SNP-long data of 99 individuals in the form of 0 s, 1 s and 2 s, which is represented as a 99 × 7690 matrix. This data set is used for learning the linkage-disequilibrium and MAF statistics along with parental data generation based on the method proposed in Deznabi et al. (2018). While the dataset and the parental data generated from it are processed, HW equilibrium is assumed. These statistics are then employed in the BP algorithm for probabilistic state inference. The threshold of pairwise correlations used for the results is specified as , since the changes in results down to are insignificant. The results for other ρ values can be examined in Supplementary Appendix Section 4.6. Throughout the experiments, the length of data dl is fixed to 1000, the number of SPs (h) is fixed to 20, and wl values vary between 10 and 100. In exceptional cases, watermarks with are also tested, too.
5.2 Evaluation metrics
We evaluate the proposed scheme via precision, which corresponds to the percentage of attackers correctly identified as malicious, utility lost using entropy and kinship coefficients, and ϵ-LDP achieved for various attack types and parameter configurations. In collusion attacks, two SP collusion scenario contains all 190 pairs of 20 SPs since, h is fixed to 20 and . This number increases rapidly as the number of collusion SPs increases. To keep the computational cost low, we took the number of malicious SP scenarios as 190 unique random sets for each case. Besides, we kept the number of malicious SPs up to k = 10 since we assume that we know the parameter k and k > 10 increases our detection results back. We find the malicious set of SPs by checking the watermark patterns. For the details of the detection algorithms, please see Section 4.1 in Supplementary Appendix.
5.3 Results of attacks
We evaluate the proposed scheme for the attack model described in Section 3.3. The robustness of the watermark is evaluated against collusion and removal attacks, in which the knowledge of single SP and correlation attacks are incorporated to reflect the worst-case scenario. For the details and results of single SP attacks that have the highest precision results, please see Supplementary Appendix Section 4.2. In these experiments, we assume worst-case scenarios to create lower bounds. The assumptions that give maximum malicious SP information are as follows.
Malicious SPs know the exact value of watermark length (wl).
For every SP, Ik is identical . It means all SPs have the same set of indices of data.
Malicious SPs have all the population information e.g., correlations, MAFs, frequency of states.
Malicious SPs know the SNPs of the data owner’s father and mother.
Malicious SPs know all the observable (phenotypical) features of the data owner and correspondent SNP states.
5.3.1 Collusion attack
In collusion attacks, multiple SPs collude and bring their data together to detect and modify the watermarked indices. Firstly, the states different for the same SNP are identified as watermarked because the watermark pattern is unique for each SP. The maximum number of indices that can be identified as watermarked by malicious SPs is , where k is the number of malicious SPs. Secondly, when the malicious SPs find fewer points in collusion attack than , they target additional indices as watermarked using the prior information on the data, e.g. MAFs and LD correlations, similar to the single SP attack. These indices are usually the least likely states when prior information is considered. At the end of the collusion attacks, malicious SPs change the states of data in two ways. The states that are not the same across all malicious SPs are modified to the most frequent ones. Then, the states that are the same across all malicious SPs but having the least likelihoods are modified to the most likely states possible. Our initial detection method, modification setups and assumptions on the collusion attack are similar to those of a single SP attack. Since collusion attacks contain much more information about the probability of a state than the single SP attack, we expect the precision results of collusion attacks to be lower than single SP attacks. We expect a decrease in precision with the increasing number of malicious SPs.
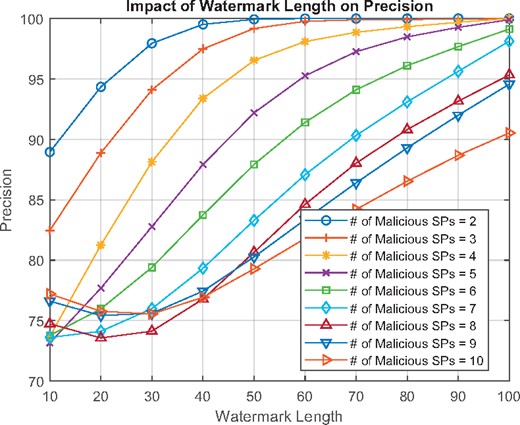
Figure 2 shows the impact of watermark length on precision for . Supplementary Appendix Section 4.3 shows the impact of detection methods and ϵ on precision. Similar to single SP attacks, ϵ has no significant impact on precision. Among 20 SPs, gives the worst results. Precision decreases as the number of malicious SPs increases, but after , almost all k malicious SP scenarios are detected with precision rates higher than 80%. With , precision increases up to 90% even for the worst case of 10 malicious SPs. We conducted experiments for and in some cases achieved precision up to 98% for .
The impact of watermark length on precision for a collusion attack ()
5.3.2 Removal attack
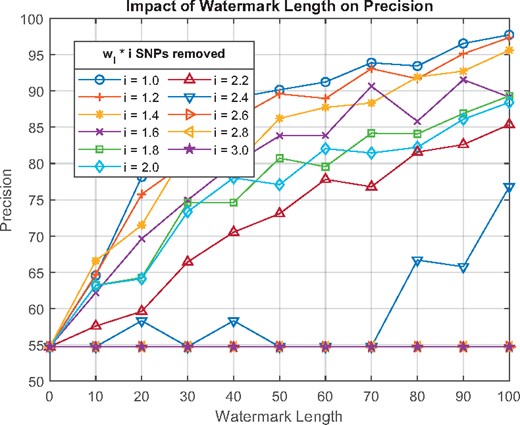
In the removal attack, index sets longer than wl are targeted and the multiplier factor of wl for removal is denoted by i. For instance, results for i = 1.6 on wl = 100 means that indices are removed. Detection method, modification and identification of the indices to be removed are same as the collusion attack.
Figure 3 shows the impact of watermark length and the multiplier factor i for 10 colluding malicious SPs. Apart from a few inconsistencies, the precision decreases as i increases, but for , the results are almost identical. Figure 3 also demonstrates that our proposed system is robust against removal attacks with 85% precision when malicious SPs remove up to 120% more data than wl. For , the entire watermark is distorted and the precision drops to the level of randomness. It is important to note that these removals are performed in the conditions most optimal to the attackers with the combined knowledge of colluding SPs and all the prior information available used in the watermarking algorithm. Therefore, results reflect the worst-case scenario. Supplementary Appendix Section 4.4 shows the results of the removal attacks of 2 and 6 malicious SPs.
The impact of watermark length on precision for a removal attack (# of malicious SPs = 10)
5.4 Utility evaluation
Besides the number of indices changed, we evaluate the utility using two methods. The first method calculates the utility loss using entropy. We calculate the total entropy of SNPs using their MAF values and HW equilibrium. The watermark-embedded SNPs lose their utility. In this method, percentage-wise changes in the total entropy between embedded and non-embedded versions are reflected as the utility loss. The second method calculates the kinship coefficients for the embedded and non-embedded versions using the equations from (Kale et al., 2017) and tables of interpretation from (Manichaikul et al., 2010). The details of the second method can be found in Supplementary Appendix Section 4.5.
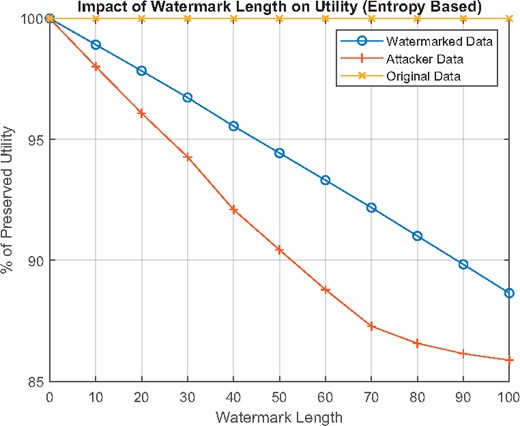
5.4.1 Entropy-based utility
In this method, for wl = 100 which corresponds to 10% of dl, utility loss is linear with respect to wl, and it is around 12%, as can be seen in Figure 4. This value may seem high, but robustness against collusion attacks dictates that SNPs with high variance (entropy) should be favored for robust watermarking with low wl. For the malicious SP-generated data used for calculating precision against attacks, utility loss is even higher. As the number of malicious SPs increases, the uncertainty of SPs about the states of SNPs decreases. Hence, they produce data with similar utility loss to the watermarked versions.
6 Conclusion
We propose a novel watermarking scheme for sequential genome data employing BP with ensured ϵ-LDP. This system is designed for use between the data owner and SPs some of whom are assumed to be malicious. We implemented the algorithm against the worst-case scenario of malicious SPs. We assume that SPs know almost all the statistics of the data. Therefore, we tested the robustness of watermarks against single SP attacks, collusion attacks and removal attacks. The BP algorithm greatly mitigates the risk of unauthorized sharing. The algorithm secure high precision rates even against the worst-case scenarios when all potential prior information that malicious SPs can use are considered. We keep the changes on data minimum for preserving the utility of data by using a short watermark length (wl). Our experiments show that when wl is kept higher than 50, even for a high number of malicious SPs, robustness is preserved more than 80% and utility is preserved up to 95%. We observe that ϵ does not significantly affect precision. Privacy is preserved without disturbing precision, which addresses a potential liability issue from data being known and offers a privacy measure of plausible deniability needed, especially for rare SNPs.
Acknowledgements
Research reported in this publication was supported by the National Library Of Medicine of the National Institutes of Health under Award Number R01LM013429.
Conflict of Interest: none declared.
References
CMS (
IGSR (