-
PDF
- Split View
-
Views
-
Cite
Cite
Loane Danès, Nicolas Tchitchek, Christophe Bécavin, Bacnet: a user-friendly platform for building multi-omics websites, Bioinformatics, Volume 37, Issue 9, May 2021, Pages 1335–1336, https://doi.org/10.1093/bioinformatics/btaa828
Close - Share Icon Share
Abstract
To face up to the exponential growth of heterogeneous datasets of various organisms, we developed a user-friendly platform for building multi-omics websites, which is named Bacnet. This platform helps bioinformaticians to construct four key web interfaces: (i) an interactive genome viewer; (ii) an expression and protein atlas; (iii) an interface for analysis of co-expression network; (iv) an interface for exploring homolog presence. We believe our platform will help the bioinformaticians to construct personalized user interfaces dedicated to biologists studying non-reference organisms.
https://github.com/becavin-lab/bacnet; Java; Eclipse RAP; Eclipse RCP.
1 Introduction
There are many databases for each type of -omics data (Athar et al., 2019; Cunningham et al., 2019; Harrison et al., 2019; Kapushesky et al., 2010; Perez-Riverol et al., 2019). The development of these databases was essential to allow a shared and reproducible science. However, most of these databases are only repositories for raw data and metadata information. Extensive bioinformatic works are then needed to analyze and interpret these data. Consequently, most of the biologists and non-bioinformatician experts are not be able to access or analyze these data because of a lack of time or knowledge. There is a real need to develop rich user web-interfaces that would allow biologists to go through these datasets.
We developed Bacnet a Java-based platform for the rapid development of ‘personalized’ multi-omics websites. Bacnet is easy to deploy on web servers and can be modified at willing. It was already used for the development of four websites, two of them being already published Listeriomics (Bécavin et al., 2017), and CrisprGo (Rousset et al., 2018). We believe Bacnet is a unique platform that allows sharing and publishing scientific results of specific organisms, opening the era of ‘personalized –omics’.
2 Materials and methods
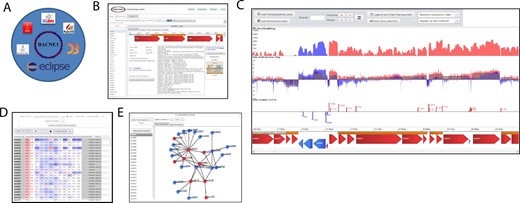
Bacnet is based on Java API named Eclipse e4 RAP. This technology is designed for rapid prototyping of websites with rich client interfaces (Fig. 1A). To help new users to deploy their own multi-omics website, we created several wiki pages in which the user can recreate a lighter version Listeriomics website (Bécavin et al., 2017). They provide more information on the architecture of Bacnet and how it was designed.
Overview of Bacnet platform. (A) Bacnet dependencies. (B) Gene panel. (C) Visualization of RNASeq, transcription start site and transcription termination site datasets. (D) Heatmap of expression atlas. (E) Co-expression network tool
3 Results
3.1 Genomic and phylogenomic interface
First, users have to provide a simple tab-delimited text file (genomics.txt) containing genome names, metadata information and RefSeq (O'Leary et al., 2016) database link. Using bacnet.e4.rap.setup first panel, the user can download the genome (.fna and .gtf files) and serialize it to allow Bacnet platform to quickly load all genomes. Then one can create a phylogenomics tree, using JolyTree (Criscuolo, 2019), of the different genomes. A homolog search tool has been implemented to complete the phylogenomics study. Users can run BlastP search (Camacho et al., 2009), for comparison of all annotated proteins in all genomes. In the multi-omics website, users will be able to display all homologs of a gene through all listed genomes in the gene visualization panel (Fig. 1B).
3.2 Transcriptomic and proteomic interface
Prior to any transcriptomic or proteomic integration, one has to provide a tab-delimited text file (bioconditions.txt). This file referenced all biological conditions which will be integrated into the multi-omic website. It can be either metadata information on transcriptomic datasets from ArrayExpress (Athar et al., 2019) or proteomic datasets found on a specific publication. Bacnet allows integrating microarray, tiling and RNA-Seq datasets, through the ArrayExpress (Athar et al., 2019) database. Using the MAGE-TAB format (IDF, SDRF, ADF files) describing each differential expression experiment, all differential expression tables will be imported. At this point, a manual curation is usually needed to correctly name the dataset and correct the metadata information. The normalized tables will then be used by the expression atlas interface implemented in Bacnet. Proteomics processed datasets are usually provided in supplementary information of scientific papers. For that reason, no automatic downloading system is provided by Bacnet. The user has to download and format each proteomic expression dataset.
Once all omics datasets are integrated, the user can deploy the multi-omics website and, either visualize every dataset on the genome viewer (Fig. 1C), or visualize their differential expression in heatmap tables (Fig. 1D). For every corresponding genes and proteins, the expression and proteome atlas will be available. Allowing the user to directly see which genomic element is highlighted in one of the omic datasets integrated into the database.
3.3 Co-expression network interface
If transcriptomic datasets are available, users can create a co-expression network to discover potentially new molecular regulations. Users can automatically select the datasets which will be taken into account for the network inference. Once selected the correlation between all genes and genome elements will be calculated and saved in a table for export. Users will be then able to visualize the network of co-expression for each gene dynamic graphical interface (Fig. 1E).
3.4 Website and desktop application deployment
Prior to Bacnet deployment, users can create an initial view of the website and select the visualization tools to add. The choice of Eclipse RAP architecture was made because of its easy to use interface named WindowBuilder. One can also add existing graphical API based on Javascript. Providing all datasets have been integrated into the database the multi-omics website is ready to publish on Apache Tomcat server. The database made of flat files should be uploaded onto the server, no SQL like architecture is needed. This makes the personalized website easy to deploy and update on every server type. If needed the website can also be exported in an executable file to be installed locally for private communication within one laboratory.
Acknowledgements
The authors thank Pascale Cossart for supporting the early development of the Bacnet platform, and all the users of Listeriomics and CrisprGo for their multiple feedbacks.
Funding
C.B. received support from the ANR [BACNET 10-BINF-02-01] and Fondation pour la Recherche Médicale [DEQ20180339158]. N.T. was supported by fellowships from the ANRS (France Recherche Nord&Sud Sida-HIV Hépatites).
Conflict of Interest: none declared.
References