-
PDF
- Split View
-
Views
-
Cite
Cite
Neil Swainston, Mark Dunstan, Adrian J Jervis, Christopher J Robinson, Pablo Carbonell, Alan R Williams, Jean-Loup Faulon, Nigel S Scrutton, Douglas B Kell, PartsGenie: an integrated tool for optimizing and sharing synthetic biology parts, Bioinformatics, Volume 34, Issue 13, July 2018, Pages 2327–2329, https://doi.org/10.1093/bioinformatics/bty105
Close - Share Icon Share
Abstract
Synthetic biology is typified by developing novel genetic constructs from the assembly of reusable synthetic DNA parts, which contain one or more features such as promoters, ribosome binding sites, coding sequences and terminators. PartsGenie is introduced to facilitate the computational design of such synthetic biology parts, bridging the gap between optimization tools for the design of novel parts, the representation of such parts in community-developed data standards such as Synthetic Biology Open Language, and their sharing in journal-recommended data repositories. Consisting of a drag-and-drop web interface, a number of DNA optimization algorithms, and an interface to the well-used data repository JBEI ICE, PartsGenie facilitates the design, optimization and dissemination of reusable synthetic biology parts through an integrated application.
PartsGenie is freely available at https://parts.synbiochem.co.uk.
1 Introduction
The computational design of synthetic biology parts is greatly aided by the increasing availability of open access tools, standards and data repositories. A number of excellent tools are available to synthetic biologists for designing novel parts, including the ribosome binding site (RBS) Calculator (Salis, 2011) and RedLibs (Jeschek et al., 2016) for an RBS optimization, and numerous academic and commercial packages for codon optimization of coding sequences (CDSs) for expression in a given host (Angov, 2011). In addition to this, the introduction of community-developed standards for representation of synthetic biology designs, such as Synthetic Biology Open Language (SBOL; Roehner et al., 2016) and SBOL Visual (Quinn et al., 2015) have greatly facilitated the sharing and reuse of synthetic DNA. Indeed, this journal now recommends the sharing of genetic designs and has a dedicated repository to which synthetic biologists are encouraged to submit synthetic DNA designs to support submitted manuscripts (Hillson et al., 2016).
Despite these advantages, practical problems exist for the synthetic biologist in utilizing these packages. First, there is still as yet little integration of tools, such that separate tools are required for an RBS optimization, codon selection and the optimization of designs to enable cost-effective synthesis by manufacturers. Second, tools that support the assembly of synthetic biology parts adhering to data standards (Zhang et al., 2017) typically prioritize the re-use of existing ‘features’, such as RBSs, CDSs, promoters and so on and do not interface with tools that design novel features. Finally, upon submitting designs to manufacturers for synthesis, a further optimization step is typically required to comply with vendor-specific synthesis constraints. Following such a step-wise approach, which is heavily reliant on the use of numerous software packages and manual cut-and-pasting of DNA sequences between tools, is unnecessarily time consuming and potentially prone to user error.
PartsGenie is therefore introduced for the design of synthetic biology parts, integrating many of the above challenges in a single web application, which can subsequently be extended to cover a more comprehensive pipeline for applications such as metabolic engineering. Driven by an intuitive drag-and-drop interface, the user can assemble multiple DNA parts from a range of synthetic DNA features, which can include a mixture of fixed, user-defined sequences, and variable sequences that are optimized simultaneously by the algorithm. The final results are directly exportable to the well-used data repository JBEI ICE (Ham et al., 2012), following journal recommendations. Such a system, bridging the gaps between multiple design algorithms and standardized data repositories, benefits both the designer in terms of simplification, and the synthetic community as a whole through data sharing.
2 Materials and methods
PartsGenie is written as a two-tier, single-page web application, using Python, Javascript and the Bootstrap and AngularJS web development libraries. The multi-objective optimization algorithm follows a simulated annealing approach. PartsGenie uses a performance-optimized version of the RBS Calculator. The system interfaces with ICE through its RESTful API. PartsGenie is hosted on Google Compute Engine and is distributed as a Docker file with source code available under MIT License at https://github.com/synbiochem/PathwayGenie.
3 Results
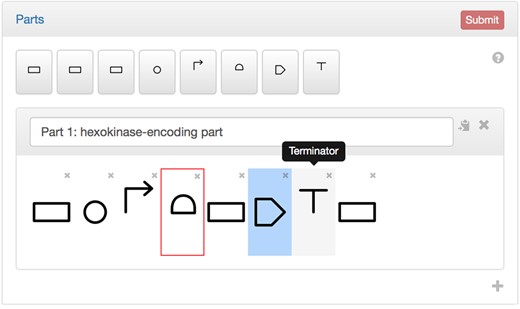
PartsGenie allows for the flexible design of multiple synthetic biology parts through the use of a simple web interface (Fig. 1). Designs can be assembled through the arrangement of multiple features, which can be drag-and-dropped from a palette that include RBSs, CDSs, promoters and so on, represented as glyphs in SBOL Visual format. Parameters required for their optimization can be specified by selecting the feature and filling in the resulting form. Examples of such parameters are a translation initiation rate (TIR) for RBSs and desired amino acid sequence for CDSs. In specifying amino acid sequences, PartsGenie offers an integrated UniProt search tool, allowing sequences, ids, NCBI Taxonomy terms and enzyme classification (EC) numbers to be automatically extracted and associated with the CDS. Extracted sequences can subsequently be edited directly in the Feature panel, to introduce mutations and truncations if necessary. A host organism is needed for designs that include the RBS or CDS features, in order to determine the optimum Shine–Dalgarno sequence for RBSs, and preferred codon usage for CDSs. (Supported prokaryotic organisms are selected from a simple auto fill field, and the backend algorithm extracts Shine–Delgarno sequences and codon usage tables automatically from the RBS Calculator and Codon Usage Database (Nakamura et al., 2000) respectively.) Multiple designs can be submitted as a single optimization task, and designs may be copied in the interface to facilitate the submission of multiple, related designs.
The PartsGenie drag-and-drop design interface. The Parts panel allows designs to be assembled from the palette of feature buttons above. Missing parameters and incorrectly ordered features (in this instance, an RBS not directly followed downstream by a CDS) are highlighted in a box, and the job Submit button disabled until such errors are corrected, preventing the submission of invalid designs to the optimisation algorithm
Filters can also be specified, which are applied to prevent undesired subsequences appearing in the optimized sequence. These include limits on the number of repeating nucleotides, the exclusion of selected codons from CDS features, and the removal of unwanted restriction sites.
Upon submission, the user is presented with a progress panel, indicating the status of the running multi-objective optimization, a simulating annealing algorithm that optimizes all objectives simultaneously. The progress panel indicates the codon adaptation index (CAI; Sharp et al., 1987) of any supplied CDS features, indicating codon suitability for the target host organism; a measure of the difference between desired and actual TIR for RBSs; the number of invalid sequences in the design, as defined by the filters; and the number of ‘rogue’ RBSs, that is, sites that resemble RBSs with a sufficiently high TIR, which may therefore reduce the efficiency of translation initiation at the intended RBS. Invalid sequences and rogue RBSs are removed from the design through optimization before designed parts are returned.
Optimized designs can be viewed in an interactive Results panel, showing the result of each submitted design and allowing each individual feature to be investigated. Designs may be saved to any JBEI ICE repository. PartsGenie therefore supports data sharing with standards such as SBOL and Genbank via its integration with ICE, from which these formats may be exported.
PartsGenie has been designed to be sufficiently generic to be of use in a range of synthetic biology studies. However, it is envisaged that over time PartsGenie will be integrated into a larger computational design pipeline for metabolic engineering. Such a pipeline will include a number of existing and novel software tools, including RetroPath (Carbonell et al., 2014) for pathway elucidation and Selenzyme (Carbonell et al., 2018) for enzyme selection. Downstream of PartsGenie, automated methods for the assembly of genetic constructs through interfacing with robotics liquid handling systems will be introduced.
Funding
This work was supported by the Biotechnology and Biological Sciences Research Council (BBSRC) and the Engineering and Physical Sciences Research Council (EPSRC) under grant ‘Centre for synthetic biology of fine and speciality chemicals (SYNBIOCHEM)’ [BB/M017702/1].
Conflict of Interest: none declared.
References