-
PDF
- Split View
-
Views
-
Cite
Cite
K Anton Feenstra, Sanne Abeln, Johan A Westerhuis, Filipe Brancos dos Santos, Douwe Molenaar, Bas Teusink, Huub C J Hoefsloot, Jaap Heringa, Training for translation between disciplines: a philosophy for life and data sciences curricula, Bioinformatics, Volume 34, Issue 13, July 2018, Pages i4–i12, https://doi.org/10.1093/bioinformatics/bty233
Close - Share Icon Share
Abstract
Our society has become data-rich to the extent that research in many areas has become impossible without computational approaches. Educational programmes seem to be lagging behind this development. At the same time, there is a growing need not only for strong data science skills, but foremost for the ability to both translate between tools and methods on the one hand, and application and problems on the other.
Here we present our experiences with shaping and running a masters’ programme in bioinformatics and systems biology in Amsterdam. From this, we have developed a comprehensive philosophy on how translation in training may be achieved in a dynamic and multidisciplinary research area, which is described here. We furthermore describe two requirements that enable translation, which we have found to be crucial: sufficient depth and focus on multidisciplinary topic areas, coupled with a balanced breadth from adjacent disciplines. Finally, we present concrete suggestions on how this may be implemented in practice, which may be relevant for the effectiveness of life science and data science curricula in general, and of particular interest to those who are in the process of setting up such curricula.
Supplementary data are available at Bioinformatics online.
1 Introduction
Computational approaches have become essential for research initiatives aiming to solve societal issues such as health, agriculture, nutrition, biotechnology and bio-based economy (Brazas et al., 2014, 2017a, b; Greene et al., 2016; Yanai and Chmielnicki, 2017). This necessity is mirrored at the educational level, but training programmes in computational biology, bioinformatics and systems biology (BioSB) seem to be lagging behind this development (Alves et al., 2011; Attwood et al., 2015), despite continued efforts, such as the PLoS Computational Biology Education collection which has already been available for more than a decade (Lewitter, 2007, 2013). Providing an appealing masters’ curriculum is a complicated puzzle, particularly in an extensive and challenging research discipline as BioSB. Such a programme needs to deliver students with the worldview, knowledge and practical abilities to conduct bioinformatics and/or systems biology research in a wide range of application areas and working environments.
The challenge is to provide sufficient depth for students from widely varying backgrounds (Abeln et al., 2013) while at the same time encouraging and training them to be able to bridge between the fields of computational methods and biological or biomedical application areas, which may range from research, to industry, to policy making, amongst others. This need, for translation, is seen in many interdisciplinary fields, and in particular throughout the data sciences (Altman, 2018; Dunn and Bourne, 2017; Manyika et al., 2011; Pournaras, 2017; Seidl et al., 2018). It is thus not unique to computational biology; nevertheless, due to the neigh unfathomable complexity of biological systems, it is arguably most strongly felt in the life sciences (Altman, 2018; Greene et al., 2016; Stefan et al., 2015).
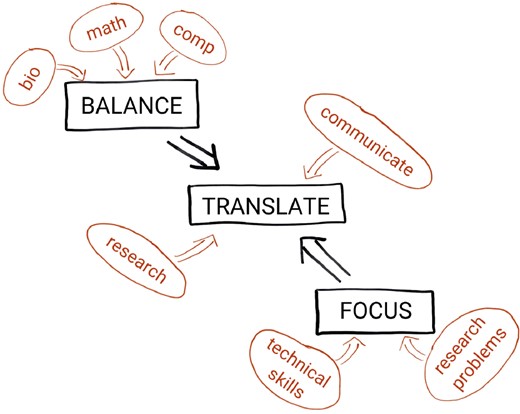
Students therefore need to obtain an overview of multiple fields, but in order to enable them to build bridges, sufficient in-depth knowledge, or focus, is vital. It is obviously impossible to provide this depth across the whole width of several fields of study. Moreover, providing sufficient depth for students from widely varying backgrounds can be challenging. By choosing several concrete questions from open biological and biomedical problems, and showing how these can be coupled to computational methods to provide an answer to these problems, one can foster awareness and a frame of mind from which these bridges may grow (Abeln et al., 2013). While setting out a guide to bioinformatics education for the life sciences, Via et al. (2013) describe this as ‘using interactivity, problem-solving exercises and cooperative learning to substantially enhance training quality’. Cvijovic et al. (2016), covering systems biology education, emphasize the ‘ability to phrase and communicate research questions [such] that they can be solved by the integration of experiments and modelling, as well as to communicate and collaborate productively across different […] disciplines’. Emery and Morgan (2017) highlight the need to provide interesting research questions in a realistic biological context. Thus, one needs a focus on open and challenging research problems to solve, and the technical skills to solve them; one needs a balance between basic skills, such as biology, mathematics (modelling) and computational tools (programming); then, building on this focus and balance, one will be able to develop the ability to translate between different disciplines. This is summarized in Figure 1.
Conceptual organization of bioinformatics and systems biology education along three key elements: balance, translate and focus
Balance should exist at many levels. Most importantly, individuals should be trained to have a balanced understanding of biological, mathematical and computational disciplines. Since, typically, students are taught with different (BSc) backgrounds, it is important that they come out of the MSc with a balanced set of skills. A requirement for any successful data science programme is teaching students how to translate research problems from one discipline to another. Programmes can foster this skill by stimulating peer support and collaboration, encouraging students to communicate with people from different scientific and cultural backgrounds. Lastly, it is crucial that focus exists within each course both on technical skills such as programming, algorithm design, experimental design, statistical techniques and mathematical modelling and on the application of these skills to complex problems within the life science domain. Here it is important that students, despite coming from a broad range of bachelors, get enough depth in the problems they encounter in the programme.
In this paper, we aim to describe this philosophy and how it is implemented in our master programme. We give an overview of how aspects of translation, focus and balance are integrated throughout the course work, and give examples from several of the courses. Various examples are given out of our own experience over the years of what problems occur when either of translation, balance and focus are underrepresented or appear too late in the curriculum. Finally, some concrete advice on practical implementation is given. According to mastersportal.com, there currently are 1970 master programmes in bioinformatics and/or systems biology. We believe the overview and guidelines provided here may be useful for those who are currently setting up a similar curriculum in life- or data science, or are adapting their existing curriculum to address the challenges of data science teaching.
2 Curriculum
The 2-year BioSB Masters’ joint programme at the Vrije Universiteit, Amsterdam (VU) and University of Amsterdam (UvA), aims to provide such a curriculum. The programme has grown out of a Master in Bioinformatics at the VU, a separate Master in Life Sciences at the UvA, both starting in 2003, and a track Systems Biology existing since 2009 in the Master Biomolecular Sciences at the VU. The current joint programme started in 2010. It offers core techniques and formalisms to every student, while allowing sufficient differentiation and choice in research projects and practical applications. For more detailed examples of course content, please refer to Abeln et al. (2013). Further detail may be obtained from the study guide (studiegids.uva.nl/xmlpages/page/2017-2018-en/search-programme/programme/2984; www.vu.nl/en/study-guide/2017-2018/master/a-b/bioinformatics/.), which is also included as Supplementary Material.
An intensive start to the programme helps to set an exemplary pace and to establish basic knowledge. In our programme, students have 24 contact hours per week in the first 2 months of the programme. During this period, while being introduced into the main topics of BioSB, depending on their background, students are taught mathematics, biology and programming to allow them to focus on technical skills (Table 1, top rows). Thus they can start filling gaps in their knowledge and start with a balanced repertoire of background knowledge.
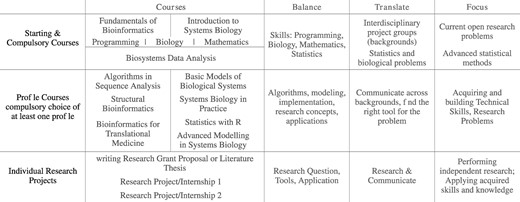
Implementation of balance-translate-focus throughout the bioinformatics and systems biology curriculum
 | |
 | |
Top rows: the starting and compulsory courses, which provide a balance of basic skills, translate between the topic areas and an introduction to open research problems in selected focus areas. Middle rows: core courses for the two profiles, bioinformatics and systems biology; students have to select one of these profiles, but typically end up doing at least a few courses from the other profile as well (balance). Translation here is from tool & method to application & problem. Bottom rows: individual projects, which are research oriented. Here, a balance between the research problem, appropriate tools and their application is key. Translation is on connecting tool to problem, as before, but more importantly here also encompasses interaction with other disciplines in the internship working environment, and communication of results to a broader audience. Focus is on becoming an independent researcher.
Implementation of balance-translate-focus throughout the bioinformatics and systems biology curriculum
 | |
 | |
Top rows: the starting and compulsory courses, which provide a balance of basic skills, translate between the topic areas and an introduction to open research problems in selected focus areas. Middle rows: core courses for the two profiles, bioinformatics and systems biology; students have to select one of these profiles, but typically end up doing at least a few courses from the other profile as well (balance). Translation here is from tool & method to application & problem. Bottom rows: individual projects, which are research oriented. Here, a balance between the research problem, appropriate tools and their application is key. Translation is on connecting tool to problem, as before, but more importantly here also encompasses interaction with other disciplines in the internship working environment, and communication of results to a broader audience. Focus is on becoming an independent researcher.
The aim of these first 2 months is to provide students with enough of a primer into programming, calculus, linear algebra and molecular biology to allow them to start working on the remaining gaps in their background more or less independently. In consultation with their mentor, and depending on their background, the student has several options:
For programming the path is most straightforward. Students without programming background, follow the ‘programming conversion class’ during the first 2 months, which entails about 2 weeks of work on the very basics of programming in python (variables, loops, if-then-else, dictionaries, lists, file IO), which is enough for them to understand what the programmer is doing in their group in the ‘Fundamentals of Bioinformatics’ (FoB) course. In the second period, they may enrol in the bachelor course ‘Introduction to Python’, which offers a more rigorous foundation for developing their programming skills.
For calculus, we expect students to have an advanced ‘high-school’ level understanding of calculus and linear algebra. The ‘maths conversion class’ during the first 2 months serves as a refresher for students who need that, and aims to get them to the level where they can comfortably participate in the basic modelling exercises of ‘Introduction to Systems Biology’ (ISB). Students who wish to enter the master with limited prior exposure to mathematics, will enrol in a bachelor course basic calculus in the first two periods, postponing any systems biology courses to the second year. However, this option will be difficult to accommodate within the nominal 2-year period for the full programme, and is therefore not generally advised.
For molecular biology, the ‘biology conversion class’ is a succinct treatment of molecular cell biology by Alberts (2015), which we find to be sufficient for students without a biology background to follow the biological parts of both starting courses. The other compulsory and profile courses contain a sufficient minimum level of biology integrated in the contents to meet the learning outcomes. Nevertheless, we also encourage these students to select more biology-oriented optional courses.
Our policy is to accept students into the programme who are versed in at least one of the above three subjects. Therefore, students are expected to complete conversion classes addressing the other two subjects, which most students can do successfully in practice.
2.1 Translate—implementation in the curriculum
BioSB rely critically on cross-talk between scientists from different backgrounds and expertizes. Throughout all courses and internships we focus on training the ability to communicate with colleagues outside the students’ own specialization. The programme thus stimulates collaboration between students, and fosters a spirit of respectful recognition of each other’s knowledge and experience (translate). From a student perspective, working and studying with peers from different BSc backgrounds is not only an invaluable experience, but is also highly regarded in every field of employment (Abeln et al., 2013; Auerbach, 2012; Cvijovic et al., 2016; Goodman and Dekhtyar, 2014; Machanick and Tastan Bishop, 2015; Manyika et al., 2011). To create such a learning environment, we mix students with different backgrounds in groups right from the start of their masters. In these heterogeneous groups, they tackle complicated research tasks and discover gaps in their own knowledge. Owing to early mixing, students from different backgrounds are encouraged to establish bonds which tend to support them throughout the year (Abeln et al., 2013).
2.2 Focus—implementation in the curriculum
On the other hand, the programme also stimulates and ensures a level of independent work by the students through assignments in compulsory courses (Table 1, middle rows). These focus on technical skills, and complex problem solving (modelling and/or algorithmics). Moreover, each student needs to carry out two internships, or research projects, in which they engage in and contribute to cutting-edge research at a research institute, university or research hospital (Table 1, bottom rows). Here, in particular, independence is expected and stimulated. On the practical side, it should however be noted that this setup is labour intensive in terms of supervision by the teaching staff. No matter to what extent the supervisory task can be reduced for our teachers, this scales linearly with student numbers.
2.3 Balance—implementation in the curriculum
One of the main characteristics of our programme is the influx of students from widely differing backgrounds. We use this mix of backgrounds to mutual advantage of students and ourselves, by creating from it a stimulating learning environment for the students. Mixing students from different backgrounds in a group to solve a complex task creates an opportunity for the students to reflect on their own abilities. This cross-disciplinary approach characterizes the teaching–learning environment in the master. Compulsory courses ensure all students receive a balanced set of skills and knowledge, and build a common frame of reference on the topics of BioSB. For more detail about the design of the curriculum and the main courses, please refer to Abeln et al. (2013).
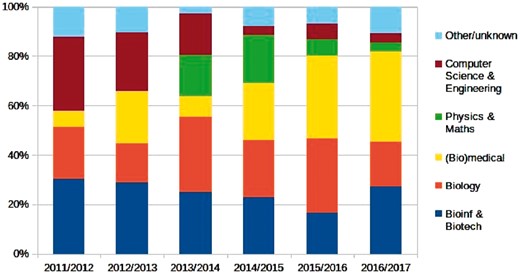
Because we want our student population to be highly diverse in topic and type of education, the programme is open to students from (almost) any science bachelor programme. The resulting diversity brings in cross-disciplinarity from the start, as illustrated in Figure 2. This ensures students learn to collaborate and translate across disciplines. For several main subjects students are accepted ‘as is’: computer science and biology from Dutch research university BSc programmes, and some related BSc programmes from the VU and UvA, for which we have had good experiences in previous years. All other students, i.e. other research-oriented BSc’s, University College graduates, vocational BSc’s, foreign diplomas, are assessed individually. Key selection elements are sufficiently high grades (i.e. focus) in at least one of the main topics, biology, computer science or mathematics, coupled with an active interest in solving biological questions with algorithmic or modelling approaches (i.e. translation).
Diversity of incoming students by topic area, over six cohorts from 2011 to 2016; the vertical axis shows fraction of students. Counting individual programme topics, bioinformatics bachelors are the largest fraction (here counted with together ‘biotech’), but on average bioinformaticians account for only 12% of all students. In recent years, biomedical bachelors are starting to dominate the influx, recently accounting for about one-third of new students
As can already be seen in Figure 2, over the last few years the distribution of incoming students has shifted. Now, a majority of students enter from a biological background, often combined with a medical, technical or bioinformatics component, which is in line with growing demands for bioinformatics and general data science skills in the life sciences and medical research, as already noted in the introduction (Brazas et al., 2014, 2017a, b; Greene et al., 2016; Yanai and Chmielnicki, 2017). We have still to find out how this may impact our programme in the long term, since we strive for a balance in student backgrounds. This trend, if ongoing, may therefore mean a lack students with a sufficiently quantitative background (i.e. computer science, statistics, mathematics). On the other hand, it might well be that biology curricula will start incorporating more quantitative and data orientated components (so delivering more ‘exact’ biologists).
Over the years, we have observed that motivation is the most predictive factor in student success, more than the background of the student. Assessing motivation, therefore, also plays a large role in the intake procedures. To some extent, these factors, background and motivation, are exchangeable in the sense that a larger gap between background and requirements may successfully be balanced by a sufficiently high motivation (Murayama et al., 2013). Expectation management is very important here, and from the intake interview we ensure students know what is expected from them, and what they should do to start addressing gaps in their backgrounds. This makes them take ownership of the responsibility to address these gaps. We have seen that providing translational aspects and balance from early on in the programme keeps motivation up in challenged students, because they get a sense of the importance of gaining complex skills as they see the wider picture.
Grosso modo, all students entering our master are deficient to some extent, for the reasons outlined above and simply because suitable bachelor programmes remain scarce. Therefore, most students are lacking in two out of the three subjects mentioned at the start of Section 2: programming, mathematics or molecular biology/biochemistry, which we address during the first two courses, and by assigning additional courses if necessary. Since 2012 we are also offering a BSc Minor programme ‘Bioinformatics and Systems Biology’ (half a year in total), which contains elementary courses on bioinformatics and on systems biology, and a combination of programming, calculus and biology. Depending on the optional courses selected by the students, the minor programme may deliver students who are set up to start our master without deficiencies.
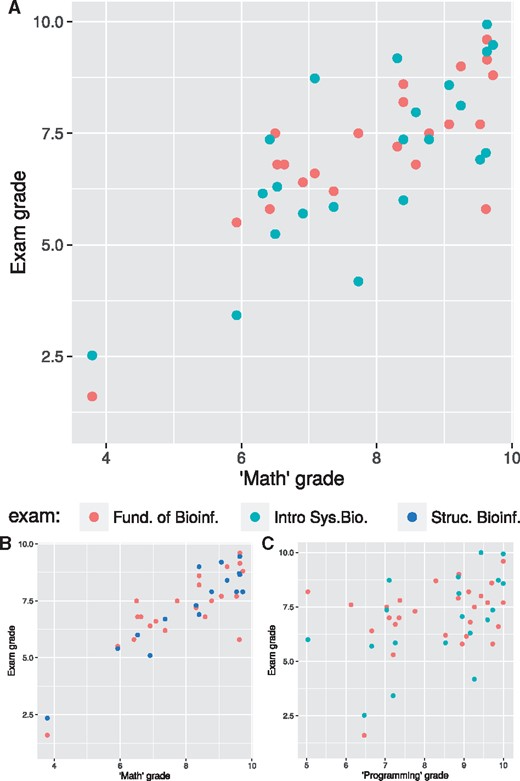
We have found that among the three deficiency topics, calculus plays a key role. First, the grades of the maths class (calculus and linear algebra) are predictive not only for the exam grades of ISB (Fig. 3A), which should come as no surprise, but also for exam grades of FoB (Fig. 3A) and even for the later structural bioinformatics course (Fig. 3B). What is more, the grades for the programming class appear much less predictive for both starting courses (Fig. 3C). Figure 3 is based on grades of 2015–2016, Table 2 summarizes the same analysis for the year 2016–2017, where the same patterns become visible. Apart from this, we have also observed that students who struggle to improve their calculus skills, typically incur delays in most other courses they enrolled in.
Correlation between grades in the maths and programming classes, and subsequent course exam grades, based on grades from 75 students in 2016–2017
| . | Maths . | Program. . | ISB exam . | FoB exam . | ASA exam . | SB exam . |
|---|---|---|---|---|---|---|
| Maths | — | 0.29 | 0.77 | 0.66 | 0.66 | 0.71 |
| Program. | 0.29 | — | 0.51 | 0.47 | 0.58 | 0.69 |
| ISB exam | 0.77 | 0.51 | — | 0.78 | 0.76 | 0.67 |
| FoB exam | 0.66 | 0.47 | 0.78 | — | 0.76 | 0.60 |
| ASA exam | 0.66 | 0.58 | 0.76 | 0.76 | — | 0.72 |
| SB exam | 0.71 | 0.69 | 0.67 | 0.60 | 0.72 | — |
| . | Maths . | Program. . | ISB exam . | FoB exam . | ASA exam . | SB exam . |
|---|---|---|---|---|---|---|
| Maths | — | 0.29 | 0.77 | 0.66 | 0.66 | 0.71 |
| Program. | 0.29 | — | 0.51 | 0.47 | 0.58 | 0.69 |
| ISB exam | 0.77 | 0.51 | — | 0.78 | 0.76 | 0.67 |
| FoB exam | 0.66 | 0.47 | 0.78 | — | 0.76 | 0.60 |
| ASA exam | 0.66 | 0.58 | 0.76 | 0.76 | — | 0.72 |
| SB exam | 0.71 | 0.69 | 0.67 | 0.60 | 0.72 | — |
The number of grades per correlation varies between 27 and 48, on average 37. High correlations (0.70) are shown in bold. ISB: introduction to systems biology; FoB: fundamentals of bioinformatics; ASA: algorithms in sequence analysis; SB: structural bioinformatics.
Correlation between grades in the maths and programming classes, and subsequent course exam grades, based on grades from 75 students in 2016–2017
| . | Maths . | Program. . | ISB exam . | FoB exam . | ASA exam . | SB exam . |
|---|---|---|---|---|---|---|
| Maths | — | 0.29 | 0.77 | 0.66 | 0.66 | 0.71 |
| Program. | 0.29 | — | 0.51 | 0.47 | 0.58 | 0.69 |
| ISB exam | 0.77 | 0.51 | — | 0.78 | 0.76 | 0.67 |
| FoB exam | 0.66 | 0.47 | 0.78 | — | 0.76 | 0.60 |
| ASA exam | 0.66 | 0.58 | 0.76 | 0.76 | — | 0.72 |
| SB exam | 0.71 | 0.69 | 0.67 | 0.60 | 0.72 | — |
| . | Maths . | Program. . | ISB exam . | FoB exam . | ASA exam . | SB exam . |
|---|---|---|---|---|---|---|
| Maths | — | 0.29 | 0.77 | 0.66 | 0.66 | 0.71 |
| Program. | 0.29 | — | 0.51 | 0.47 | 0.58 | 0.69 |
| ISB exam | 0.77 | 0.51 | — | 0.78 | 0.76 | 0.67 |
| FoB exam | 0.66 | 0.47 | 0.78 | — | 0.76 | 0.60 |
| ASA exam | 0.66 | 0.58 | 0.76 | 0.76 | — | 0.72 |
| SB exam | 0.71 | 0.69 | 0.67 | 0.60 | 0.72 | — |
The number of grades per correlation varies between 27 and 48, on average 37. High correlations (0.70) are shown in bold. ISB: introduction to systems biology; FoB: fundamentals of bioinformatics; ASA: algorithms in sequence analysis; SB: structural bioinformatics.
Correlation between grades in the maths and programming classes, and subsequent course exam grades, based on 2015–2016 grades. A—Grades of the maths class correlate well with exam grades of fundamentals of bioinformatics (FoB) and introduction to systems biology (ISB). B—Correlation of maths grades with bioinformatics courses, FoB and the later structural bioinformatics course, is also high. C—For the programming class grades, this correlation is much lower, here shown for FoB and ISB
From this we can conclude that too large a gap in the calculus background is prohibitive for students to be successful in our masters’ programme. This may be a direct result from dependence of the curriculum on calculus skills, but could also be due to an underlying factor, such as a students’ ability to make abstractions, which would help both in calculus, but also in other key topic areas in various courses. One may nevertheless expect such skills to be further developed by focussing on learning calculus (Murayama et al., 2013). This is a clear example where, although breath of knowledge, i.e. balance, is vital in an interdisciplinary setting, equally crucial is sufficient depth in several key areas, i.e. focus.
For this reason, we have over the past years put increasingly strong emphasis on this important role of maths in our masters, from the first information meetings for new students. Since the intake of 2017 we explicitly advise students to remedy their gap in calculus before the start of the programme, if needed. Ideally this would be done during their bachelor, either by selecting appropriate separate courses, or by enrolling into our bachelor minor programme (mentioned above), or with a summer course as a last resort. Students may start the master, but until they attain a basic level in calculus, they are excluded from the systems biology courses, in favour of a bachelor course on (basic) calculus. Initial results of this policy are very promising: the number of failing grades (below 5.5) scored in the maths classes of Fall 2017 was 10%, down by about half compared to the preceding three years (average 19%). It should be noted that during the same period, enrolment has nearly doubled, from just above 30 to almost 60. Therefore, while time per student has decreased for the teachers, raising awareness amongst students about the importance of sufficient depth and focus on basic skills, so far has resulted in improved student performance.
2.4 Integrating balance—translate—focus
During the taught curriculum, our key idea is to show complex and current problems within systems biology and bioinformatics to cover all necessary topics and skills (focus). Typically, there will be a couple of larger assignments in which students use the theory provided to tackle such problems. This way all the materials can be covered, and skills can be trained, using examples from current research. Hence, within the compulsory profile courses biology, programming and modelling are not separated, but follow an integrated approach (balance), as discussed above and illustrated in Figure 1. We here give an example from two of our courses.
The course Algorithms for Sequence Analysis is loosely based on the book by Durbin (Durbin et al., 1998), with many additions based on our own research (e.g. Bawono et al., 2017). The course focuses on three well-known algorithms, coupled to concrete biological problems, with an assignment offered for each one:
Dynamic programming for pairwise sequence alignment,
Hidden Markov models for sensitive profile-based sequence searching and
De Bruijn graphs and Burrows–Wheeler transform (BWT) for read assembly in high-throughput sequencing.
Further topics include Blast, PSI-blast and various sequence and alignment analysis and visualizations, ensuring a balanced coverage of the field. Assignments and final exam focus on student’s understanding of the translation between biological problem and algorithmic solution, which can go both ways.
The course structural bioinformatics is not based on an existing book; we have recently published two first chapters of what we hope will grow into a full introductory textbook on the topic (Abeln et al., 2017a, b, manuscript in preparation; Feenstra and Abeln, 2018). The course focuses on three distinct areas of protein structure analysis and prediction:
Calculation of dihedral (phi/psi) angles from atomic coordinates,
Constructing a predicted protein structure using homology modelling and
Calculating the binding free energy of two interacting proteins.
Throughout the lectures, these methods are coupled to the overarching question of deciphering protein function (translation). Concrete questions include: How to use protein structure to learn about protein function; what is the impact of a mutation on the function of the protein and how are protein function and interaction related? Basic concepts are covered, including structure prediction, protein folding, statistical thermodynamics and molecular simulations, providing a balanced background on which students’ understanding of methods and application can grow.
This approach means students can reach the level of performing independent research by the end of the taught courses, where the ability to translate research problems from one discipline to another, and communication with peers from different scientific backgrounds are key components. This culminates in the research projects in the second year, which serve as proof of competence and concludes their master work. Table 3 provides an overview of how the intended learning outcomes of the programme are aligned to support the integration of balance, translate and focus at all levels of the programme.
The intended learning outcomes of the programme are structured along the three principle elements: balance, translate and focus
| After completing the programme, the student has: | |
|
|
| After completing the programme, the student has: | |
|
|
The intended learning outcomes of the programme are structured along the three principle elements: balance, translate and focus
| After completing the programme, the student has: | |
|
|
| After completing the programme, the student has: | |
|
|
3 Discussion
3.1 Multi-disciplinary work field
Worldwide, an increasing number of BioSB master programmes exist (Pattin et al., 2014; Sainani, 2015), although unified benchmarks are only just being developed (e.g. Rosenwald et al., 2016; Welch et al., 2014). This is to be expected given the inter- and multidisciplinary nature of BioSB. An additional complicating factor is that BioSB research and education are typically embedded in biology or computer science departments, or within medical centres. Combining systems biology and bioinformatics into one master programme, fits well with developments in the field where both are generally considered integral parts of computational biology (Bare and Baliga, 2014; Kappler et al., 2017; Korcsmaros et al., 2013; Yang et al., 2009). In the Netherlands, at the post-graduate level the Netherlands Consortium for Systems Biology and the Netherlands Bioinformatics Centre merged into the BioSB Research School in 2015 (www.biosb.nl) (van Gelder et al., 2017).
The curriculum frameworks of Welch et al. (2012, 2014) and Rosenwald et al. (2016) explicitly mention the combination of BioSB as an essential component in providing a core curriculum; most notably programming and modelling. Greene et al. (2016) describe our programme as an innovative approach in interdisciplinary education which ‘are one way that programs may adapt their curricula to train students more broadly in the big data deluge’. All our students start working on these interdisciplinary and data-rich aspects already in the first 2 months of their curriculum.
Thus, the programme, as implemented in our masters’ in Amsterdam, yields students with a transferable skill set (translate), enabling students to find employment both inside the life sciences and in the more general data sciences, both in academia and industry. Instilling a balance between the things learned by students, sufficient depth and focus in selected key topics and translation capability in applying knowledge and skills to varying applications areas, helps prepare master students well for the job market. The programme provides ample opportunity for the student to select a particular domain of interest and build (focus) on experience in the target subdomain. This also helps optimize their odds to find work in the area of interest. Indeed, as far as we have been able to monitor, our students thus far all managed to find good positions after graduating.
Chang (2015) analysed a year and a half of projects in their bioinformatics core facility, and found that 46 data analysis projects had required over 34 different types of analysis methods. The vast majority of projects thus required unique, one-off approaches that were tailor-made for the task at hand, had not been used before, and likely would not be used again. In other words, there is no routine, and each analysis project becomes a research project in itself, requiring staff at PhD level to perform effectively. Note, that the lack of generalizability of such methods may be another hurdle to publication for these researchers. It should be emphasized that translation, here between project requirement and method capability, seems to have been a key element of success.
When collaborating with experimental groups, often crucial for obtaining validation, it may be difficult for bioinformatics students to obtain a sufficiently prominent position in the author list of publications. Without experimental validation, it may be hard to publish the work in a sufficiently high-impact venue (Chang, 2015). We have a limited view on the further career paths of our alumni, but from anecdotal evidence we recognize many of the concerns expressed by Chang (2015).
Yanai and Chmielnicki (2017) more recently observed a more positive trend, which we also occasionally noticed among alumni and in collaborations, that increasingly bioinformatics research is appreciated on its own merit. This realization will make it easier for bioinformatics PhD students to claim first authorship, even on papers with experimental work.
We share their view that computational biologists should develop a feel for the lab work, and, conversely, that biology curricula urgently need to incorporate computational approaches in their core curricula (Yanai and Chmielnicki, 2017). We find it difficult to see, however, that in the foreseeable future, this would lead to computational biology labs where each member both operates computational and experimental facilities. Though this view on the future of computational biology is quite appealing, taking translation to an extreme, this may nevertheless not be a feasible option in all situations. The heterogeneity of required skill sets and knowledge to span both computational and experimental biology may be too great, which would lead to a lack of balance as well as focus. We therefore expect that computational biologists, computer scientists and biologists—while learning and appreciating more about each other’s fields, and integrating more of each other’s approaches, methods and techniques into their own work—may likely continue their current course of intensive collaborations for some time to come.
3.2 Problem solving using the right tool set
BioSB are challenging research disciplines, which require practitioners to have a well-developed concept of the art of doing science, a high ability for mathematical and algorithmic abstraction, a broadly developed knowledge (balance), an ability to quickly absorb and integrate novel concepts (translate) and well-developed modelling, engineering and practical skills (focus). These aspects are all emphasized as critically important for the job market in the life sciences (Greene et al., 2016; Via et al., 2013; Welch et al., 2014), the data sciences (Dunn and Bourne, 2017; Lyon and Mattern, 2017; Pournaras, 2017; Seidl et al., 2018) and both (Brazas et al., 2017a; Greene et al., 2016).
To be successful as an educational programme in life and/or data sciences means that all three aspects—translate, balance and focus—need to be integrated in the programme from the outset. From our own experience, there are several issues that may arise if integration is not achieved optimally, which may serve to illustrate the importance of all three aspects.
3.3 Lessons learnt
3.3.1 When balance comes too late
When the two starting courses, FoB and ISB, were run for the first time, the ‘conversion classes’ on biology, programming and maths were not yet implemented. The idea was that students would experience their deficiencies during the work done in these first courses, which was supposed to instil into them a sense of importance of these topics for their studies. During subsequent courses, these skills would then gradually be built up. What actually happened was that students felt unjustly and repeatedly confronted with their inabilities, with no time reserved for them to repair their faults, as they saw it. As this was highly demotivating to the students, developing a sufficiently broad and balanced skill set should start straight away.
3.3.2 When translation comes too late
In the first few rounds of ISB, the focus of this course was on providing an overview of the field. Modelling exercises were reserved for the ensuing courses, Basic Models of Biological Systems, and further deepened in Advanced Modelling in Systems Biology. What happened was that students did not experience the need for developing their mathematical skills, even though the relevance of mathematics to systems biology was explained repeatedly in class. Interestingly, students also found that by the end of the course, they still were not sure what systems biology was about, the overview lectures had managed to transfer a feeling of the problems that needed solving, but not so much the role of systems biology in those solutions. So, introducing students to how research methodologies can be translated to a solution of an open problem, or, conversely, how to find the appropriate method to solve your current problem, should start straight away.
3.3.3 When focus comes too late
Before the current format of the two introductory courses was developed, the master programme started with one out of two pre-existing courses which were borrowed from other master programmes, one for students from a non-biology background, and one for those with a non-computer science background. While this setup addressed balance to some degree, it did postpone learning the main topic of the programme. Students expressed experiencing a lack of focus, and found the course materials fragmented without much cohesion. In an early accreditation round, the committee in their report judged our programme harshly on this point: ‘The learning objectives and the structure of the courses are not always clear to the students […] which is to the detriment of the effectiveness of their study.’ Moreover, without focused research projects, part of the students will be unmotivated for the hard work of balancing their skills, not having experienced why these skills are essential.
From previous experiences from working in industrial research, some of us noticed from the start of the programme that the practical ability to work with complex research data is a highly desired skill. It is too late when the research project (internship) is the first time students encounter such complexity in their data analysis. Thus, complex data is already introduced in the project work in the starting course FoB. The compulsory course Biosystems Data Analysis, and the optional courses Systems Biology in Practice, Statistics with R and Bioinformatics for Translational Medicine further prepare students for exactly this purpose. Supervisors of internships where data analysis is required, recognize the independence of our students in this area, and appreciate the quality of their work.
3.4 Practical implications
Having a highly diverse student population brings in cross-disciplinarity from the start. The diversity of student backgrounds ensures that students learn to collaborate and translate their research across disciplines. This leads to several practical requirements.
Most importantly, a focused, balanced and translational curriculum consisting of mainly custom-built and topic-specific courses taught by collaborating researchers will need to be developed. For example, our programme would benefit from a more advanced statistics course to cater for students who wish to specialize in this area, since available courses from the master statistics are currently ill fitted to the background of our students.
An additional requirement in a practical sense is to actively bring diverse students together, for example, in small diverse multi-background student groups to work on course group assignments (Abeln et al., 2013; Shapiro et al., 2013). This instils a sense of ‘respectful collaboration of different experts’.
Furthermore, a mechanism should be provided to make sure that the open questions with which students are challenged, remain relevant. The ever increasing pace of development of experimental techniques in the life sciences drives BioSB to be fast changing fields, which in turn accelerate innovation in the life sciences. In the research projects during the second year, relevance is therefore achieved in a natural way by ensuring placement of students in cutting-edge research environments. Additionally, during the academic part (course work) of the programme, students are already exposed to state-of-the-art research, taught by active researchers. In this way, we want to ensure that our students remain as up-to-date with the latest developments as possible.
3.5 Publishing on bioinformatics education
We note that in the developing community of computational biology education, there seems to be the need to foster a culture of publishing about our education programmes. There are 1970 master programmes in bioinformatics and/or systems biology worldwide, but despite our best efforts we have been able to locate only a relative handful of descriptions of educational programmes in peer-reviewed literature (Abeln et al., 2013; Alves et al., 2011; Cvijovic et al., 2016; Machanick and Tastan Bishop, 2015; Pattin et al., 2014; Sahinidis et al., 2005; Stefan et al., 2015; Via et al., 2013).
In addition to publishing about education programmes to support an exchange of best practices, we should also explicitly aim for citing each other comprehensively. For example, the papers in the 2013 Briefings in Bioinformatics (BiB) special issue on education, are read about three times per month, but the number of shares is only three per year and citations on average are only about once per year; note the large discrepancy with the BiB impact factor, which has been between five and nine in the same period.
A different issue is the publication of comprehensive textbooks. The current practice of contributing book chapters leaves something to be desired. As an alternative that may also increase overall quality (Bourne, 2013), PLoS Computational Biology launched a first open book, on Translational Bioinformatics with an introduction by Russ Altman (Altman, 2012). Chapters have been individually reviewed and are indexed and citable (e.g. Altman, 2012).
3.6 Comparison with other curricula
Many other programmes in bioinformatics and/or systems biology exist, and it will be instructive to see where similarities and differences arise. The papers on education programmes we have been able to trace (see above) all mention organizational difficulties typical to cross-disciplinary education, and the challenge of dealing with diverse student backgrounds. However, they do not allow detailed insight into organizational matters. Nonetheless, although in line with what we argue in this paper, a few further observations are noteworthy:
Sahinidis et al. (2005) (in Illinois, USA) describe their data-oriented programme, which is separated into a chemical biology and a computer science-oriented ‘concentration’. They emphasize the integration of teaching and research, and translation between the two, albeit they offer little concrete advice on how to achieve this.
Machanick and Tastan Bishop (2015) (South Africa) emphasize the importance of ‘social construction’ as the way in which knowledge is built, which may be especially important when crossing disciplinary boundaries as the social structures may differ. They hence focus explicitly on translation, also by fostering collaboration between students.
Cvijovic et al. (2016) give an overview of master’s programmes in systems biology across Europe. They highlight much the same issues, and conclude translation is essential, here also between theoretical/computational and experimental approaches. The authors’ focus is on skill requirements and curriculum components, rather than concrete practical implementations.
Several other papers describe the implementation of bioinformatics courses in life science programmes (Cummings and Temple, 2010; Stefan et al., 2015; Via et al., 2013), or in undergraduate programmes (Alves et al., 2011). Here, also keeping up with the fast-paced development of methods in the areas of BioSB is mentioned as a general challenge.
4 Conclusion
Teaching a dynamic, multidisciplinary topic like bioinformatics, systems biology or data science, requires continuous translation between methods and problems, between algorithms and application. Translation can only be achieved by providing sufficient depth and focus in key areas of the multidisciplinary field itself, coupled with a balanced foundation and contribution from the adjacent disciplines.
In summary, for a successful programme in bioinformatics, systems biology or data science, one needs:
Focus, balance and translation which are crucial and should each be in place from the start of the programme,
Balance in students background and skill sets,
Focus on key topics from the programme’s main area,
Translation to current open research problems and
To teach students to translate their knowledge and skills to other disciplines (and also to future developments within the disciplines).
From these essential ingredients, a nurturing and stimulating learning environment may develop, which will yield highly motivated graduates who are able to exploit their skills and knowledge in a demanding and dynamic working and research environment.
From the experiences in our broad BioSB programme, we hope that these recommendations will be instructive to organizers of computational biology programmes, including BioSB, and may also be useful in a wide range of life science and data sciences programmes.
Acknowledgements
We would like to thank all current and former staff in our research groups, Punto Bawono, Frank Bruggeman, Nicola Bonzanni, Mohammed El-Kebir, Bas Stringer, Chao Zhang, Maurits Dijkstra, Klaas Hellingwerf, Hannes Hettling, Ali May, Erik van Dijk, Bart van Houte, Jens Kleinjung, Remco Kort, Pinar Ozturk, Joost Teixeira De Mattos, Age Smilde, Dicle Hasdemir, Gunnar Klau, Bernd Brand, René Pool, Leendert Hamoen, Sandra Smit, Eunice Kleinjan, Meike Wortel, Sandra Monsonis Centelles, Annika Jacobsen, Walter Pirovano, Qingzhen Hou and Thomas Binsl, for their part in setting up and running the curriculum for the Bioinformatics and Systems Biology MSc in Amsterdam. We thank all the teaching assistants for offering their support in our busiest courses. We also would like to thank our education committee, in particular Rob van Spanning, and also the various student members who have been instrumental in shaping up the curriculum of our MSc programme throughout the years. Finally, we thank all our former and current students for their enthusiasm, dedication and trust.
Funding
This work is partly financed by the Netherlands Organization for Scientific Research (NWO).
Conflict of Interest: none declared.
References