-
PDF
- Split View
-
Views
-
Cite
Cite
Jianhao Peng, Olgica Milenkovic, Idoia Ochoa, METHCOMP: a special purpose compression platform for DNA methylation data, Bioinformatics, Volume 34, Issue 15, August 2018, Pages 2654–2656, https://doi.org/10.1093/bioinformatics/bty143
Close - Share Icon Share
Abstract
DNA methylation is one of the most important epigenetic mechanisms in cells that exhibits a significant role in controlling gene expressions. Abnormal methylation patterns have been associated with cancer, imprinting disorders and repeat-instability diseases. As inexpensive bisulfite sequencing approaches have led to significant efforts in acquiring methylation data, problems of data storage and management have become increasingly important. The de facto compression method for methylation data is gzip, which is a general purpose compression algorithm that does not cater to the special format of methylation files. We propose METHCOMP, a new compression scheme tailor-made for bedMethyl files, which supports random access.
We tested the METHCOMP algorithm on 24 bedMethyl files retrieved from four randomly selected ENCODE assays. Our findings reveal that METHCOMP offers an average compression ratio improvement over gzip of up to 7.5x. As an example, METHCOMP compresses a 48 GB file to only 0.9 GB, which corresponds to a 98% reduction in size.
METHCOMP is freely available at https://github.com/jianhao2016/METHCOMP.
Supplementary data are available at Bioinformatics online.
1 Introduction
DNA methylation is one of the most common mechanisms of epigenetic modification and a key element in controlling vertebrate gene function and cell differentiation (Razin and Riggs, 1980). Recent years have seen a surge in the number of projects focused on determining methylation abnormalities in carcinogenesis (Das and Singal, 2004). DNA methylation metrics were also found to be important in early detection of tumors and in determining the prognosis of the disease. In another direction, targeted DNA methylation has beed used to re-express erroneously silenced genes in cancer cells (Baylin, 2005). The recent survey (Robertson, 2005) lists a number of other diseases currently known to be caused by improperly regulated DNA methylation and details the underlying aberration mechanisms.
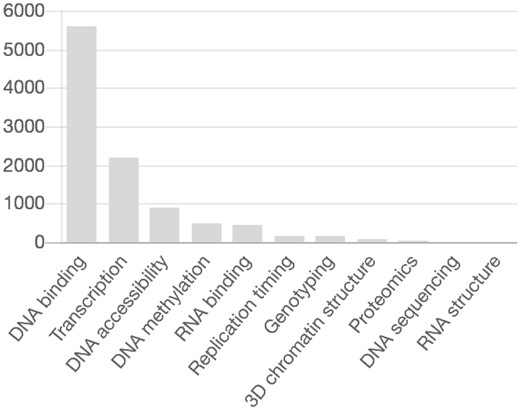
Given its importance in fundamental biological and medical research, DNA methylation has been the subject of many large-scale projects including MethylomeDB (Galperin and Cochrane, 2011), DiseaseMeth (Lv et al., 2011), NGSmethDB (Hackenberg et al., 2010) and MethBase (Song et al., 2013). In particular, over 800 ENCODE project assays are DNA methylation-related (https://www.encodeproject.org/matrix/? type=Experiment), amounting to roughly 10% of the total assays (Fig. 1, credit: ENCODE); 200 additional assays involve methylation state data. These files take around 78 TB of space and have to be stored for years. Hence, one needs to address this problem by designing efficient specialized compression algorithms for methylation data.
The number of assays in the ENCODE database (y-axis) for different experimental categories in Humans. Methylation category is ranked fourth
Methylation data from these projects is almost exclusively generated from whole-genome shotgun bisulfite sequencing (WGBS) (Yang et al., 2004) coupled with a reduced representation bisulfite sequencing (RRBS) pipeline. The raw data is converted into BED format (https://genome.ucsc.edu/FAQ/FAQformat.html#format1), which in the ENCODE database is referred to as a bedMethyl file (https://www.encodeproject.org/wgbs/#outputs). The bedMethyl files keep all methylation states such as CHG, CpG and CHH in order to enable maximal information content, and are readable as plain text files. Consequently, bedMethyl files have an average size of 15 GB and often exceed 40 GB. As a result, a large volume of DNA methylation data files has to be stored and transferred online for analysis, learning and data mining purposes. Until now, only traditional gzip software has been used to reduce the footprint of methylation data. Unfortunately, gzip compressors are universal methods designed to operate on diverse types of redundancy, and are hence not specialized for repetitive patterns encountered in bedMethyl files. This leads to significantly compromised compression performance. To address this problem, we developed a new, specialized compression method for bedMethyl files, termed METHCOMP. METHCOMP relies on a carefully integrated processing structure which encompasses de-interleaving different columns of bedMethyl files to optimize the corresponding differential and runlength coding schemes and it includes a highly efficient collection of arithmetic encoders in addition to random access support. As a result, METHCOMP offers almost an order of magnitude improvement in the compression ratio of bedMethyl files when compared to gzip, and roughly compacts the files to 2% of their original size.
2 Methods and experimental results
The METHCOMP encoder uses different compression strategies tailored to each data column in bedMethyl files, including adaptive arithmetic, runlength and differential encoding. In addition, columns that contain redundant information are discarded during compression. Compression and decompression may be executed in a single or multiblock setting. The multiblock setting enables parallel compression and random access, without a significant degradation in the compression ratio. Furthermore, parallel decompression is possible, hence greatly increasing the speed of data retrieval. See Supplementary Material for details.
To test the performance of our compression method, we ran the METHCOMP compression and decompression procedure on four randomly selected assays of the ENCODE project, and all the bedMethyl files within these assays. The codes of the corresponding ENCODE WGBS assays are ENCSR835OJU, ENCSR888JFA, ENCSR351IPU and ENCSR656TQD (Supplementary Material). The selected assays contain 6 bedMethyl files each. The bedMethyl files belonging to each assay may be retrieved from: http://www.encodeproject.org/experiments/ prepended to the project code (e.g. ENCSR835OJU). Due to space constraints, we provide individual results for bedMethyl files belonging to the first assay ENCSR835OJU only, and combined results for all files belonging to each of the remaining assays. The original sizes of the tested bedMethyl files are listed in Table 1: they vary in value from 2.6 GB to 48 GB. In all simulations, we used the multiblock setting with each block containing 5 million lines of the original file.
Comparison of compression performance of gzip and METHCOMP
| File name . | Original size (GB) . | Compressed size (GB) . | Compression ratio . | Improvement . | ||
|---|---|---|---|---|---|---|
| . | . | gzip . | METHCOMP . | gzip . | METHCOMP . | . |
| 167OJH | 13 | 2.30 | 0.315 | 5.65 | 41.21 | 7.29 |
| 327MVH | 48 | 7.30 | 0.969 | 6.58 | 49.55 | 7.53 |
| 428AXW | 2.6 | 0.47 | 0.086 | 5.50 | 30.25 | 5.50 |
| 677YTO | 13 | 2.30 | 0.317 | 5.65 | 40.96 | 7.25 |
| 751DLO | 2.6 | 0.47 | 0.086 | 5.50 | 30.25 | 5.50 |
| 945JPE | 48 | 7.3 | 0.970 | 6.58 | 49.50 | 7.52 |
| ENCSR1 | 128.2 | 20.14 | 2.745 | 6.37 | 46.70 | 7.33 |
| ENCSR2 | 139 | 21.82 | 3.124 | 6.37 | 44.50 | 6.99 |
| ENCSR3 | 138.9 | 21.93 | 3.115 | 6.33 | 44.59 | 7.04 |
| Average | 6.34 | 45.48 | 7.17 | |||
| File name . | Original size (GB) . | Compressed size (GB) . | Compression ratio . | Improvement . | ||
|---|---|---|---|---|---|---|
| . | . | gzip . | METHCOMP . | gzip . | METHCOMP . | . |
| 167OJH | 13 | 2.30 | 0.315 | 5.65 | 41.21 | 7.29 |
| 327MVH | 48 | 7.30 | 0.969 | 6.58 | 49.55 | 7.53 |
| 428AXW | 2.6 | 0.47 | 0.086 | 5.50 | 30.25 | 5.50 |
| 677YTO | 13 | 2.30 | 0.317 | 5.65 | 40.96 | 7.25 |
| 751DLO | 2.6 | 0.47 | 0.086 | 5.50 | 30.25 | 5.50 |
| 945JPE | 48 | 7.3 | 0.970 | 6.58 | 49.50 | 7.52 |
| ENCSR1 | 128.2 | 20.14 | 2.745 | 6.37 | 46.70 | 7.33 |
| ENCSR2 | 139 | 21.82 | 3.124 | 6.37 | 44.50 | 6.99 |
| ENCSR3 | 138.9 | 21.93 | 3.115 | 6.33 | 44.59 | 7.04 |
| Average | 6.34 | 45.48 | 7.17 | |||
Notes: The compression ratio is computed as (original size/compressed size). Individual file names correspond to assay ENCSR835OJU. ENCSR1, ENCSR2 and ENCSR3 stand for the combined results for assay ENCSR888JFA, ENCSR351IPU and ENCSR656TQD, respectively.
Comparison of compression performance of gzip and METHCOMP
| File name . | Original size (GB) . | Compressed size (GB) . | Compression ratio . | Improvement . | ||
|---|---|---|---|---|---|---|
| . | . | gzip . | METHCOMP . | gzip . | METHCOMP . | . |
| 167OJH | 13 | 2.30 | 0.315 | 5.65 | 41.21 | 7.29 |
| 327MVH | 48 | 7.30 | 0.969 | 6.58 | 49.55 | 7.53 |
| 428AXW | 2.6 | 0.47 | 0.086 | 5.50 | 30.25 | 5.50 |
| 677YTO | 13 | 2.30 | 0.317 | 5.65 | 40.96 | 7.25 |
| 751DLO | 2.6 | 0.47 | 0.086 | 5.50 | 30.25 | 5.50 |
| 945JPE | 48 | 7.3 | 0.970 | 6.58 | 49.50 | 7.52 |
| ENCSR1 | 128.2 | 20.14 | 2.745 | 6.37 | 46.70 | 7.33 |
| ENCSR2 | 139 | 21.82 | 3.124 | 6.37 | 44.50 | 6.99 |
| ENCSR3 | 138.9 | 21.93 | 3.115 | 6.33 | 44.59 | 7.04 |
| Average | 6.34 | 45.48 | 7.17 | |||
| File name . | Original size (GB) . | Compressed size (GB) . | Compression ratio . | Improvement . | ||
|---|---|---|---|---|---|---|
| . | . | gzip . | METHCOMP . | gzip . | METHCOMP . | . |
| 167OJH | 13 | 2.30 | 0.315 | 5.65 | 41.21 | 7.29 |
| 327MVH | 48 | 7.30 | 0.969 | 6.58 | 49.55 | 7.53 |
| 428AXW | 2.6 | 0.47 | 0.086 | 5.50 | 30.25 | 5.50 |
| 677YTO | 13 | 2.30 | 0.317 | 5.65 | 40.96 | 7.25 |
| 751DLO | 2.6 | 0.47 | 0.086 | 5.50 | 30.25 | 5.50 |
| 945JPE | 48 | 7.3 | 0.970 | 6.58 | 49.50 | 7.52 |
| ENCSR1 | 128.2 | 20.14 | 2.745 | 6.37 | 46.70 | 7.33 |
| ENCSR2 | 139 | 21.82 | 3.124 | 6.37 | 44.50 | 6.99 |
| ENCSR3 | 138.9 | 21.93 | 3.115 | 6.33 | 44.59 | 7.04 |
| Average | 6.34 | 45.48 | 7.17 | |||
Notes: The compression ratio is computed as (original size/compressed size). Individual file names correspond to assay ENCSR835OJU. ENCSR1, ENCSR2 and ENCSR3 stand for the combined results for assay ENCSR888JFA, ENCSR351IPU and ENCSR656TQD, respectively.
Table 1 lists the compression results obtained using METHCOMP, and shows that it offers, on average, a 7-fold improvement compared to gzip. Table 2 describes the compression and decompression speeds achieved by both algorithms. Although gzip has twice the compression and three times the decompression speed of METHCOMP, both complete a run within minutes. In addition, only METHCOMP supports efficient random access of specific data blocks. More results are provided in the Supplementary Material.
Comparison of compression speeds of gzip and METHCOMP
| File name . | Original size (GB) . | Compression speed (MB/s) . | Decompression speed (MB/s) . | ||
|---|---|---|---|---|---|
| . | . | gzip . | METHCOMP . | gzip . | METHCOMP . |
| 167OJH | 13 | 23.48 | 11.08 | 369.78 | 98.61 |
| 327MVH | 48 | 23.88 | 11.49 | 390.10 | 106.85 |
| 428AXW | 2.6 | 21.82 | 10.96 | 332.80 | 88.75 |
| 677YTO | 13 | 23.27 | 11.09 | 350.32 | 97.17 |
| 751DLO | 2.6 | 20.80 | 11.05 | 380.34 | 88.75 |
| 945JPE | 48 | 23.86 | 11.16 | 387.02 | 103.92 |
| ENCSR1 | 128.2 | 22.11 | 12.02 | 331.71 | 100.60 |
| ENCSR2 | 139 | 23.67 | 11.47 | 368.10 | 94.08 |
| ENCSR3 | 138.9 | 23.02 | 11.74 | 356.51 | 96.63 |
| Average | 22.91 | 11.61 | 356.18 | 98.31 | |
| File name . | Original size (GB) . | Compression speed (MB/s) . | Decompression speed (MB/s) . | ||
|---|---|---|---|---|---|
| . | . | gzip . | METHCOMP . | gzip . | METHCOMP . |
| 167OJH | 13 | 23.48 | 11.08 | 369.78 | 98.61 |
| 327MVH | 48 | 23.88 | 11.49 | 390.10 | 106.85 |
| 428AXW | 2.6 | 21.82 | 10.96 | 332.80 | 88.75 |
| 677YTO | 13 | 23.27 | 11.09 | 350.32 | 97.17 |
| 751DLO | 2.6 | 20.80 | 11.05 | 380.34 | 88.75 |
| 945JPE | 48 | 23.86 | 11.16 | 387.02 | 103.92 |
| ENCSR1 | 128.2 | 22.11 | 12.02 | 331.71 | 100.60 |
| ENCSR2 | 139 | 23.67 | 11.47 | 368.10 | 94.08 |
| ENCSR3 | 138.9 | 23.02 | 11.74 | 356.51 | 96.63 |
| Average | 22.91 | 11.61 | 356.18 | 98.31 | |
Note: The speed is computed according to (original size / time taken by the task).
Comparison of compression speeds of gzip and METHCOMP
| File name . | Original size (GB) . | Compression speed (MB/s) . | Decompression speed (MB/s) . | ||
|---|---|---|---|---|---|
| . | . | gzip . | METHCOMP . | gzip . | METHCOMP . |
| 167OJH | 13 | 23.48 | 11.08 | 369.78 | 98.61 |
| 327MVH | 48 | 23.88 | 11.49 | 390.10 | 106.85 |
| 428AXW | 2.6 | 21.82 | 10.96 | 332.80 | 88.75 |
| 677YTO | 13 | 23.27 | 11.09 | 350.32 | 97.17 |
| 751DLO | 2.6 | 20.80 | 11.05 | 380.34 | 88.75 |
| 945JPE | 48 | 23.86 | 11.16 | 387.02 | 103.92 |
| ENCSR1 | 128.2 | 22.11 | 12.02 | 331.71 | 100.60 |
| ENCSR2 | 139 | 23.67 | 11.47 | 368.10 | 94.08 |
| ENCSR3 | 138.9 | 23.02 | 11.74 | 356.51 | 96.63 |
| Average | 22.91 | 11.61 | 356.18 | 98.31 | |
| File name . | Original size (GB) . | Compression speed (MB/s) . | Decompression speed (MB/s) . | ||
|---|---|---|---|---|---|
| . | . | gzip . | METHCOMP . | gzip . | METHCOMP . |
| 167OJH | 13 | 23.48 | 11.08 | 369.78 | 98.61 |
| 327MVH | 48 | 23.88 | 11.49 | 390.10 | 106.85 |
| 428AXW | 2.6 | 21.82 | 10.96 | 332.80 | 88.75 |
| 677YTO | 13 | 23.27 | 11.09 | 350.32 | 97.17 |
| 751DLO | 2.6 | 20.80 | 11.05 | 380.34 | 88.75 |
| 945JPE | 48 | 23.86 | 11.16 | 387.02 | 103.92 |
| ENCSR1 | 128.2 | 22.11 | 12.02 | 331.71 | 100.60 |
| ENCSR2 | 139 | 23.67 | 11.47 | 368.10 | 94.08 |
| ENCSR3 | 138.9 | 23.02 | 11.74 | 356.51 | 96.63 |
| Average | 22.91 | 11.61 | 356.18 | 98.31 | |
Note: The speed is computed according to (original size / time taken by the task).
Acknowledgement
The authors are grateful to Minji Kim and Mikel Hernaez for many fruitful discussions during the software implementation stage.
Funding
This work was supported in part by an NIH BD2K grant for Targeted Software Development and the NSF IUCRC Center for Computational Biotechnology and Genomic Medicine.
Conflict of Interest: none declared.
References