-
PDF
- Split View
-
Views
-
Cite
Cite
Anliang Wang, Xiaolong Yan, Zhijun Wei, ImagePy: an open-source, Python-based and platform-independent software package for bioimage analysis, Bioinformatics, Volume 34, Issue 18, September 2018, Pages 3238–3240, https://doi.org/10.1093/bioinformatics/bty313
Close - Share Icon Share
Abstract
This note presents the design of a scalable software package named ImagePy for analysing biological images. Our contribution is concentrated on facilitating extensibility and interoperability of the software through decoupling the data model from the user interface. Especially with assistance from the Python ecosystem, this software framework makes modern computer algorithms easier to be applied in bioimage analysis.
ImagePy is free and open source software, with documentation and code available at https://github.com/Image-Py/imagepy under the BSD license. It has been tested on the Windows, Mac and Linux operating systems.
1 Introduction
Images with unprecedented resolution and complexity appear in modern biological research (Legland, 2016; Meijering, 2016). Analysis software for those images has thus received special attention. Basically, biologists can take advantage of such software to gain deeper insight into biological structures and processes (Murphy, 2014; Schneider, 2012). However, biologists normally lack programming skills, whereas computer experts usually lack background information about biology (Tomancak and Cardona, 2012). As a result, it is difficult to apply novel algorithms designed by computer scientists to biological research. Therefore, it is a challenge to develop user-friendly, interoperable and extensible software to bridge the gap between computer science and biological research.
Open-source software has achieved worldwide fame to overcome this challenge because of its transparency, extensibility and interoperability (Tomancak and Cardona, 2012). The advantages can help scientists easily understand and improve the algorithms to meet personalized needs. As the most popular image-analysis tool, ImageJ uses those advantages to not only provide robust software but also establish a valuable paradigm (Rueden, 2017; Schneider, 2012). Its plugin framework, macro language and community-driven development have helped both developers and end users. In particular, the new generation of ImageJ has recently advanced many features such as the decoupled data model and redesigned plugins mechanism (Rueden, 2017). However, partly due to the Java-based core, these improvements do not fully guarantee the bridge between biologists and computer-vision experts. There are still obstacles to incorporating some novel algorithms and excellent frameworks such as scikit-image and OpenCV into ImageJ (Rueden, 2017).
Here, we develop a bioimage analysis software package based on Python because Python, a very high-level language, represents a shallower learning curve for biologists and holds richer third-part extensions designed by computer scientists (Pérez, 2011). The software particularly references the architecture of ImageJ (Rueden, 2017; Schneider, 2012) and emphasizes high extensibility and interoperability.
2 Materials and methods
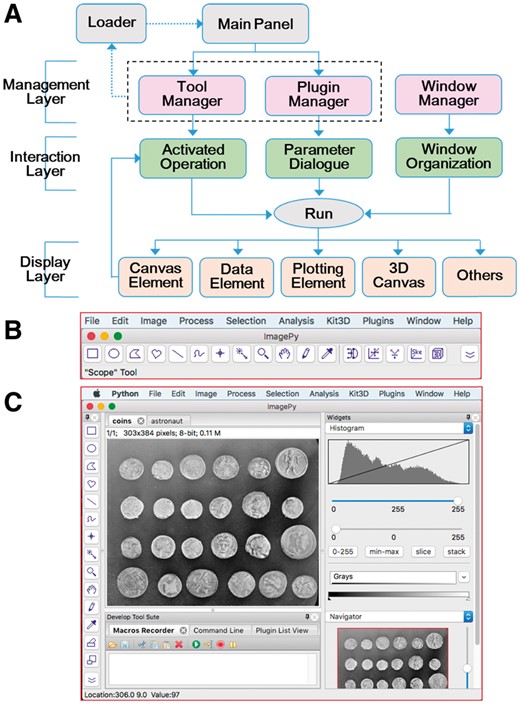
The plugin philosophy is one of the key reasons for the success of ImageJ (Rueden, 2017). ImagePy also adopts this merit in building its architecture, which mainly consists of the management, interaction and window layers (Fig. 1A). To ensure better interoperability, ImagePy basically manipulates the image data by using Numpy, which is the de facto standard for scientific computing in Python (Millman and Aivazis, 2011; Wart, 2011). The basic user interface (UI) is, to some degree, like that of ImageJ and implemented by using wxPython, which is a cross-platform UI toolkit for the Python language.
Software architecture and UI of ImagePy. (A) Three layers mainly comprise the architecture, and Run performs the main function. The dotted-line-arrow means that Loader is automatically implemented only one time after starting the software. The main panel has two modes: (B) the classical style, which imitates ImageJ and (C) the AUI, which serves individuals’ requirements
The management layer is the key and fundamental part in shaping the framework of ImagePy. All plugins, tools and windows are globally organized under this layer (Fig. 1A). The plugin manager takes charge of the basic engines and their sub-classes, such as Filter and Macro, the tool manager undertakes the arrangement of the tools and the window manager dispatches order and information to the active windows, such as closing and opening. The engine of the loader maps the components both from plugin manager and tool manager as the menubar and toolbar of the main panel (Fig. 1A and B), respectively. Following the manager layer, the interaction layer combines the commands of operation, dialogue and organization into the run function, which then generates the data for the display layer. This layer consists of the canvas element, data element, plotting element, etc. If necessary, the interaction layer can obtain feedback from the canvas element. This feedback actually helps perform the interaction between the mouse and the window layer. Overall, this framework separates the data model from the UI and thus improves the extensibility for different levels of ImagePy.
3 Results
Based on the framework, the major features of ImagePy are defined, including the basic UI, mathematical operations, filters, pixel statistics, characteristic extraction, 3D reconstruction and other functions. Actually, we offer two types of UIs for selection (Fig. 1B–C). The classical one is for users who are familiar with ImageJ. The other one is designed based on the advance user interface (AUI), which is customized via widgets plugin for users who prefer a more interactive display. The framework basically displays images with a maximum of four dimensions, but it can manipulate image data with more than four dimensions. ImagePy takes advantage of the Python dictionary to record the macro commands, which are just log messages that include a series of operations and parameters used by customer. The framework provides a user-friendly interface to accomplish the recording process (Fig. 1C). More importantly, the command codes are intrinsically decoupled from the UI and then conveniently developed as a plugin. This design benefits the extensibility of ImagePy.
4 Discussion
ImagePy is an open-source, Python-based, platform-independent software package for bioimage analysis. Inheriting from Python (Pérez, 2011), ImagePy is also particularly adept at interoperating with multiple languages and especially good for programming with a syntax that is approachable for biological scientists who are not professional programmers. Moreover, ImagePy decouples the data model from its UI and establishes a plugin framework together with easy-to-use macro commands. Intrinsically and conveniently, ImagePy can easily incorporate functions from existing libraries of the Python ecosystem, such as SciPy(Van Der Walt, 2011), scikit-image(Van Der Walt, 2014), OpenCV(Kaehler and Bradski, 2017) and Matplotlib (Hunter, 2007). Those achievements substantially promote the ability of extending and developing new plugins based on existing ones. Given the importance of data exploration and machine learning capabilities for biologists and data scientists (Dao, 2016), we will push forward with integrating those aspects into ImagePy. Therefore, ImagePy is not only an image-processing program but also a highly scalable framework. Our contribution can promise biologists and computer experts a seamless connection that makes modern computer algorithms available to analyse complex images both for biological and medical fields.
Acknowledgement
The authors thank two anonymous reviewers for useful advice.
Funding
This work was supported by the National Key Research and Development Program of China [2016YFC1401500], the National Natural Science Foundation of China [41506109, 41676189 and 11602051], the fundamental research funds for the central universities [DUT16LK27] and the China Postdoctoral Science and Foundation [2016M591433].
Conflict of Interest: none declared.
References