-
PDF
- Split View
-
Views
-
Cite
Cite
Leonardo Martins, Ramakanth Neeli-Venkata, Samuel M D Oliveira, Antti Häkkinen, Andre S Ribeiro, José M Fonseca, SCIP: a single-cell image processor toolbox, Bioinformatics, Volume 34, Issue 24, December 2018, Pages 4318–4320, https://doi.org/10.1093/bioinformatics/bty505
Close - Share Icon Share
Abstract
Each cell is a phenotypically unique individual that is influenced by internal and external processes, operating in parallel. To characterize the dynamics of cellular processes one needs to observe many individual cells from multiple points of view and over time, so as to identify commonalities and variability. With this aim, we engineered a software, ‘SCIP’, to analyze multi-modal, multi-process, time-lapse microscopy morphological and functional images. SCIP is capable of automatic and/or manually corrected segmentation of cells and lineages, automatic alignment of different microscopy channels, as well as detect, count and characterize fluorescent spots (such as RNA tagged by MS2-GFP), nucleoids, Z rings, Min system, inclusion bodies, undefined structures, etc. The results can be exported into *mat files and all results can be jointly analyzed, to allow studying not only each feature and process individually, but also find potential relationships. While we exemplify its use on Escherichia coli, many of its functionalities are expected to be of use in analyzing other prokaryotes and eukaryotic cells as well. We expect SCIP to facilitate the finding of relationships between cellular processes, from small-scale (e.g. gene expression) to large-scale (e.g. cell division), in single cells and cell lineages.
Supplementary data are available at Bioinformatics online.
1 Introduction
Cells, including bacteria, are efficient at parallel task processing. To understand the functioning of each process, aside from information about that process, one also needs information on related processes. E.g. knowledge of gene expression profiles is needed to understand the cell division process. Similarly, information on the mechanisms of detection of environment shifts is needed to explain the emergence of major changes in the gene expression profile.
Recent advances in fluorescence microscopy and synthetic fluorescent proteins engineering now allow observing in vivo many cellular processes. However, it remains mostly unknown how the different cellular processes relate and how information flows across scales. It is critical to overcome these knowledge gaps, since complex cellular processes rely on the timely execution of many simple tasks, such as time counting, signal activation, etc. These simpler processes occur in parallel, via semi-independent systems, such as signalling cascades and small gene circuits, operating at various scales.
To help addressing this, we propose a new toolbox, ‘SCIP’, which is able to extract information at the single cell, single molecule level, from time-lapse, multi-modal microscopy images. While most of its tools are automatic, we also implemented manual support systems to overcome, when needed, e.g. deficiencies in the segmentation and tracking algorithms. Further, while SCIP is designed for non-specialists in image analysis and programming, it is publicly available source code, to allow tuning and further development of methods not considered here.
We created a specialized analysis tool for the following biological processes: i) cell segmentation, cell tracking and lineage construction, ii) gene expression (including at the single-RNA level), iii) nucleoid morphology and nucleoid partitioning prior to cell division, iv) Z-ring assembly, v) Min system dynamics, vi) dynamics of protein aggregates and vii) detection of inclusion bodies. Finally, we added a tool to segment and track any undefined visible cellular structure (in some cases, this may require manual support). Importantly, all tools above can be used to analyze images from a single experiment (i.e. from the same cells), allowing a fast combination of data from multiple sources, not presently possible. Further, in all cases, the tools can be used in images of cell populations, or in time series of cells with single-cell tracking.
To achieve this, SCIP performs segmentation on images with Morphological information, such as Phase-Contrast, Bright-Field and Differential Interference Contrast images (segmenting not only cell borders but also inclusion bodies within). Then, it aligns the segmentation results with the information obtained from, e.g. confocal microscopy with functional information microscopy confocal images. From the latter ones, SCIP segments structures tagged with fluorescent proteins, such as the nucleoid, the Z-Ring, MS2-GFP-tagged RNA molecules and any other fluorescent proteins. Not only SCIP outputs spatial information, but it also combines the results from time-lapse images, to provide time-lapse based information on each feature observed. In addition, using all the information, it allows determining if and when there are correlations between the various morphological and functional features observed.
Finally, while there are tools and plugins (Danuser, 2011; Häkkinen et al., 2013) to perform several tasks performed by SCIP [e.g. cell segmentation (Lamprecht et al., 2007)], to the best of our knowledge, no other tool currently performs all SCIP’s tasks and integrates them to, among other, find correlations between the dynamics of parallel cellular processes (particularly using temporal information) from multi-mode sets of images. Further, no current tool can manually improve the results of all these tasks (as SCIP does), with the added value of being able to analyze any visible structure with no predefined shape or temporal behaviour.
2 Materials and methods
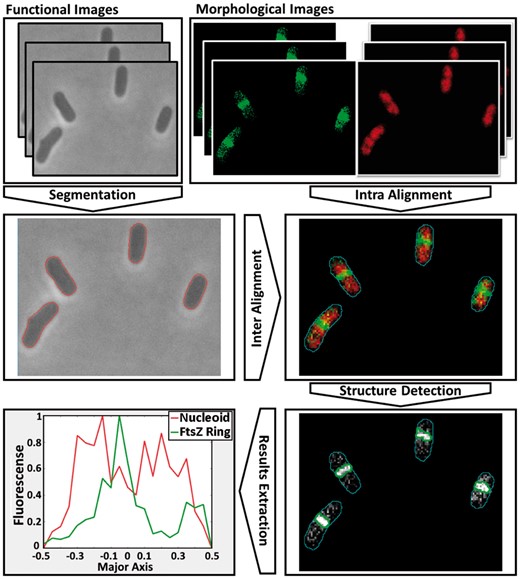
Figure 1 shows the workflow of SCIP. The steps, methods and Graphical User Interface (GUI) are described in Supplementary Material, along with examples of usage. Two cell segmentation methods are available in SCIP. One is based on a gradient segmentation combined with Machine Learning techniques (Häkkinen et al., 2013). The second uses median filters followed by a Multilevel Otsu threshold (Otsu, 1979). Two methods for cellular structures segmentation are also available. The first is based on two-dimensional Gaussian approximation while the second, ‘TreshMorph’, is a novel algorithm based on different threshold methods followed by Morphological Operations. Using the GUI, images are loaded, and then each of the tasks can be selected. In the end, it produces an output (images and tables in MATLAB format). Also in Supplementary Material is the evaluation of each tool’s efficiency and a description of the empirical methods and the microscopy techniques employed. Finally, a manual for each tool, and how to combine them, is provided, along with benchmark results of the tools.
SCIP Workflow. The GUI segments morphological images and inter-aligns them with functional images, from which information is obtained
3 Discussion
We expect several future endeavours in single-cell biology to focus on the internal spatio-temporal organisation of intracellular components and on better understanding the combined functioning of multiple cellular processes. SCIP was engineered for assisting such endeavours in a simple, automated fashion, by producing objective and reproducible analyses of multi-modal data. Previous successful examples of such approaches include recent findings on how immortal cell lineages dispose of unwanted protein aggregates (Oliveira et al., 2016), how serine chemoreceptors are placed at the cell poles (Neeli-Venkata et al., 2016), and on how Z-rings are formed at midcell (Tsukanov et al., 2011). These findings result from studying how the timing of ‘simple’ parallel cellular processes contributes to the success of complex, dynamic cellular processes. It is this multiple information that SCIP gathers and combines.
Overall, we expect SCIP to provide assistance to present and future single-cell biology studies, as these are based on microscopy and are becoming ever more multi-modal. Also, we expect it to contribute to an increased understanding of relationships between parallel dynamic cellular processes due to its modular-based multi-tasking abilities.
Funding
We thank Portuguese Foundation for Science and Technology FCT/MCTES - SFRH/BD/88987/2012 (LM), TUT Graduate Programme (RN-V), Vilho, Yrjo and Kalle Vaisala Foundation (SO), Academy of Finland - 295027 and 305342 (ASR), Jane & Aatos Erkko Foundation 610536 (ASR), FCT Strategic Program - UID/EEA/00066/203 (JMF). The funders had no role in the study design, data collection and analysis, decision to publish, or manuscript preparation.
Conflict of Interest: none declared.
References




Comments
https://www.nature.com/articles/s41598-018-24916-9
https://github.com/AngeloTorelli/AutoCellSeg