-
PDF
- Split View
-
Views
-
Cite
Cite
Pierre Salvy, Georgios Fengos, Meric Ataman, Thomas Pathier, Keng C Soh, Vassily Hatzimanikatis, pyTFA and matTFA: a Python package and a Matlab toolbox for Thermodynamics-based Flux Analysis, Bioinformatics, Volume 35, Issue 1, January 2019, Pages 167–169, https://doi.org/10.1093/bioinformatics/bty499
Close - Share Icon Share
Abstract
pyTFA and matTFA are the first published implementations of the original TFA paper. Specifically, they include explicit formulation of Gibbs energies and metabolite concentrations, which enables straightforward integration of metabolite concentration measurements.
High-throughput analytic technologies provide a wealth of omics data that can be used to perform thorough analyses for a multitude of studies in the areas of Systems Biology and Biotechnology. Nevertheless, most studies are still limited to constraint-based Flux Balance Analyses (FBA), neglecting an important physicochemical constraint: thermodynamics. Thermodynamics-based Flux Analysis (TFA) in metabolic models enables the integration of quantitative metabolomics data to study their effects on the net-flux directionality of reactions in the network. In addition, it allows us to estimate how far each reaction operates from thermodynamic equilibrium, which provides critical information for guiding metabolic engineering decisions.
We present a Python package (pyTFA) and a Matlab toolbox (matTFA) that implement TFA. We show an example of application on both a reduced and a genome-scale model of E. coli., and demonstrate TFA and data integration through TFA reduce the feasible flux space with respect to FBA.
Documented implementation of TFA framework both in Python (pyTFA) and Matlab (matTFA) are available on www.github.com/EPFL-LCSB/.
Supplementary data are available at Bioinformatics online.
1 Introduction
Constraint-based analysis on genome-scale metabolic models (GEMs) is a popular method to study metabolism and cellular physiology. Flux Balance Analysis (FBA), in particular, has been used to predict network-level behaviors, such as specific growth rate, gene essentiality, etc. The MATLAB-based COBRA toolbox (Schellenberger et al., 2011) and its Python counterpart COBRApy (Ebrahim et al., 2013) are today the most popular tools to perform such studies, and offer an intuitive interface to model GEMs using a linear programming formulation.
However, FBA-derived approaches often lead to flux distributions that are contradicting with physiology and bioenergetics due to the lack of thermodynamic constraints in their formulation (Ataman and Hatzimanikatis, 2015; Soh and Hatzimanikatis, 2010). We present here an implementation of Thermodynamics-based Flux Analysis (TFA; Henry et al., 2007; Soh and Hatzimanikatis, 2014), a framework to constrain GEMs or any metabolic network with thermodynamics. This framework allows to reduce the feasible flux solution space and eliminate thermodynamically-infeasible flux distributions, thus increasing the predictive accuracy of these models.
Previous works have been based on (Henry et al., 2007) to embed thermodynamic information in GEMs. However, they either require additional assumptions (Fleming and Thiele, 2011; Fleming et al., 2009), or calculate the thermodynamics feasibility decoupled from the FBA problem (Zamboni et al., 2008). TFA integrates the thermodynamics feasibility in the same MILP problem as FBA, and can unbiasedly account for all allowed thermodynamic profiles.
Our framework is provided under the form of a MATLAB toolbox as well as a Python package. It supported the publication of several studies integrating metabolomics in genome-scale models (Andreozzi et al., 2016; Chakrabarti et al., 2013; Chiappino-Pepe et al., 2017; Kiparissides and Hatzimanikatis, 2017; Miskovic et al., 2017; Savoglidis et al., 2016).
2 Materials and methods
2.1 Embedding thermodynamics constraints
The first step towards building constraint-based models utilizing thermodynamics with TFA is to ensure a proper thermodynamic curation of the model. In particular, TFA requires the information on (i) Compartment-specific pH, ionic strength and membrane potentials; (ii) Elemental and charge balance of every reaction; (iii) , the Gibbs free energy of formation of metabolic compounds in aqueous phase, pH 7 and 0 M ionic strength, all concentrations held at 1 M, at . (i) Is obtained from literature data. If this is missing, data on phylogenetically close species can be assumed, if available. (ii) Is dependent on the quality of the genome scale model used as an input. TFA will however take care of adjusting the dominant protonation state of metabolites depending on their pKa and the pH of their compartment. We then perform a correction according to the Debye–Huckel equation (Debye and Huckel, 1923) to adjust the energies to the relevant ionic strength in the compartment. For (3), can be obtained using literature data, or estimation methods like group contribution method (Jankowski et al., 2008). If a metabolite does not have , the reactions that include this metabolite in their stoichiometry will not be constrained with thermodynamics. It is not possible to solely add a reaction , as the Gibbs energy needs to be linked to metabolite concentrations in order to propagate the thermodynamic constraints throughout the network. The pKa of a compound can be calculated with ChemAxon (Szegezdi and Csizmadia, 2007).
The Gibbs free energy of reactions are then transformed with respect to cellular physiology by applying the transformation as proposed in (Alberty, 2005), as well as in this context by (Soh and Hatzimanikatis, 2014), using the given compartment-specific parameters: pH and ionic strength. Concentrations are used directly to integrate quantitative metabolomics data into the model. Upon thermodynamic curation of the model, we can formulate it as an MILP problem as explained in the Supplementary Material and (Soh and Hatzimanikatis, 2014).
The different types of analysis that can be performed are detailed in the Supplementary Material, and both packages include tutorials on how to perform them.
2.2 Implementation
The Python package pyTFA is built to integrate with COBRApy (Ebrahim et al., 2013), and takes advantage of Optlang (Jensen et al., 2017) for solver agnosticism and model operations.
The MATLAB implementation matTFA is built on top of The COBRA Toolbox (Schellenberger et al., 2011)
In the current implementation, the code uses SEED IDs (Overbeek et al., 2005) to match metabolites with a table of thermodynamic information taken from (Jankowski et al., 2008). It is also possible to input additional manually.
3 Usage
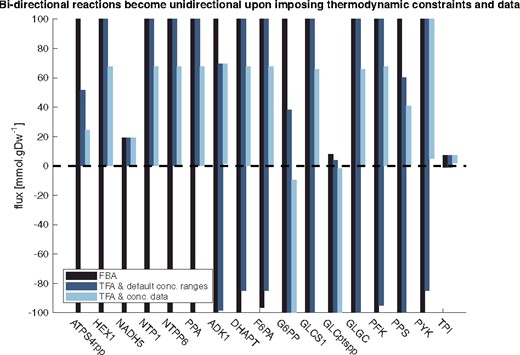
The software packages come with a tutorial that demonstrates the effects of integrating thermodynamic information as well as concentration data. A reduced Escherichia coli (Ataman et al., 2017), as well as the genome-scale model [iJO1366, (Orth et al., 2011)] used for its generation are provided. Figure 1 shows the output for a typical use case: A FBA model is constrained with thermodynamics, and then additional concentration data is added. Supplementary Figure S1 illustrates that the more constrained the model is, the more reduced the allowed ranges of fluxes are. Both packages detail how to reproduce this figure.
Variability analysis for reactions whose directions are not constrained by FBA. By subsequently adding thermodynamics constraints and concentration data, all the reaction directionalities are determined
4 Conclusion
We propose the software package to add thermodynamic information to constraint-based metabolic models. The resulting formulation is amenable to different types of analysis with high value for the Metabolic Engineering and Systems Biology communities. We demonstrated it with a case study of a reduced system for E. coli focusing on glycolysis, as well as the original GEM. Our package is available for MATLAB, and Python 3, on GitHub: respectively, https://github.com/EPFL-LCSB/matTFA and https://github.com/EPFL-LCSB/pytfa.
Funding
This work has received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No 722287, RobustYeast within ERA net project via SystemsX.ch and the European Union’s Horizon 2020 research and innovation programme under grant agreement No 686070.
Conflict of Interest: none declared.
References