-
PDF
- Split View
-
Views
-
Cite
Cite
Henry J Martell, Stuart G Masterson, Jake E McGreig, Martin Michaelis, Mark N Wass, Is the Bombali virus pathogenic in humans?, Bioinformatics, Volume 35, Issue 19, October 2019, Pages 3553–3558, https://doi.org/10.1093/bioinformatics/btz267
Close - Share Icon Share
Abstract
The potential of the Bombali virus, a novel Ebolavirus, to cause disease in humans remains unknown. We have previously identified potential determinants of Ebolavirus pathogenicity in humans by analysing the amino acid positions that are differentially conserved (specificity determining positions; SDPs) between human pathogenic Ebolaviruses and the non-pathogenic Reston virus. Here, we include the many Ebolavirus genome sequences that have since become available into our analysis and investigate the amino acid sequence of the Bombali virus proteins at the SDPs that discriminate between human pathogenic and non-human pathogenic Ebolaviruses.
The use of 1408 Ebolavirus genomes (196 in the original analysis) resulted in a set of 166 SDPs (reduced from 180), 146 (88%) of which were retained from the original analysis. This indicates the robustness of our approach and refines the set of SDPs that distinguish human pathogenic Ebolaviruses from Reston virus. At SDPs, Bombali virus shared the majority of amino acids with the human pathogenic Ebolaviruses (63.25%). However, for two SDPs in VP24 (M136L, R139S) that have been proposed to be critical for the lack of Reston virus human pathogenicity because they alter the VP24-karyopherin interaction, the Bombali virus amino acids match those of Reston virus. Thus, Bombali virus may not be pathogenic in humans. Supporting this, no Bombali virus-associated disease outbreaks have been reported, although Bombali virus was isolated from fruit bats cohabitating in close contact with humans, and anti-Ebolavirus antibodies that may indicate contact with Bombali virus have been detected in humans.
Data files are available from https://github.com/wasslab/EbolavirusSDPsBioinformatics2019.
Supplementary data are available at Bioinformatics online.
1 Introduction
Ebolaviruses represent a serious public health concern. The past few years have seen multiple outbreaks in Africa, including an epidemic between 2013 and 2016, which resulted in more than 28 000 cases and 11 000 deaths (Coltart et al., 2017; Lo et al., 2017; Michaelis et al., 2016). Until recently, only five species of Ebolavirus had been identified. Four of these Ebolavirus species, Ebola virus, Sudan virus, Bundibugyo virus and Taï forest virus are known to be pathogenic to humans, while the fifth, Reston virus, is not (Baseler et al., 2017; Cantoni et al., 2016; Michaelis et al., 2016; Miranda and Miranda, 2011). In August 2018, a new species of Ebolavirus, Bombali ebolavirus, was identified in the Bombali region of Sierra Leone (Goldstein et al., 2018). Currently, it is not known if Bombali virus causes disease in humans.
To investigate why Reston virus is not pathogenic in humans and the other four Ebolaviruses are, we have previously identified amino acid positions that are differentially conserved between these two groups (specificity determining positions; SDPs; Rausell et al., 2010) and analysed their effects on protein structure and function together with the changes associated with Ebola virus adaptation to new species (Pappalardo et al., 2016, 2017a). The results indicated that certain SDPs in the karyopherin-binding region of the Ebolavirus protein VP24 are critical determinants of species-specific Ebolavirus pathogenicity (Pappalardo et al., 2016, 2017b). Here, we first update our comparison of human pathogenic and non-human pathogenic Ebolaviruses by including the many Ebolavirus genome sequences that have become available in the last few years. Then we use this dataset to analyse the Bombali virus sequence at amino acid positions that are associated with human pathogenicity.
2 Results
2.1 Identifying determinants of Ebolavirus pathogenicity
Our original study was based on a set of 196 Ebolavirus genomes. We identified 180 SDPs that were differentially conserved between Reston virus and the human pathogenic Ebolaviruses, of which 47 mapped to protein structures and eight were proposed to have an effect on protein structure and function (Michaelis et al., 2016; Pappalardo et al., 2016). Here, we have expanded the dataset to 1408 Ebolavirus genomes (those retained after filtering an initial set of 2076 genomes for quality and completeness—see Supplementary Methods). This represents 7.5 times more sequences than used in the original study and also includes an increase in the number of Reston virus sequences from 17 to 27.
Phylogenetic analysis of the whole genome sequence and for each of the seven Ebolavirus proteins clearly separated each of the Ebolavirus species (Supplementary Fig. S1). However, the phylogenetic trees did not separate Reston virus from the human pathogenic Ebolavirus species (Supplementary Fig. S1).
High levels of conservation were observed within each species (Supplementary Fig. S2). Comparison of Reston virus proteins to the proteins of the other four human pathogenic species showed that there is greater divergence in GP, NP, VP30 and VP35, with conservation between 58 and 69%, whereas VP24, L and VP40 have a higher level of conservation (74–81%; Supplementary Fig. S2H).
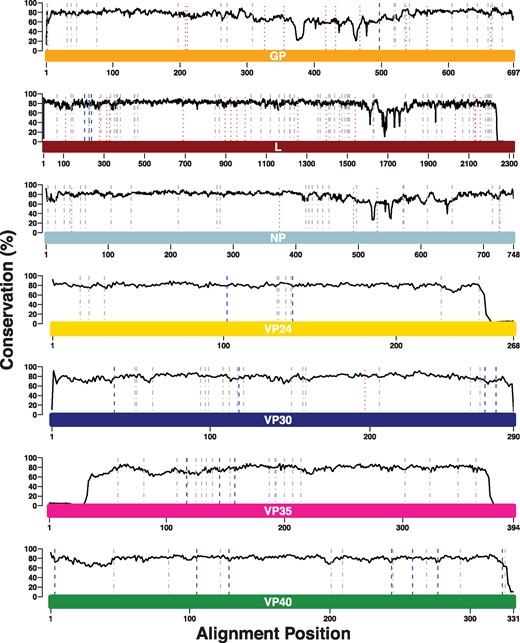
The increased number of Ebolavirus genomes resulted in a slight reduction of SDPs from 180 (originally reported as 189 but SDPs in sGP and GP were identical as they share a common N-terminus) to 166 in the seven Ebolavirus proteins (Fig. 1, Table 1 and Supplementary Tables S1–S7). Overall, 146 SDPs were retained, 34 were lost and 20 new SDPs were identified. No SDPs were lost in VP24 or VP35, and only a single SDP was lost in VP30. New SDPs were identified for each of these proteins ranging from two for VP24 to seven for VP40 (Fig. 1 and Table 1). More SDPs were lost in NP, GP and L, ranging from five for NP to 17 for L. At the same time, no SDPs were gained in NP, one was gained in GP and three in L (Fig. 1 and Table 1).
SDPs identified between human-pathogenic Ebolaviruses and Reston virus. The coloured bars represent the lengths of the protein sequence alignments, and each bar is labelled with the name of the protein that it represents. The solid black line represents the Jensen-Shannon conservation score. Dotted red lines represent SDPs. Previously identified SDPs that were lost in the updated analysis are shown by dotted lines (red), dashed-dot lines (grey) represent SDPs that were retained and dashed lines (blue) represent new SDPs that have been identified Note: x-axes differ in their scales between subplots.
Summary of the number of SDPs lost, retained and gained in the updated set of SDPs
| Protein . | Original SDPs . | SDPs lost . | SDPs retained . | SDPs gained . | Updated SDPs . |
|---|---|---|---|---|---|
| NP | 29 | 5 | 24 | 0 | 24 |
| VP35 | 19 | 0 | 19 | 3 | 22 |
| VP40 | 9 | 0 | 9 | 7 | 16 |
| GP | 30 | 11 | 19 | 1 | 20 |
| VP30 | 17 | 1 | 16 | 4 | 20 |
| VP24 | 9 | 0 | 9 | 2 | 11 |
| L | 67 | 17 | 50 | 3 | 53 |
| Protein . | Original SDPs . | SDPs lost . | SDPs retained . | SDPs gained . | Updated SDPs . |
|---|---|---|---|---|---|
| NP | 29 | 5 | 24 | 0 | 24 |
| VP35 | 19 | 0 | 19 | 3 | 22 |
| VP40 | 9 | 0 | 9 | 7 | 16 |
| GP | 30 | 11 | 19 | 1 | 20 |
| VP30 | 17 | 1 | 16 | 4 | 20 |
| VP24 | 9 | 0 | 9 | 2 | 11 |
| L | 67 | 17 | 50 | 3 | 53 |
Summary of the number of SDPs lost, retained and gained in the updated set of SDPs
| Protein . | Original SDPs . | SDPs lost . | SDPs retained . | SDPs gained . | Updated SDPs . |
|---|---|---|---|---|---|
| NP | 29 | 5 | 24 | 0 | 24 |
| VP35 | 19 | 0 | 19 | 3 | 22 |
| VP40 | 9 | 0 | 9 | 7 | 16 |
| GP | 30 | 11 | 19 | 1 | 20 |
| VP30 | 17 | 1 | 16 | 4 | 20 |
| VP24 | 9 | 0 | 9 | 2 | 11 |
| L | 67 | 17 | 50 | 3 | 53 |
| Protein . | Original SDPs . | SDPs lost . | SDPs retained . | SDPs gained . | Updated SDPs . |
|---|---|---|---|---|---|
| NP | 29 | 5 | 24 | 0 | 24 |
| VP35 | 19 | 0 | 19 | 3 | 22 |
| VP40 | 9 | 0 | 9 | 7 | 16 |
| GP | 30 | 11 | 19 | 1 | 20 |
| VP30 | 17 | 1 | 16 | 4 | 20 |
| VP24 | 9 | 0 | 9 | 2 | 11 |
| L | 67 | 17 | 50 | 3 | 53 |
Analysis of the SDPs at the codon level revealed that for the 27 Reston virus sequences, only ten SDPs showed any variation in codon usage, and for those ten positions there were always two codons present that represented synonymous changes. For five of these SDPs, only a single sequence contained a different codon and for the other five the codon usage was more closely balanced (Supplementary Table S8). For the pathogenic species, most amino acids at SDPs were encoded by multiple codons, with only 12 SDPs where a single codon was present (Supplementary Tables S9–S15). One Hundred and fifteen SDPs have only synonymous changes, while 39 SDPs also have non-synonymous changes (35 of these 39 also have synonymous changes; Supplementary Tables S9–S15). The synonymous changes largely (106 of 115) represent differences in the codon usage between the different pathogenic species (Supplementary Tables S9–S15). Twenty three of the non-synonymous changes are due to different codon usage between the species, while the remaining 16 non-synonymous changes occur in Ebola viruses. This shows that while variation occurs at the codon level, the amino acids encoded at SDPs are highly conserved.
2.2 Structural analysis of SDPs
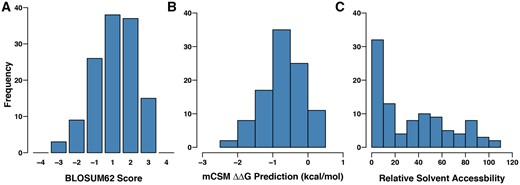
It was possible to map 92 of the 166 SDPs onto protein structures or models (Supplementary Methods; Table 2; Supplementary Tables S17 and S18), compared to 47 SDPs in the previous study (Pappalardo et al., 2016). This was partly due to greater structural coverage of the proteins, with a structure of the N terminal region of VP35 (Chanthamontri et al., 2018; Zinzula et al., 2019) now available and also a template to model the structure of L (Supplementary Fig. S3). Overall, the amino acid changes at SDPs represent conservative changes, with the majority of BLOSUM62 substitution score values being one or greater (Fig. 2A). Most are predicted to be slightly destabilizing to the protein structure (Fig. 2B), although this analysis only considered individual SDPs in isolation. One quarter of the SDPs (42) are located in the interior of the protein with the remaining three quarters having more than 20% relative solvent accessibility (Fig. 2C). These observations are consistent with the majority of SDPs having minor effects on protein structure and function.
Characteristics of the SDPs between human pathogenic Ebolaviruses and Reston virus. (A) BLOSUM62 scores for the whole set of SDPs. (B) mCSM predicted stability changes for the whole set of SDPs. (C) Relative solvent accessibility for the whole set of SDPs
Summary of SDPs per ebolavirus protein, and the predicted functional impacts
| Protein . | SDPs . | SDPs modelled . | Probable integrity . | Probable interface . | Possible integrity . | Possible interface . |
|---|---|---|---|---|---|---|
| NP | 24 | 8 | 0 | 0 | 1 | 0 |
| VP35 | 22 | 15 | 0 | 1 | 0 | 0 |
| VP40 | 16 | 13 | 1 | 1 | 0 | 0 |
| GP | 20 | 10 | 0 | 0 | 0 | 3 |
| VP30 | 20 | 5 | 0 | 1 | 0 | 0 |
| VP24 | 11 | 10 | 1 | 4 | 0 | 0 |
| L | 53 | 31 | 0 | 0 | 0 | 0 |
| Protein . | SDPs . | SDPs modelled . | Probable integrity . | Probable interface . | Possible integrity . | Possible interface . |
|---|---|---|---|---|---|---|
| NP | 24 | 8 | 0 | 0 | 1 | 0 |
| VP35 | 22 | 15 | 0 | 1 | 0 | 0 |
| VP40 | 16 | 13 | 1 | 1 | 0 | 0 |
| GP | 20 | 10 | 0 | 0 | 0 | 3 |
| VP30 | 20 | 5 | 0 | 1 | 0 | 0 |
| VP24 | 11 | 10 | 1 | 4 | 0 | 0 |
| L | 53 | 31 | 0 | 0 | 0 | 0 |
Note: SDPs were assessed to have an effect on the protein stability/integrity or protein-protein interactions. These were classed as ‘probable’ or ‘possible’ depending on the strength of evidence supporting the effect of the number of SDPs lost, retained and gained in the updated set of SDPs.
Summary of SDPs per ebolavirus protein, and the predicted functional impacts
| Protein . | SDPs . | SDPs modelled . | Probable integrity . | Probable interface . | Possible integrity . | Possible interface . |
|---|---|---|---|---|---|---|
| NP | 24 | 8 | 0 | 0 | 1 | 0 |
| VP35 | 22 | 15 | 0 | 1 | 0 | 0 |
| VP40 | 16 | 13 | 1 | 1 | 0 | 0 |
| GP | 20 | 10 | 0 | 0 | 0 | 3 |
| VP30 | 20 | 5 | 0 | 1 | 0 | 0 |
| VP24 | 11 | 10 | 1 | 4 | 0 | 0 |
| L | 53 | 31 | 0 | 0 | 0 | 0 |
| Protein . | SDPs . | SDPs modelled . | Probable integrity . | Probable interface . | Possible integrity . | Possible interface . |
|---|---|---|---|---|---|---|
| NP | 24 | 8 | 0 | 0 | 1 | 0 |
| VP35 | 22 | 15 | 0 | 1 | 0 | 0 |
| VP40 | 16 | 13 | 1 | 1 | 0 | 0 |
| GP | 20 | 10 | 0 | 0 | 0 | 3 |
| VP30 | 20 | 5 | 0 | 1 | 0 | 0 |
| VP24 | 11 | 10 | 1 | 4 | 0 | 0 |
| L | 53 | 31 | 0 | 0 | 0 | 0 |
Note: SDPs were assessed to have an effect on the protein stability/integrity or protein-protein interactions. These were classed as ‘probable’ or ‘possible’ depending on the strength of evidence supporting the effect of the number of SDPs lost, retained and gained in the updated set of SDPs.
Our previous structural analysis proposed a set of eight SDPs that were highly likely to alter protein structure and function, and a further five for which there was lower confidence (Pappalardo et al., 2016). Twelve of these 13 SDPs were retained in the current analysis, with only the lower confidence NP A705R no longer being sufficiently conserved to be identified as an SDP.
Of the 20 newly identified SDPs, ten were mapped onto protein structures (Table 2). Among these SDPs, we identified only one (VP24 R140S) that was likely to have an effect on protein structure and function. This results in nine SDPs overall with high confidence of having an effect on protein structure and function and four with lower confidence (Supplementary Table S19).
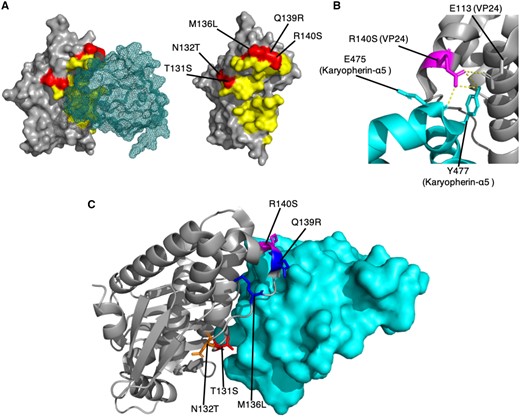
The VP24 SDP R140S is located in the VP24 interface site with human karyopherin α5 (KPNA5; Xu et al., 2014) where four other SDPs are located (T131S, N132T, M136L and Q139R). R140 can form hydrogen bonds with residues E476 (backbone) and Y477 (sidechain) in KPNA5, and also with the sidechain of E113 in VP24 (Fig. 3A and B). Reston virus S140 would still have the potential to form hydrogen bonds but not as extensively as R140. We have previously proposed that T131S, M136L and Q139R were likely to alter the binding of Reston virus VP24 to karyopherins, which may affect the ability of VP24 to inhibit the host interferon response (Pappalardo et al., 2016). The addition of R140S further supports this hypothesis, suggesting that this VP24 interface is vital to determining species-specific pathogenicity. Our hypothesis has recently been supported by experimental studies. Guito et al. (2017) showed that Reston virus VP24 is less effective at inhibiting the human interferon response. Further, histidine is present at residue 140 in Bundibugyo virus VP24 and has been implicated in reduced efficiency of downregulating interferon signalling (Schwarz et al., 2017).
SDPs in VP24 suggest that Bombali virus may not be pathogenic in humans. (A) SDPs in the VP24-Karyopherin-α5 interface. VP24 is shown in surface representation (grey) and karyopherin-α5 is shown as a mesh representation (teal). SDPs in VP24 are shown in red, and all residues within 5 Å of karyopherin-α5 are shown in yellow. (B) Hydrogen bonding of the SDP residue R140 in the Ebola virus VP24. VP24 (grey) and karyopherin-α5 (teal) are shown in cartoon format. Hydrogen bonds are represented by yellow dashed lines. (C) Agreement of Bombali virus sequences with the SDPs in the VP24-Karyopherin-α5 interface. VP24 (grey) is shown in cartoon representation, and Karyopherin-α5 (teal) is shown in surface representation. SDPs are shown in stick format, and coloured red where Bombali and Ebola virus agree, blue where Bombali virus agrees with Reston virus, orange where the Bombali virus amino acid is unique, and magenta where the amino acid present in the two Bombali virus sequences differ and one agrees with Ebola virus and the other with Reston virus
2.3 Comparison of Bombali virus with the other Ebolaviruses
Phylogenetic analysis of the genome sequences and of six of the seven Ebolavirus proteins grouped Bombali virus with Ebola, Sudan, Tai forest and Bundibugyo viruses, with Sudan and Reston viruses on a separate branch (Supplementary Fig. S1). For the seventh Ebolavirus protein, VP30, Bombali virus was grouped with Reston virus and the four known human-pathogenic species were on a separate branch (Supplementary Fig. S1k and l). While the phylogenetic analysis tends to group Bombali virus with human pathogenic Ebolavirus species, the pathogenic and non-pathogenic species are not clearly separated, making it difficult to infer from this phylogenetics analysis if Bombali virus is likely to be pathogenic in humans.
When considering the SDPs that differentiate human pathogenic and non-pathogenic Ebolaviruses, in Bombali virus the majority of amino acids at these positions (105; 63.25%) were identical to the human pathogenic Ebolaviruses, while 21 (12.65%) were shared between Bombali virus and Reston virus, and 40 (24.10%) were unique to Bombali virus (Supplementary Table S20). For the two available Bombali virus sequences, the amino acids present at SDPs agreed for all but one of the positions (VP24 R140S), where one of the sequences had the amino acid present in the human pathogenic viruses (R), while the other sequence contained the amino acid present in Reston virus (S).
With Bombali virus reported to have a 55–59% similarity to the other Ebolavirus species (Goldstein et al., 2018), the Ebola-Bombali SDP residue similarity is about 15% higher than the overall average, indicating high conservation amongst these positions, consistent with previous findings (Pappalardo et al., 2016). For all of the individual proteins, the Bombali sequences have greater agreement with the amino acids present in human pathogenic species at SDPs [30.0% (VP30) to 77.36% (L)], while the agreement with Reston virus is only 10–19% (Supplementary Table S20).
This suggests that Bombali virus is more closely aligned with the human pathogenic Ebolaviruses than with Reston virus. However, this may reflect closer relatedness among the African Ebolaviruses (Ebola virus, Sudan virus, Bundibugyo virus, Taï Forest virus, Bombali virus) compared to the Asian Reston virus, than similarities in human pathogenicity. In the phylogenetic analysis Bombali virus does group with most of the human pathogenic species (Supplementary Fig. S1).
Of the nine SDPs where we are confident that they are likely to alter protein structure and function (see above), Bombali virus has the same amino acid as the human pathogenic species at five positions and the same as Reston virus for three. The ninth position differs among the two available Bombali virus sequences (VP24 R140S; Table 3). While the majority of amino acids in Bombali virus at SDPs in VP24 agree with human pathogenic Ebolavirus amino acids (73%; Supplementary Table S20), two critical SDPs in the VP24-karyopherin binding region (M136L, Q139R) are identical to Reston virus (Fig. 3) (Pappalardo et al., 2016, 2017a). Additionally, at residue 132, an SDP which points away from the KPNA5 interface, there is a Bombali virus-specific amino acid (A132; N in EBOV, T in RESTV; Fig. 3). This may indicate that the Bombali virus is not be as pathogenic as pathogenic compared to the other Ebolaviruses that are known to cause disease.
Comparison of Bombali virus sequences with the nine SDPs identified as having a likely functional impact on human pathogenicity
| Protein . | SDP . | Bombali agreement . |
|---|---|---|
| VP24 | T131S | EBOV |
| VP24 | M136L | RESTV |
| VP24 | Q139R | RESTV |
| VP24 | R140S | EBOV/RESTV |
| VP24 | T226A | EBOV |
| VP30 | R262A | EBOV |
| VP35 | E269D | EBOV |
| VP40 | P85T | EBOV |
| VP40 | Q245P | RESTV |
| Protein . | SDP . | Bombali agreement . |
|---|---|---|
| VP24 | T131S | EBOV |
| VP24 | M136L | RESTV |
| VP24 | Q139R | RESTV |
| VP24 | R140S | EBOV/RESTV |
| VP24 | T226A | EBOV |
| VP30 | R262A | EBOV |
| VP35 | E269D | EBOV |
| VP40 | P85T | EBOV |
| VP40 | Q245P | RESTV |
Comparison of Bombali virus sequences with the nine SDPs identified as having a likely functional impact on human pathogenicity
| Protein . | SDP . | Bombali agreement . |
|---|---|---|
| VP24 | T131S | EBOV |
| VP24 | M136L | RESTV |
| VP24 | Q139R | RESTV |
| VP24 | R140S | EBOV/RESTV |
| VP24 | T226A | EBOV |
| VP30 | R262A | EBOV |
| VP35 | E269D | EBOV |
| VP40 | P85T | EBOV |
| VP40 | Q245P | RESTV |
| Protein . | SDP . | Bombali agreement . |
|---|---|---|
| VP24 | T131S | EBOV |
| VP24 | M136L | RESTV |
| VP24 | Q139R | RESTV |
| VP24 | R140S | EBOV/RESTV |
| VP24 | T226A | EBOV |
| VP30 | R262A | EBOV |
| VP35 | E269D | EBOV |
| VP40 | P85T | EBOV |
| VP40 | Q245P | RESTV |
3 Discussion
In this study, we have updated our previous analysis of amino acid positions that are differentially conserved (SDPs) between human pathogenic Ebolaviruses and the non-human pathogenic Reston virus by the inclusion of more than 1200 additional genome sequences. We have also analysed the amino acids present in Bombali virus at the SDPs to infer whether Bombali virus may cause disease in humans.
Our updated analysis of the SDPs that distinguish Reston virus from the four known human pathogenic Ebolavirus species reduced the number of SDPs from 180 to 166. The vast majority of SDPs were retained from the original analysis, including all the SDPs that we have proposed are likely to affect protein structure and function and may have a role in determining pathogenicity. This demonstrates that our initial study using only 196 genomes provided robust results. While we have identified a small subset of SDPs that we propose may be associated with pathogenicity, this reflects those SDPs that we have been able to map to protein structure and use analysis of structures to identify a likely functional effect. It is of course possible that some of the SDPs that we have not been able to propose a functional effect for may have a role in determining pathogenicity. However, our updated results also further strengthen our findings that VP24 is central to determining host-specific pathogenicity (Pappalardo et al., 2016, 2017a), a notion that is further supported by experimental evidence showing that Reston virus VP24 is less effective than the other Ebolavirus VP24 proteins at inhibiting the host immune response (Guito et al., 2017). Since the number of available Reston virus sequences remains small, particularly compared to the number of sequences across the four human pathogenic species, a larger number of Reston virus sequences would likely further refine the set of SDPs by capturing the variation within Reston viruses.
Our analysis of the Bombali virus sequence at the SDPs identified overall greater agreement with the human pathogenic Ebolaviruses. This could be the consequence of the common African origin of the human pathogenic Ebolaviruses and the Bombali virus, in contrast to the Asian Reston virus. The amino acids at SDPs in VP24 that we propose are most important in determining human pathogenicity are the same in Bombali virus and Reston virus. This suggests that Bombali virus may not be pathogenic, or have reduced pathogenicity, in humans. This is supported by the fact that Bombali virus was isolated from fruit bats, which were cohabitating in houses and other populated areas (Goldstein et al., 2018) and although this makes human contact highly likely, no disease outbreaks have been reported. Further, a study in the Bombali region detected anti-Ebola virus NP antibodies in humans without reports of disease (Mafopa et al., 2017). Although originally interpreted as evidence for asymptomatic Ebola virus infection, it is possible that this test actually detected antibodies against the then unknown Bombali virus that cross-reacted with Ebola virus antigen. Hence, antibodies directed against Ebolavirus proteins may indicate exposure of humans to low- or non-human pathogenic Bombali virus in the Bombali region.
In conclusion, based on our findings Bombali virus may be non-pathogenic or of low pathogenicity in humans. However, since few mutations seem to be sufficient for Ebolavirus adaptation to a new species (Pappalardo et al., 2017a), human pathogenic Bombali viruses may emerge, in particular as the Bombali virus shares many more conserved amino acid positions with human pathogenic Ebolaviruses than the non-human pathogenic Reston virus and further human contact with Bombali virus is likely to occur.
Conflict of Interest: none declared.
References
Author notes
The authors wish it to be known that, in their opinion, Henry J. Martell and Stuart G. Masterson authors should be regarded as Joint First Authors.