-
PDF
- Split View
-
Views
-
Cite
Cite
Wen Yao, Fangfang Huang, Xuehai Zhang, Jihua Tang, ECOGEMS: efficient compression and retrieve of SNP data of 2058 rice accessions with integer sparse matrices, Bioinformatics, Volume 35, Issue 20, October 2019, Pages 4181–4183, https://doi.org/10.1093/bioinformatics/btz186
Close - Share Icon Share
Abstract
We proposed to store large-scale genotype data as integer sparse matrices, which consumed much fewer computing resources for storage and analysis than traditional approaches. In addition, the raw genotype data could be readily recovered from integer sparse matrices. Utilizing this approach, we stored the genotype data of 1612 Asian cultivated rice accessions and 446 Asian wild rice accessions across 8 584 244 SNP sites in the ECOGEMS database with 310 MB of disk usage. Graphical interface for visualization, analysis and download of SNP data were implemented in ECOGEMS, which made it a valuable resource for rice functional genomic studies.
The code and data of ECOGEMS are freely available at https://github.com/venyao/ECOGEMS. ECOGEMS is deployed at http://ecogems.ncpgr.cn and http://150.109.59.144: 3838/ECOGEMS/ for online use.
Supplementary data are available at Bioinformatics online.
1 Introduction
Population level SNP (single nucleotide polymorphism) data are of great value to the identification of natural or artificial selection signatures, haplotype network analysis, determination of linkage disequilibrium blocks etc. (Huang et al., 2012; Yan et al., 2013). Currently, large scale genotype data are mostly stored as SQL tables or compressed text files in the server side, and are accessed by users at the client side through interactive web browsers (Zhao et al., 2015). The large size of SQL tables or text files and the remote connection between the server and the client side has restrained the distribution and utilization of the whole dataset. In addition, many URLs of servers and web links of datasets rapidly become broken, which further hindered the exploitation of genotype data.
In rice, next-generation sequencing data of several thousand accessions of Asian cultivated rice (Oryza sativa) and its progenitor (Oryza rufipogon, Asian wild rice), were reported (Huang et al., 2012; Xie et al., 2015). However, existing databases including RiceVarMap and SNP-Seek contain only SNP data among accessions of Asian cultivated rice (Mansueto et al., 2017; Zhao et al., 2015). Database for SNPs among Asian cultivated rice and Asian wild rice is not yet available.
Here, we propose to store genotype data using integer sparse matrix, in which most of the elements are zero. Utilizing this approach, we were able to store the genotypes of 2058 rice accessions at 8 584 244 SNP sites with only 310 MB of disk usage in the ECOGEMS database.
2 Implementation and functionalities
We developed a new approach to store large SNP dataset by converting character genotype matrices of ‘A’, ‘T’, ‘C’ and ‘G’ to integer sparse matrices of ‘0’ and ‘1’ (Supplementary Fig. S1) (Supplementary Material). The character genotype data could be efficiently recovered from the integer sparse matrices. We applied this approach to 8 584 244 SNPs among 446 O.rufipogon accessions and 1612 Asian cultivated rice accessions (Supplementary Table S1; Huang et al., 2012; Xie et al., 2015). Within the character genotype matrices, 70 226 533 entries were missing values (0.4%) while 1 222 358 529 were minor alleles (6.9%), indicating a sparsity of 92.7% (Supplementary Figs S2–S4). By converting the character matrices into sparse matrices, we stored this dataset as compressed R data files with only 310 MB of disk usage (Fig. 1A).
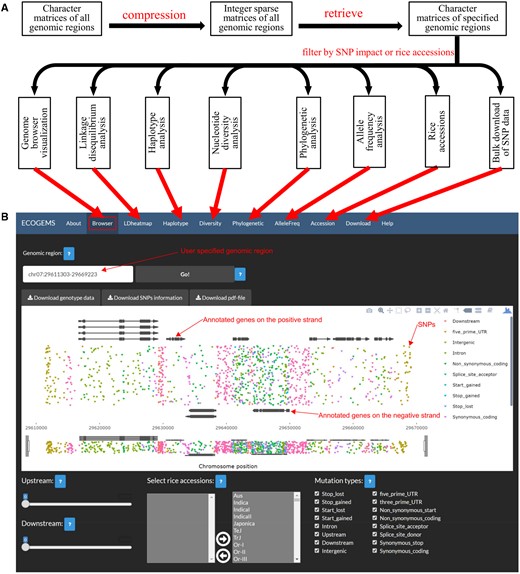
Construction and overview of the ECOGEMS database. (A) The process to construct the ECOGEMS database and the functionalities implemented in ECOGEMS. (B) SNPs in the genomic region chr07: 29611303-29669223 are shown as inverted triangles with different colors in the genome browser to demonstrate the interface of the ECOGEMS database. Structure of annotated genes is shown on top or bottom of the SNP region. The ‘Upstream’ and the ‘Downstream’ widgets allow extending the specified genomic region to its upstream and downstream respectively. The ‘Select rice accessions’ widget allows choosing rice accessions among which SNP sites are extracted. The ‘Mutation type’ widget allows filtering SNP sites by the impact of SNPs to gene structures. The three ‘Download’ widgets on the top allow downloading the genotype data and the visualization results of SNPs site
We further showed that the computing resources consumed by the new approach were much fewer than that of traditional approaches (Supplementary Figs S5–S9). With the help of the R Shiny package, we were able to build a genotype database named ECOGEMS using this dataset (Fig. 1). Several functionalities were implemented in the database with graphical user interface and were elaborated as follows. Moreover, all the functionalities were implemented as R functions in separated scripts, which can also be run in command-line mode of the R environment to generate associated R objects.
2.1 Genome browser visualization
For a specified genomic region or gene model, all the SNPs among user-selected rice accessions were extracted and subjected to genome browser visualization (Fig. 1B) (Supplementary Material). The detailed information of selected SNP sites and the genotypes of chosen rice accessions at the selected SNP sites could be readily extracted as text files for further analysis.
2.2 Linkage disequilibrium analysis
For a specified genomic region or gene model, a heat map could be created to display the pairwise linkage disequilibrium between different SNP sites (Supplementary Fig. S10). Gene models in the specified genomic region could also be displayed on top of the heat map.
2.3 Haplotype analysis
The 2058 rice accessions could be categorized into varying haplotype groups based on the genotype data in a specified genomic region. Haplotype network could then be built, which is helpful to demonstrate the relationship between individual genotypes at the population level (Supplementary Figs S11 and S12; Yan et al., 2013). Geographical distribution of rice accessions of different haplotype groups could be displayed on the world map (Supplementary Fig. S13).
2.4 Nucleotide diversity analysis
Functionalities were provided to calculate and demonstrate nucleotide diversities among subgroups of rice accessions in specified genomic regions. PROG1 is a key gene regulating the transition from prostrate to erect growth in rice domestication (Tan et al., 2008). Using the functionality, we showed that the nucleotide diversity in the genomic region harboring PROG1 was significantly reduced in cultivated rice compared with that of wild rice, in accordance with previous results (Supplementary Fig. S14; Huang et al., 2012).
2.5 Phylogenetic analysis
For a specified genomic region or gene model, a neighbor-joining (NJ) tree could be built based on a pairwise distance matrix derived from the simple matching distance for all SNP sites in this region (Supplementary Material). Using this functionality, we constructed a NJ tree based on SNPs within the 40-kb genomic region around PROG1 (Supplementary Fig. S15). The structure of this tree is in accordance with the results of previous study (Huang et al., 2012).
2.6 Rice accessions
The detailed information and the geographical distribution of user-chosen rice accessions were shown in tables and figures respectively, available for download (Supplementary Fig. S16).
2.7 Bulk download of SNP data
The ECOGEMS database also provide the functionality for bulk download of SNP data including the detailed information of all SNP sites and the genotypes of selected rice accessions at all SNP sites in large genomic regions.
For all the above analyses, SNP sites used in the analysis can be further screened by filtering rice accessions or by limiting the impact of SNPs to gene structures. In addition, diverse widgets were provided in ECOGEMS to tune the appearance of output figures and tables with simple mouse-click.
3 Discussion and conclusion
We proposed a new approach to store, retrieve and analyze population level SNP data with integer sparse matrices, which consumed much fewer computing resources than traditional approaches. We implemented this approach in the ECOGEMS database using pure R, a popular programing language used in biological studies. This approach is very useful to R users, as it requires no knowledge of SQL or other server-side programing languages. The code of ECOGEMS is publicly available, which could be readily applied to other organisms for the construction of SNP databases. We further expand the application of this approach to organisms with heterozygous SNPs, such as maize and human (Supplementary Fig. S17). For genotype data composed of triallelic SNP sites (or even more alleles) in addition to biallelic SNP sites, multiple sparse matrices needed to be created. The algorithm to compress and retrieve genotype data composed of triallelic SNP sites using sparse matrix was illustrated in Supplementary Figure S18. The sparse matrix approach and the code of ECOGEMS could also be integrated with other database systems to construct specific SNP databases.
ECOGEMS is the first database containing population level SNP data of Asian cultivated rice and its progenitor, which is of great value to future functional genomics studies of rice. ECOGEMS could be easily installed on personal computers for self-use or be installed on servers to provide online use of ECOGEMS to other users.
Funding
This research was supported by the Key Grant Science and Technique Foundation of Henan Province [161100110500-0102] and the Research Start-Up Fund to Topnotch Talents of Henan Agricultural University [30500581].
Conflict of Interest: none declared.
References