-
PDF
- Split View
-
Views
-
Cite
Cite
S Hollizeck, S Q Wong, B Solomon, D Chandrananda, S-J Dawson, Custom workflows to improve joint variant calling from multiple related tumour samples: FreeBayesSomatic and Strelka2Pass, Bioinformatics, Volume 37, Issue 21, November 2021, Pages 3916–3919, https://doi.org/10.1093/bioinformatics/btab606
Close - Share Icon Share
Abstract
This work describes two novel workflows for variant calling that extend the widely used algorithms of Strelka2 and FreeBayes to call somatic mutations from multiple related tumour samples and one matched normal sample. We show that these workflows offer higher precision and recall than their single tumour-normal pair equivalents in both simulated and clinical sequencing data.
Source code freely available at the following link: https://atlassian.petermac.org.au/bitbucket/projects/DAW/repos/multisamplevariantcalling and executable through Janis (https://github.com/PMCC-BioinformaticsCore/janis) under the GPLv3 licence.
Supplementary data are available at Bioinformatics online.
1 Introduction
Joint variant calling methods are routinely used to call germline variants by leveraging population-wide information across multiple related samples (DePristo et al., 2011; Toptas et al., 2018). This concept is also advantageous for somatic variant calling to potentially overcome the challenges of spatial heterogeneity and low tumour purity. However, there is a critical lack of robust algorithms that allow multi-sample somatic calling. Most studies still rely on variant calling of separate tumour-normal pairs, subsequently combining the results across a sample cohort (Hu et al., 2019; Leong et al., 2019; Wang et al., 2019).
There are two major pitfalls for combining variants called from individual tumour samples. First, it is very difficult to differentiate between a false negative result due to ‘missing data’ versus the true absence of a variant. Second, there is limited sensitivity for low allele frequency variants thus, decreasing the ability to detect minor clones, particularly in samples with low tumour purity.
Currently, only three algorithms claim to have the functionality to jointly analyze multiple samples: multiSNV (Josephidou et al., 2015), SuperFreq (Flensburg et al., 2020) and Mutect2 (Benjamin et al., 2019), each presenting different limitations. For instance, multiSNV cannot call indels and along with SuperFreq, is not optimized for analysis of deep coverage whole-genome sequencing (WGS) data. Mutect2 has previously been shown to be disadvantageously conservative as well as computationally inefficient (Chen et al., 2020).
To enable highly sensitive, fast and accurate variant detection from multiple related tumour samples, we have developed joint variant calling extensions to two widely used single-sample algorithms, FreeBayes (Garrison and Marth, 2012) and Strelka2 (Kim et al., 2018). Using both simulated and clinical sequencing data, we show that these workflows are highly accurate and can detect variants at much lower variant allele frequencies than commonly used methods.
2 Materials and methods
2.1 FreeBayesSomatic workflow
A variant is classified as somatic when both somaticLOD as well as somatic VAF pass the criteria somaticLOD (Equation 3) and somaticVAF (Equation 5).
The thresholds chosen for both LOD and VAF calculations were previously fitted by the blue-collar bioinformatics workflow for the DREAM synthetic 3 dataset using the SpeedSeq likelihood difference approach (Chapman et al., 2020) and were selected to identify high confidence variants.
2.2 Strelka2Pass workflow
In contrast to FreeBayes, whilst Strelka2 has a multiple-sample mode for germline analysis and tumour-normal pair somatic variant calling capabilities, it cannot jointly analyze multiple related tumour samples. We enable this feature by adapting a two-pass strategy previously used for RNA-seq data (Veeneman et al., 2016). First, somatic variants are called from each tumour-normal pair. All detected variants across the cohort are then used as input for the second pass of the analysis where we re-iterate through each tumour-normal pair but assess allelic information for all input genomic sites.
This allows the summation (Equation 8) of the SomEVS score across all supporting variants to assign a ‘PASS’ filter, if it reached a joint SomEVS score threshold. This threshold can be set by the user and is 20 by default, which corresponds to an estimated error rate of 1%. These ‘recovered’ variants need to pass a set of additional quality metrics related to depth of coverage, mapping quality and read position rank sum score.
As an additional improvement, we also built multiallelic support into Strelka2 which originally only reports the most prevalent variant at a specific site. Within the two-pass analysis, we reconstruct the available evidence for a multiallelic variant at a called site from the allele-specific read counts and report the minor allele at this site, if there is sufficient support from other samples. This method allows recovery of minor alleles only if another sample has this variant called by Strelka2, as SomEVS scores are not available for minor alleles.
3 Validation
3.1 Simulated data
We first simulated a phylogeny with somatic and germline variants from ten tumour samples and one normal (Fig. 1A, Supplementary Fig. S1A and B) (Supplementary Methods). Germline variants were simulated at a uniform allele frequency of 0.5. Somatic VAFs were sampled from a custom distribution, modelled to favour low allele frequency variants to closely represent real world data (min VAF: 0.001; max VAF: 1; Supplementary Fig. S1C and D). Paired-end sequencing reads with realistic error profiles were simulated for WGS data at 160× average coverage using the ART-MountRainier software (Huang et al., 2012). The simulated reads were aligned to GRCh38 and both germline and somatic variants from the phylogeny were spiked into the aligned reads using Bamsurgeon (Ewing et al., 2015). We compared the workflows for FreeBayes and Strelka2 with and without our extensions for joint variant calling on the simulated datasets. The performance of Mutect2 joint variant calling was also assessed using its proposed best practice workflow. As both Mutect2 and FreeBayes do not return a verdict for each individual sample, we needed to assign each sample in the multi-sample VCF its own FILTER value. We called a somatic variant as present in a sample, if there were at least two reads supporting it for this sample and the overall FILTER showed a ‘PASS‘, which was the same cut-off used in the refiltering step in the Strelka2-pass workflow.
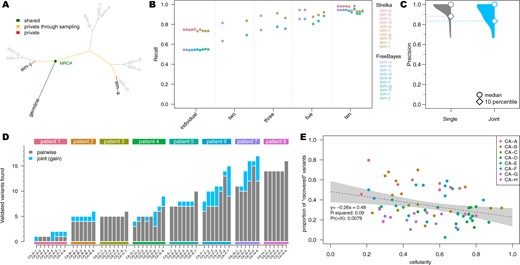
Comparison of joint multi-sample variant calling and single tumour-normal paired calling methods; (A) simulated phylogeny highlighting two samples with high evolutionary distance (sim-a and sim-j) where MRCA denotes the most recent common ancestor. (B) Recall estimates of FreeBayes and Strelka2, run in individual tumour-normal paired and joint calling configurations using two (sim-a and sim-j), three (sim-a, sim-g and sim-j), five (sim-a, sim-c, sim-f, sim-h and sim-j) and all ten tumour samples. (C) Precision of Strelka2 and (D) number of variants called by Strelka2 run in both tumour-normal paired (grey) and added with joint calling configurations (blue), which have been validated by targeted amplicon sequencing (TAS). (E) Correlation between cellularity and proportion of variants found only with joint calling using Strelka2Pass for clinical samples; grey area shows the ‘95%’ confidence interval for the linear model fit (dotted line)
While the precision of each method without our extensions was greater than 99.8%, they all missed at least 25% of all variants in the samples (i.e. recall ). In contrast, the recall of the modified workflows increased to with only a minute decrease in the precision for both FreeBayes and Strelka2 (Supplementary Fig. S2). Mutect2 however, had virtually no change in precision, but the recall actually decreased from to when analyzing the samples jointly (Supplementary Fig. S2B). Additionally, with our modified workflows, true positive variants were called with VAFs as low as 0.008 (median detected VAF for joint sample analysis and for single tumour-normal pair analysis), enabling improved distinction between true variants and technical errors (Supplementary Fig. S3). This improvement in performance for Strelka2 is only achieved after the refiltering step and not just a result of the second pass (Supplementary Fig. S4) (Supplementary Methods).
The performance of joint variant calling in Mutect2 was inferior compared to all other methods (Supplementary Fig. S2A and B). This was primarily due to the ‘clustered_events’ filter in Mutect2, which excluded the majority of false negative variants, with negligible contribution to the exclusion of true negative variants (Supplementary Fig. S5A and B). This result was unexpected as the simulated variants were evenly distributed along the genome and the corresponding allele frequencies were sampled randomly (Supplementary Fig. S1D).
Since the extent of the improvement in our joint calling workflows is bound by the number of shared variants between samples, we sub-sampled the simulated dataset, to show the effect of incomplete sampling on our methods, which is more likely in clinical settings. Furthermore, the evolutionary distance between the related samples in addition to the number of samples, has a major impact on the number of shared variants, as only variants acquired between the germline and the most recent common ancestor (MRCA), will benefit from the joint analysis. Therefore, we selected three sample subsets which included two, three and five samples with high evolutionary distance to show the minimum expected improvement (Fig. 1A and B). There was a clear linear improvement for both FreeBayesSomatic and Strelka2Pass when increasing the number of samples even if they had a distant evolutionary relationship. In contrast, when using only two samples with a small evolutionary distance, the increase in performance was almost as large as when jointly analyzing all ten available samples. This shows that samples with a high number of shared variants will perform better in joint calling workflows (Supplementary Fig. S6).
3.2 Clinical data
To validate the performance of our new workflows, we then analyzed WGS and whole-exome sequencing (WES) data of multi-region tumour samples from eight patients, with multiple tumour sites (average 7 samples per patient; total number of samples 55), enrolled in a rapid autopsy program conducted at the Peter MacCallum Cancer Centre (Supplementary Table S1 and Supplementary Methods) (Solomon et al., 2020; Vergara et al., 2021). The published studies had multiple somatic variants from the clinical samples orthogonally validated through targeted amplicon sequencing (TAS). We used these TAS-validated variants as the gold standard to evaluate the performance of different workflows, acknowledging that the technical biases inherent to TAS data are different to those present in WGS and WES (Supplementary Fig. S7) and that there would be sampling biases depending on different tumour cells analyzed in each data type.
In concordance with the results of the simulated data, our improved workflows found additional variants in all but one patient (Fig. 1D, Supplementary Fig. S8) (total additional variants Strelka2Pass: 64; FreeBayesSomatic: 85) with only a slight drop in precision for FreeBayesSomatic (mean: 0.94 versus 0.88) and Strelka2Pass (mean: 0.97 versus 0.92). Since the panel of variants validated by TAS was limited (7108 bp for patients CA-B through -H), this increase in detected variants suggests that a high number of shared variants in samples are missed with current approaches, which in turn leads to an overestimation of tumour heterogeneity between samples, as these variants are thought to not be present rather than undetected.
Even though the number of shared variants is a major influencing factor when jointly calling variants, low cellularity samples benefit more from the joint calling, as conventional methods cannot reliably distinguish low allele frequency variants from noise. Through a joint analysis approach, the number of recovered variants is higher in low cellularity samples, which indicates, that especially for clinical samples with variable tumour purity, joint analysis can have a major impact on improving performance (Fig. 1E, Supplementary Fig. S9).
Mutect2 in contrast, did not show significant improvement in any sample in its joint calling configuration, but showed inferior performance compared to the tumour-normal pairwise approach in two samples (Supplementary Fig. S8E), similar to its decreased performance in the simulated data (Supplementary Fig. S2). This was due to true variants being removed by the internal filters of the tool (Supplementary Fig. S5C and D). This is in stark contrast to our novel workflows, where the joint analysis preserves all called sites from the pairwise method and finds additional variants. Overall, Mutect2 found less validated variants in all patients than both Strelka2Pass (mean: 2.2) and FreeBayesSomatic (mean: 2.5) with comparable levels of precision (Supplementary Figs S8 and S10) but longer run times (Supplementary Table S2).
Our improved workflow also enabled the discovery of multiallelic variants with Strelka2, which led to the discovery of on average 42 additional variants (min: 1; max: 535) in the analyzed WES and 987 additional variants in the WGS (min: 81; max 2329). These variants are strong indicators of sub clonal structure and could be invaluable for the study of evolutionary trajectories in cancer.
4 Discussion
Here, we present an extension to two widely used variant callers, enabling them to analyze multiple related tumour samples and improve the sensitivity of detecting low allele frequency variants. This is highly relevant in clinical settings where low tumour purities in samples is a common occurrence. These workflows are an important step to satisfy the current unmet need for multi-sample tumour variant calling. While we have showcased their improvements in patient sequencing data, additional validation on larger clinical datasets is warranted to ensure the methodology performs robustly in real world settings. Importantly, these workflows are fully containerized and can be run through Janis (Lupat et al., 2021) on almost any high-performance computing environment, as well as cloud services. Each workflow is highly optimized and parallelized to facilitate the analysis of the large amount of data joint variant calling requires. The workflow specification also allows the easy adjustment of parameters to enable customization for the user’s needs and priorities, whereas building an ensemble workflow using multiple callers is up to the discretion of the user (Supplementary Fig. S11).
Acknowledgements
The authors thank all patients who provided tissue samples utilized in this study. The authors acknowledge Dr Lavinia Tan for assistance provided with the collection of patient clinical samples.
Funding
This work was supported by the National Health and Medical Research Council [1196755 to S.-J.D., 1158345 to S.-J.D. and B.J.S., 1194783 to S.Q.W., 1173450 to B.J.S.]; and CSL Centenary Fellowship to S.J.D; Victorian Cancer Agency [19008 to D.C., 19002 to S.Q.W.].
Conflict of Interest: S.-J.D. was a member of advisory boards for AstraZeneca and Inivata. The S.-J.D. lab received funding from Cancer Therapeutics CRC and Roche-Genentech. B.J.S. was a member of advisory boards for AstraZeneca, Roche-Genentech, Pfizer, Novartis, Amgen, Bristol Myers Squibb and Merk.
Data availability
The simulated data and the respective final variant calling files underlying this article are available from Figshare at https://melbourne.figshare.com, and can be accessed with https://doi.org/10.26188/13635186 for the dataset and https://doi.org/10.26188/13635187 for the called variants.
The biological data underlying this article are available at the European Genome-Phenome Archive (EGA) at https://ega-archive.org/, and can be accessed with study id EGAS00001004023 and EGAS00001004950.
References
Author notes
The authors wish it to be known that, in their opinion, the last two authors should be regarded as Joint Last Authors.