-
PDF
- Split View
-
Views
-
Cite
Cite
Sebastiaan Valkiers, Max Van Houcke, Kris Laukens, Pieter Meysman, ClusTCR: a python interface for rapid clustering of large sets of CDR3 sequences with unknown antigen specificity, Bioinformatics, Volume 37, Issue 24, December 2021, Pages 4865–4867, https://doi.org/10.1093/bioinformatics/btab446
Close - Share Icon Share
Abstract
The T-cell receptor (TCR) determines the specificity of a T-cell towards an epitope. As of yet, the rules for antigen recognition remain largely undetermined. Current methods for grouping TCRs according to their epitope specificity remain limited in performance and scalability. Multiple methodologies have been developed, but all of them fail to efficiently cluster large datasets exceeding 1 million sequences. To account for this limitation, we developed ClusTCR, a rapid TCR clustering alternative that efficiently scales up to millions of CDR3 amino acid sequences, without knowledge about their antigen specificity.
Benchmarking comparisons revealed similar accuracy of ClusTCR as compared to other TCR clustering methods, as measured by cluster retention, purity and consistency. ClusTCR offers a drastic improvement in clustering speed, which allows the clustering of millions of TCR sequences in just a few minutes through ultraefficient similarity searching and sequence hashing.
ClusTCR was written in Python 3. It is available as an anaconda package (https://anaconda.org/svalkiers/clustcr) and on github (https://github.com/svalkiers/clusTCR).
Supplementary data are available at Bioinformatics online.
1 Introduction
T-cells constitute a key component of the adaptive immune system and are one of the primary determinants of distinguishing self from nonself. The T-cell receptor (TCR) is responsible for the recognition of peptide antigens presented by the major histocompatibility complex (MHC). The TCR complex is a heterodimer expressed on the T-cell surface, consisting of two distinct chains (α and β) that both contribute to the recognition of the cognate antigen. High-throughput targeted sequencing technology enables sequencing of the unique and diverse TCR α and/or β nucleotide sequences in a sample, allowing quantitative mapping of the immune receptor repertoire. One of the major goals of quantitative immunology is the identification of groups of T-cells with common specificity towards an antigen. Exactly determining a TCR’s epitope specificity requires knowledge about the epitope and demands for time-consuming in vitro experiments such as MHC multimer assays (Davis et al., 2011). An alternative way of characterizing specificity groups is unsupervised clustering of TCR sequences (Meysman et al., 2019). This does not require prior knowledge of specific epitopes and allows interrogation of complete repertoire datasets by searching for sequentially similar TCRs. In spite of being a powerful approach to drastically reduce the repertoire complexity, sequence-based clustering has various complications. The most pronounced bottleneck of clustering sequence data is the scalability of pairwise distance calculations. Calculating pairwise distances scales quadratically with the number of input sequences (O(n2)). Current methods for TCR clustering such as TCRDist (Dash et al., 2017), iSMART (Zhang et al., 2020) and GLIPH2 (Huang et al., 2020), rely on demanding pairwise distance calculations, from which clusters can be determined. Other methods like TCRNET and ALICE can also be used to partition repertoires into epitopes-specific groups. These methods apply an enrichment strategy in which they identify clones that show stronger enrichment as compared to their theoretical generation probabilities. Although these algorithms have been successfully used to cluster individual repertoires, they are not designed to partition large groups of repertoires spanning multiple disease states. With ClusTCR, we created an unsupervised clustering procedure that can efficiently classify large sets of CDR3 sequences into specificity groups by drastically limiting the number of required pairwise comparisons.
2 Approach
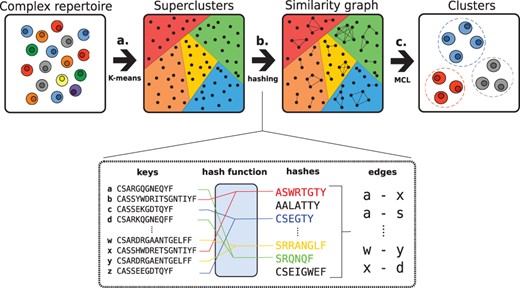
The complete workflow of ClusTCR is illustrated in Figure 1. In brief, the clustering approach works in two steps, one to allow fast and efficient clustering and a second to perform accurate clustering.
Workflow of ClusTCR. (a) Sequences are first roughly categorized into superclusters through efficient K-means clustering. (b) Within each supercluster, a hash function is applied to sort sequences. Sequence pairs with a maximum edit distance of 1 are selected from each hash. These sequence pairs are used to construct a graph. (c) MCL is used to find dense network substructures.
In the first step, ClusTCR queries the search space in order to roughly divide the dataset into what we defined as ‘superclusters’. To determine these superclusters, ClusTCR utilizes the Faiss Clustering Library (Johnson et al., 2019), which is specifically developed for rapid clustering of dense vectors through efficient indexing. For this, input sequences are mapped to an embedding that reflects their physicochemical properties. No specific combination of physicochemical features was found to significantly outperform others (Supplementary Fig. S1). Next, ClusTCR applies a very efficient K-means implementation to compute n centroids, generating an index. Next, a similarity search is performed on the index to assign each sequence to its nearest centroid. The default cluster size is 5000 (Supplementary Fig. S2A).
In the second step, ClusTCR reclusters each individual supercluster to accurately identify specificity groups within. Adaptive immune receptor repertoires can be represented as graphs in which nodes represent the sequences and the edges represented similarity between sequences (Madi et al., 2017). To create this graph, ClusTCR uses efficient sequence hashing to determine each pair of sequences with a maximum edit distance of 1. Here, the Hamming distance (HD) is used as the default metric to express similarity between TCRs, implying equal length of sequences within a single cluster. From our observations, this restriction does not impact the clustering quality (Supplementary Fig. S4A–D). Moreover, the use of HD drastically improves runtime over Levenshtein distance (Supplementary Fig. S4E). Next, ClusTCR uses the corresponding similarity-grouped graph to identify potential epitope-specific clusters. Evaluating the graph structure allows the interrogation of sequence-based relationship in the repertoire because similar sequences will share edges within the graph. To this end, ClusTCR applies the Markov clustering algorithm (MCL) for the identification of dense network substructures (Enright et al., 2002), representing dense groups of CDR3 sequences with similar sequential characteristics. MCL simulates stochastic flow inside the graph, thereby identifying dense network substructures where flow is high. These network substructures represent the clusters in ClusTCR’s output. This allows efficient and accurate clustering of relatively small sets of CDR3 sequences (up to 50 000). Combined with the first step, the clustering can be made efficient for any TCR dataset (Supplementary Fig. S3).
3 Performance
The efficient K-means implementation of the Faiss library allows for rapid subdivision of large repertoire files into rough clusters of CDR3 sequences with some degree of similarity. The second step involves the construction of a similarity graph and subsequent clustering with MCL. Combining both steps allows for substantial clustering flexibilty, while still being significantly faster than a simple greedy network clustering approach (Supplementary Fig. S5). To speed up the second part of the algorithm, ClusTCR applies multiprocessing, parallelizing subclustering with MCL. By balancing the MCL hyperparameters, additional improvements in speed could be realized without sacrificing clustering accuracy (Supplementary Fig. S2B). In conjunction, this results in a fast clustering methodology that outcompetes other TCR clustering approaches, while retaining comparable clustering accuracy (Supplementary Figs S6 and S7). To benchmark our algorithm, we compared it to three existing clustering methods for TCRs or CDR3 sequences: GLIPH2 (Huang et al., 2020), iSMART (Zhang et al., 2020) and TCRDist (Glanville et al., 2017; Mayer-Blackwell et al., 2020). Clustering accuracy was measured by means of cluster retention, purity and consistency (see Supplementary Materials). Benchmarking was performed on a 64-bit machine with 15.3 GB RAM, and an Intel®Core™i7-10875H CPU @ 2.30 GHz, running Ubuntu 20.04.2.LTS. We illustrate that ClusTCR offers a 50× performance increase (at 1 million sequences) compared to GLIPH2, which was found to be the second-fastest clustering algorithm (Supplementary Fig. S8). At the same time, ClusTCR shows comparable clustering accuracy compared to other TCR clustering methods as determined by the percentage of clusters with purity > 90%, both for α and β chain clustering (Supplementary Figs S6 and S7, respectively).
There does exist a trade-off between all TCR clustering methods between the number of clusters and the size or quality of the clusters. As illustrated, ClusTCR achieves 20–25% retention. This is lower compared to GLIPH2, but equal to iSMART (Supplementary Fig. S5C). However, GLIPH2’s increased cluster retention comes at the cost of clustering consistency (Supplementary Fig. S5D). ClusTCR’s consistency is higher than GLIPH2, but lower than iSMART. Collectively, these results indicate that there is no single approach that clearly outperforms all others with regards to the final cluster quality.
4 Downstream clustering analysis
Along with clustering functionality, ClusTCR provides tools for downstream analysis of the clustering results. These include calculation of cluster features such as cluster entropy, physicochemical properties and generation probability. Cluster features are particularly useful for downstream machine learning applications for TCR repertoire data. To this end, ClusTCR may serve as an efficient tool for generating lower-dimensional representations of highly complex immunosequencing data, while retaining the bulk of the information contained in the original dataset.
Funding
This work was supported by the Research Foundation Flanders (FWO) [Project No. 1S40321N] and the Flemish Government under the ‘Onderzoeksprogramma Artificiële Intelligentie (AI) Vlaanderen’ programme.
Conflict of Interest: none declared.