-
PDF
- Split View
-
Views
-
Cite
Cite
Nicolas Theodorakopoulos, Laureline Février, Mohamed Barakat, Philippe Ortet, Richard Christen, Laurie Piette, Sviatoslav Levchuk, Karine Beaugelin-Seiller, Claire Sergeant, Catherine Berthomieu, Virginie Chapon, Soil prokaryotic communities in Chernobyl waste disposal trench T22 are modulated by organic matter and radionuclide contamination, FEMS Microbiology Ecology, Volume 93, Issue 8, August 2017, No Pagination Specified, https://doi.org/10.1093/femsec/fix079
Close - Share Icon Share
Abstract
After the Chernobyl nuclear power plant accident in 1986, contaminated soils, vegetation from the Red Forest and other radioactive debris were buried within trenches. In this area, trench T22 has long been a pilot site for the study of radionuclide migration in soil. Here, we used 454 pyrosequencing of 16S rRNA genes to obtain a comprehensive view of the bacterial and archaeal diversity in soils collected inside and in the vicinity of the trench T22 and to investigate the impact of radioactive waste disposal on prokaryotic communities. A remarkably high abundance of Chloroflexi and AD3 was detected in all soil samples from this area. Our statistical analysis revealed profound changes in community composition at the phylum and OTUs levels and higher diversity in the trench soils as compared to the outside. Our results demonstrate that the total absorbed dose rate by cell and, to a lesser extent the organic matter content of the trench, are the principal variables influencing prokaryotic assemblages. We identified specific phylotypes affiliated to the phyla Crenarchaeota, Acidobacteria, AD3, Chloroflexi, Proteobacteria, Verrucomicrobia and WPS-2, which were unique for the trench soils.
INTRODUCTION
In the past decade, uranium mining, military activities and nuclear power plant accidents have introduced anthropogenic radioactive contaminants into the environment (Hu et al.2008). The Chernobyl nuclear power plant disaster led to the release of 13 650 × 1018 Becquerel (Bq) (Martin-Garin et al2012) and contributed significantly to the dispersal of many radioactive isotopes in the environment such as 137Cs, 90Sr, 60Co, 154Eu, 241Am, 234,235,236U and 239,240,241Pu, exhibiting different kinds of radiation (some are pure α emitters whereas others emit β− and/or γ−radiation). Due to their intrinsic properties (i.e. radioactive decay half-life, volatility and mobility), most of these radionuclides (RNs) are highly persistent in the terrestrial environment. Radiations emitted by RNs present either in the environment or accumulated inside organisms are able to ionize atoms or molecules, with potential consequent damage to living cells. Level of such biological detriment is linked to the energy deposited into organisms exposed to ionizing radiation, under the consensual assumption of additive effects. Thirty years after the accident, 137Cs is the main contributor to the current ambient external gamma dose rate in the Chernobyl exclusion zone. However, the proper assessment of the intensity of radiological exposure leading to observed effects requires to consider not only the ambient external dose rates (as often reported in most studies), but the total dose rates to which living organisms are exposed by including all exposure pathways and all RNs whatever their emission types (Garnier-Laplace et al.2015).
Soil microorganisms and RNs display complex relationships with each other. On the one hand, bacteria can interact with RNs via multiple mechanisms including bioreduction, biomineralization, bioaccumulation or biosorption (Lloyd and Macaskie 2002; Merroun and Selenska-Pobell 2008; Newsome, Morris and Lloyd 2014; Theodorakopoulos et al2015). On the other hand, RNs may exert radio- and chemotoxic effects on bacteria, thus influencing the structure and activity of microbial communities. Many previous studies have explored the molecular diversity of bacterial communities in various natural or contaminated sites containing RNs such as uranium (Selenska-Pobell et al.2001; Rastogi et al.2010; Islam and Sar 2011; Kumar et al.2013; Radeva et al.2013; Yan, Luo and Zhao 2016), uranium and 99Tc (Fields et al.2005; Akob, Mills and Kostka 2007; Barns et al.2007; Waldron et al.2009) and 137Cs and 99Tc (Fredrickson et al.2004). Diverse and complex bacterial assemblages have been evidenced in these habitats and although site-specific composition of bacterial communities is observed, Proteobacteria and Acidobacteria are frequently well represented. Several studies highlighted significant changes of bacterial communities upon RNs exposure (Geissler and Selenska-Pobell 2005; Akob, Mills and Kostka 2007; Rastogi et al.2010; Mondani et al.2011; Islam, Paul and Sar 2014; Yan, Luo and Zhao 2016).
In the case of Chernobyl, very few studies have been performed, most of which were based on cultivation approaches. These studies have shown a decrease in bacterial diversity under conditions of radioactive contamination/irradiation (Romanovskaya et al. 1998, 1999; Zavilgelsky et al.1998; Romanovskaya, Rokitko and Malashenko 2000; Czirják et al.2010). In 2011, the first study based on molecular approaches of sunlight-adapted biofilm microbial communities exposed to different ambient radiation levels (0.35–25 μSv/h) was reported from the Chernobyl area (Ragon et al.2011). Although the diversity was similar in irradiated and non-irradiated UV-adapted communities, an increase in the mutation level within some operational taxonomic units (OTUs) was correlated with ambient radiation. Niedrée et al. (2013) revealed a shift in bacterial population in soil microcosms upon 3-month exposure to Chernobyl-like contamination with 90Sr and 137Cs.
Atmospheric fallout from the CNPP accident resulted in radioactive contamination of large regions. One of the most impacted areas, located in the immediate vicinity of the nuclear power plant, is the so-called Red Forest where the pine trees died upon exposure to extreme radiation doses. Shortly after the CNPP accident, clean-up wastes from the Red Forest and other radioactively contaminated debris were buried in a large network of trenches near the CNPP, leading to highly contaminated environmental locations. After the clean-up operations, the trenches were covered with a thick layer of sand and planted with pine saplings, which are now reclaiming the area. One of these trenches (trench T22) has been studied since 1999, as a model of RN migration in the environment (Martin-Garin et al.2012). In our previous study (Chapon et al.2012), we identified the presence of complex bacterial communities in contaminated soil and control samples from this site using a community fingerprint method (DGGE). However, this method did not reveal any dominant community fingerprint related to RN content.
The goals of this study were to explore composition and diversity of soil prokaryotic communities in this understudied environment and to characterize the impact of the trench conditions (RN contamination and high organic matter content) on these communities. We hypothesized that soil prokaryotic communities inside and outside of trench T22 would differ, in particular upon radiation exposure. Therefore, we used high-throughput pyrosequencing of 16S rRNA genes to compare the prokaryotic communities in soil samples exhibiting contrasted RN contamination levels, collected inside and in the vicinity of the trench T22.
MATERIALS AND METHODS
Sample collection, isolation of culturable bacteria and DNA extraction
Sampling, soil analyses, isolation of culturable bacteria and DNA extraction procedures were previously described (Chapon et al.2012). Briefly, sandy soil samples containing various RN contents were collected between 50 and 60 cm depth from nine different positions in the area of the RN-contaminated trench T22, located in the Chernobyl exclusion zone (51°23΄N,30°04΄E; Bugai et al.2005). In April and October 2009, RN-contaminated soil samples (numbers 1, 3, 4, 8, 10 and 12) were collected inside the trench, along with low-contamination controls (numbers 13, 14 and 20) collected in the vicinity (Fig. 1). The soil samples were processed at Chernobyl within 18 h after sampling. In April, environmental DNA was extracted from a single subsample of each soil sample, whereas in October environmental DNA was extracted from triplicate subsamples (designated as a, b and c), after homogenization of the soil. DNA was typically extracted from 1g of soil using the PowerSoil DNA isolation kit (MO Bio, USA) and stored at –20°C. Culturable bacteria were recovered from soil samples in aerobic conditions on non-selective media agar plates (tryptic soy broth and AEM1 medium, see Chapon et al2012).
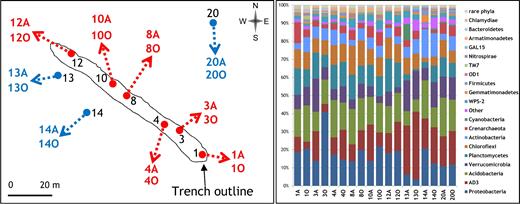
Location of the sampling points (left) and taxonomic profiles of the microbial communities (right), for soil samples collected in April (-A) and October (-O) 2009. The triplicate samples performed in October were combined.
PCR amplification and pyrosequencing
For the pyrosequencing analyses, amplicons from the V4 region of the 16S rRNA genes were generated by PCR using the universal primer set 530f (5΄-GTGCCAGCMGCNGCGG-3΄; Dowd et al.2008) and 802r (5΄-TACNVGGGTATCTAATCC-3΄; Claesson et al.2009). The coverage of this primer set was 89.1% for Bacteria and 54.1% for Archaea with no mismatch and reached 96.8% for Bacteria and 96.5% for Archaea with two mismatches as predicted with the Testprime tool of SILVA (Klindworth et al.2013). PCR amplifications were performed in triplicate in 50-μl reactions consisting of 30–50 ng of template DNA, 0.6 μM of forward and reverse primers, 0.2 mM dNTPs, 2 mM MgCl2, 1.25 U of Go Taq® Hot start polymerase (Promega, USA), and 1 X reaction buffer. For PCR, the thermocycler was programmed for an initial denaturation at 94°C for 5 min followed by 30 cycles of amplification (94°C for 30 s, 55°C for 30 s and 72°C for 45 s) and a final extension at 72°C for 5 min. Triplicate samples were pooled before purification of the amplicons. DNA from each amplicon (500 ng) was sent separately for sequencing by GATC (Konstanz, Germany) using a Roche 454 GS-FLX system (Titanium Chemistry). An adaptor and sample-specific 4-bp molecular identifier tag were added to each amplicon by an additional PCR step before mixing and pyrosequencing. Raw reads are available at the NCBI Sequence Read Archive (accession number PRJNA241298).
Processing of pyrosequencing data
The raw dataset (.sff files from two half-plates) contained 825 622 reads and was analyzed using QIIME version 1.8 (Caporaso et al.2010a) with standard parameters. Overall sequences with quality score average <25, short sequences (<200 bp) and sequences containing mismatches as well as homopolymers were removed. We conducted a denoising of chimeras, and the multiplex reads were assigned to corresponding sample according to their barcode, and we carried out OTU picking with Uclust (Edgar 2010). Representative sequences of the OTUs were aligned using the PyNAST algorithm with a minimum of 80% (Caporaso et al.2010b). These OTUs were taxonomically classified using RDP method (Wang et al. 2007) and Greengenes database (DeSantis et al.2006). The taxonomic position of bacteria and archaea was characterized using taxa summary QIIME scripts until genus level (L6). Alpha diversity parameters (number of observed OTUs, qualitative Chao1 index, Faith's Phylogenetic Diversity (PD_whole) and Shannon index) were calculated with a fixed number of 16 182 randomly picked reads for each sample. Beta-diversity was calculated using Bray-Curtis and unweighted and weighted UniFrac metrics (Lozupone and Knight 2005). PCoA plots were visualized with Emperor (Vazquez-Baeza et al.2013).
The representation of cultured bacteria (described by Chapon et al.2012) in the soil bacterial community was assessed by clustering the 16S sequences derived from these bacteria and the 454 reads in OTUs at 97% sequence similarity.
Analysis of soil physico-chemical parameters
For both sampling dates, pH, water content (WC) and NaOH-extractable organic carbon (Corg(NaOH)) were analyzed as described by Chapon et al. (2012). Briefly, soil pH was measured in a 1:5 soil:water suspension, and 2 g of soil were mixed with 10 mL of distilled water and shaken for 30 min. The pH was measured after decanting and 2 h of atmosphere equilibration. WC was determined by drying 10 g of soil at 105°C for 24 h. Corg(NaOH) was determined by dissolving 0.15 g of soil samples in 1.5 mL of 0.1 M NaOH for 15 h. After decanting and centrifugation, the organic carbon concentration in the supernatant was determined by spectrometry (absorbance measurement at 280 nm).
The 137Cs concentrations in the soil samples were measured by gamma spectrometry. In addition, 241Am, 154Eu and 60Co concentrations were measured in October 2009 by gamma spectrometry, whereas actinide concentrations (234U, 235U, 236U, 232Th, 239Pu, 240Pu and 241Pu) were measured in April 2009 by ICP-MS after dry ‘ashing’ and mineralization of the soil samples (Chapon et al.2012). 90Sr concentrations were derived from 137Cs content assuming a constant ratio of 1.87 between 137Cs and 90Sr concentrations (Kashparov et al.2001). All RN contents were significantly correlated with 137Cs content (Fig. S1, Supporting Information) except 232Th whose concentration is unvarying close to its natural value in soil (Table S1, Supporting Information). Thus, constant ratios between 137Cs and each RN were used to derive the concentrations of 241Am, 154Eu, 60Co, 234U, 235U, 236U, 239Pu, 240Pu and 241Pu when they were missing or below the detection limits (Table S1). When not measured, the 232Th concentration was considered equal to the mean of 232Th concentrations in all soil samples.
Estimation of the total absorbed dose rate by prokaryotic cell
In the absence of any method to measure the total absorbed dose rates (TADR) by prokaryotic cell, TADR values were calculated based on soil RN activities. For this, two irradiation pathways were considered in the calculation: the external irradiation from the radio-contaminated habitat of bacteria and archaea (i.e. the soil) and the internal irradiation resulting from the internalization of RN in the cells (Garnier-Laplace et al.2015).
The external and internal dose rates were calculated at each sampling point and for each sampling date by multiplying the dose conversion coefficient (DCC) for each RN by the measured RN concentration at this sampling point or in the cells, respectively. The RN concentration inside the cells was considered in equilibrium with the soil RN concentration (conservatively assuming a concentration ratio of 1 for each RN) in the absence of any other information. External and internal DCCs (Table S2A, Supporting Information) were calculated for each RN with the EDEN software version 2.3 (Beaugelin-Seiller et al.2006), assuming a different biological effectiveness for the different types of radiation (applied weighting factors were 10 for α-radiation, 3 for low β-radiation, and 1 for other β-radiation and γ-radiation) (Pröhl 2003). The prokaryotic cells were described as a sphere with a 1-μm diameter, living in an infinitely extended and uniformly contaminated soil (compositions are provided in Table S2B).
Statistical analyses
Non-parametric tests (Kruskal-Wallis) were calculated using QIIME (Caporaso et al.2010a). Redundancy analysis (RDA) was performed with R software (R Core Team 2011) on the OTUs and phylum abundance data with the principal component analysis on instrumental variables (PCAIV) function of the ade4 package (Dray and Dufour 2007). Heatmap was constructed with the gplots package of R (Bolker et al. 2011).
RESULTS
Overall prokaryotic diversity in the Chernobyl soils
This study focused on the prokaryotic communities in 36 soil samples collected inside and outside the RN-contaminated trench T22 at nine different positions in April and October 2009 (Fig. 1). The pH values ranged from 4.4 to 6.1, and soil WC ranged from 2.2% to 7.3% (Table 1). The NaOH extractable organic C content (Corg(NaOH)) was within the same range for all samples (average 2.25 ± 0.83g kg−1), except for samples 4, 13 and 14 which were lower in content (average 0.35 ± 0.30 g kg−1). Soils collected within the trench (sample numbers 1, 3, 4, 8, 10 and 12) were contaminated with RNs (137Cs activity ranging from 61 to 750 Bq g−1), whereas soils collected in the vicinity of trench T22 (sample numbers 13, 14 and 20) exhibited low RN levels (137Cs ranging from 0.35 to 1.50 Bq g−1; Table 1). Three soil subsamples from October 2009 (designated as a, b and c) were used for DNA extraction in order to assess the reproducibility of the experiment. After quality filtering, we obtained 753 703 reads with a read length average of 255 bp, ranging from 16 182 to 23 686 reads in the 36 samples (Table 1). From the reads, 8167 OTUs at 3% genetic distance were identified and after normalization of the dataset to 16 182 reads per sample and deletion of singletons, 7663 OTUs were identified with OTU counts varying between 633 and 2120 per sample (Table 1). The rarefaction curves based on PD whole tree, Chao1 and Shannon estimators tended to reach an asymptote (Fig. S2, Supporting Information). The Good's estimator of coverage varied between 95% and 98% across the samples, indicating that the depth of sequencing was nearly sufficient to detect most of the dominant OTUs (Table 1).
Primary soil characteristics of samples collected inside and outside of trench T22.
| Soil | Sampling | 137Cs | Corg(NaOH) | WC | TADR | Number of | Observed | Shannon | Good's estimator | |||
| sample | location | (Bq g−1) | pH | (g kg−1) | (%) | h−1) | Subsample | high-quality reads | OTUs | Chao1 | diversity index | of coverage |
| 1A | Inside | 420 | 5.1 | 2.56 | 5.8 | 83 | 21 279 | 1806 | 2702 | 8.6 | 0.96 | |
| 1O | Inside | 420 | 4.8 | 2.61 | 7.3 | 86 | a,b,c | 19 818 ± 1094 | 2087 ± 42 | 3030 ± 78 | 9.3 ± 0.1 | 0.95 ± 0.00 |
| 3A | Inside | 300 | 5.6 | 1.48 | 5.7 | 28 | 18 822 | 1628 | 2568 | 8.4 | 0.96 | |
| 3O | Inside | 750 | 5 | 1.77 | 5.1 | 153 | a,b,c | 20 827 ± 1469 | 1622 ± 36 | 2433 ± 29 | 8.0 ± 0.2 | 0.96 ± 0.00 |
| 4A | Inside | 61 | 5.4 | 0.49 | 3.5 | 9 | 23 083 | 1454 | 2197 | 8.3 | 0.96 | |
| 4O | Inside | 140 | 4.8 | 0.88 | 5.1 | 29 | a,b,c | 20 931 ± 489 | 1478 ± 47 | 2324 ± 51 | 8.4 ± 0.2 | 0.96 ± 0.00 |
| 8A | Inside | 640 | 5.2 | 4.12 | 6.2 | 137 | 22 159 | 1353 | 2102 | 8.1 | 0.97 | |
| 8O | Inside | 670 | 4.5 | 2.93 | 3.6 | 137 | a,b,c | 23 603 ± 90 | 1498 ± 24 | 2278 ± 54 | 8.4 ± 0.1 | 0.96 ± 0.00 |
| 10A | Inside | 290 | 4.7 | 2.15 | 4.9 | 90 | 23 613 | 1333 | 2054 | 7.9 | 0.97 | |
| 10O | Inside | 710 | 4.9 | 2.77 | 5.2 | 145 | a,b,c | 21 544 ± 2042 | 1524 ± 27 | 2275 ± 11 | 8.5 ± 0.1 | 0.96 ± 0.00 |
| 12A | Inside | 260 | 5.4 | 1.16 | 3.7 | 47 | 19 690 | 1585 | 2300 | 8.7 | 0.96 | |
| 12O | Inside | 370 | 4.4 | 1.48 | 4.8 | 76 | a,b,c | 21 515 ± 1 521 | 1452 ± 4 | 2277 ± 82 | 8.3 ± 0.0 | 0.96 ± 0.00 |
| 13A | Outside | 0.37 | 6.1 | 0.16 | 2.2 | 0.1 | 22 966 | 1145 | 1755 | 6.9 | 0.97 | |
| 13O | Outside | 1.5 | 4.3 | 0.12 | 2.7 | 0.3 | a,b,c | 17 761 ± 1626 | 639 ± 10 | 977 ± 60 | 5.8 ± 0.1 | 0.98 ± 0.00 |
| 14A | Outside | 0.91 | 6.1 | 0.1 | 2.5 | 0.4 | 22 291 | 969 | 1355 | 7.2 | 0.98 | |
| 14O | Outside | 0.79 | 4.8 | 0.34 | 4.1 | 0.2 | a,b,c | 20 118 ± 1308 | 1167 ± 16 | 1684 ± 36 | 7.8 ± 0.1 | 0.97 ± 0.00 |
| 20A | Outside | 0.35 | 5.4 | 1.99 | 4.7 | 0.3 | 23 354 | 1424 | 2108 | 8.2 | 0.97 | |
| 20O | Outside | 0.54 | 4.5 | 1.96 | 5.1 | 0.1 | a,b,c | 19 364 ± 1469 | 1427 ± 55 | 2160 ± 144 | 8.0 ± 0.1 | 0.97 ± 0.01 |
| Soil | Sampling | 137Cs | Corg(NaOH) | WC | TADR | Number of | Observed | Shannon | Good's estimator | |||
| sample | location | (Bq g−1) | pH | (g kg−1) | (%) | h−1) | Subsample | high-quality reads | OTUs | Chao1 | diversity index | of coverage |
| 1A | Inside | 420 | 5.1 | 2.56 | 5.8 | 83 | 21 279 | 1806 | 2702 | 8.6 | 0.96 | |
| 1O | Inside | 420 | 4.8 | 2.61 | 7.3 | 86 | a,b,c | 19 818 ± 1094 | 2087 ± 42 | 3030 ± 78 | 9.3 ± 0.1 | 0.95 ± 0.00 |
| 3A | Inside | 300 | 5.6 | 1.48 | 5.7 | 28 | 18 822 | 1628 | 2568 | 8.4 | 0.96 | |
| 3O | Inside | 750 | 5 | 1.77 | 5.1 | 153 | a,b,c | 20 827 ± 1469 | 1622 ± 36 | 2433 ± 29 | 8.0 ± 0.2 | 0.96 ± 0.00 |
| 4A | Inside | 61 | 5.4 | 0.49 | 3.5 | 9 | 23 083 | 1454 | 2197 | 8.3 | 0.96 | |
| 4O | Inside | 140 | 4.8 | 0.88 | 5.1 | 29 | a,b,c | 20 931 ± 489 | 1478 ± 47 | 2324 ± 51 | 8.4 ± 0.2 | 0.96 ± 0.00 |
| 8A | Inside | 640 | 5.2 | 4.12 | 6.2 | 137 | 22 159 | 1353 | 2102 | 8.1 | 0.97 | |
| 8O | Inside | 670 | 4.5 | 2.93 | 3.6 | 137 | a,b,c | 23 603 ± 90 | 1498 ± 24 | 2278 ± 54 | 8.4 ± 0.1 | 0.96 ± 0.00 |
| 10A | Inside | 290 | 4.7 | 2.15 | 4.9 | 90 | 23 613 | 1333 | 2054 | 7.9 | 0.97 | |
| 10O | Inside | 710 | 4.9 | 2.77 | 5.2 | 145 | a,b,c | 21 544 ± 2042 | 1524 ± 27 | 2275 ± 11 | 8.5 ± 0.1 | 0.96 ± 0.00 |
| 12A | Inside | 260 | 5.4 | 1.16 | 3.7 | 47 | 19 690 | 1585 | 2300 | 8.7 | 0.96 | |
| 12O | Inside | 370 | 4.4 | 1.48 | 4.8 | 76 | a,b,c | 21 515 ± 1 521 | 1452 ± 4 | 2277 ± 82 | 8.3 ± 0.0 | 0.96 ± 0.00 |
| 13A | Outside | 0.37 | 6.1 | 0.16 | 2.2 | 0.1 | 22 966 | 1145 | 1755 | 6.9 | 0.97 | |
| 13O | Outside | 1.5 | 4.3 | 0.12 | 2.7 | 0.3 | a,b,c | 17 761 ± 1626 | 639 ± 10 | 977 ± 60 | 5.8 ± 0.1 | 0.98 ± 0.00 |
| 14A | Outside | 0.91 | 6.1 | 0.1 | 2.5 | 0.4 | 22 291 | 969 | 1355 | 7.2 | 0.98 | |
| 14O | Outside | 0.79 | 4.8 | 0.34 | 4.1 | 0.2 | a,b,c | 20 118 ± 1308 | 1167 ± 16 | 1684 ± 36 | 7.8 ± 0.1 | 0.97 ± 0.00 |
| 20A | Outside | 0.35 | 5.4 | 1.99 | 4.7 | 0.3 | 23 354 | 1424 | 2108 | 8.2 | 0.97 | |
| 20O | Outside | 0.54 | 4.5 | 1.96 | 5.1 | 0.1 | a,b,c | 19 364 ± 1469 | 1427 ± 55 | 2160 ± 144 | 8.0 ± 0.1 | 0.97 ± 0.01 |
The table comprises soil physico-chemical parameters (from Chapon et al.2012), TADR by cell, OTU counts and estimators of alpha-diversity. In the soil sample name, ‘A’ stands for April and ‘O’ for October.
Primary soil characteristics of samples collected inside and outside of trench T22.
| Soil | Sampling | 137Cs | Corg(NaOH) | WC | TADR | Number of | Observed | Shannon | Good's estimator | |||
| sample | location | (Bq g−1) | pH | (g kg−1) | (%) | h−1) | Subsample | high-quality reads | OTUs | Chao1 | diversity index | of coverage |
| 1A | Inside | 420 | 5.1 | 2.56 | 5.8 | 83 | 21 279 | 1806 | 2702 | 8.6 | 0.96 | |
| 1O | Inside | 420 | 4.8 | 2.61 | 7.3 | 86 | a,b,c | 19 818 ± 1094 | 2087 ± 42 | 3030 ± 78 | 9.3 ± 0.1 | 0.95 ± 0.00 |
| 3A | Inside | 300 | 5.6 | 1.48 | 5.7 | 28 | 18 822 | 1628 | 2568 | 8.4 | 0.96 | |
| 3O | Inside | 750 | 5 | 1.77 | 5.1 | 153 | a,b,c | 20 827 ± 1469 | 1622 ± 36 | 2433 ± 29 | 8.0 ± 0.2 | 0.96 ± 0.00 |
| 4A | Inside | 61 | 5.4 | 0.49 | 3.5 | 9 | 23 083 | 1454 | 2197 | 8.3 | 0.96 | |
| 4O | Inside | 140 | 4.8 | 0.88 | 5.1 | 29 | a,b,c | 20 931 ± 489 | 1478 ± 47 | 2324 ± 51 | 8.4 ± 0.2 | 0.96 ± 0.00 |
| 8A | Inside | 640 | 5.2 | 4.12 | 6.2 | 137 | 22 159 | 1353 | 2102 | 8.1 | 0.97 | |
| 8O | Inside | 670 | 4.5 | 2.93 | 3.6 | 137 | a,b,c | 23 603 ± 90 | 1498 ± 24 | 2278 ± 54 | 8.4 ± 0.1 | 0.96 ± 0.00 |
| 10A | Inside | 290 | 4.7 | 2.15 | 4.9 | 90 | 23 613 | 1333 | 2054 | 7.9 | 0.97 | |
| 10O | Inside | 710 | 4.9 | 2.77 | 5.2 | 145 | a,b,c | 21 544 ± 2042 | 1524 ± 27 | 2275 ± 11 | 8.5 ± 0.1 | 0.96 ± 0.00 |
| 12A | Inside | 260 | 5.4 | 1.16 | 3.7 | 47 | 19 690 | 1585 | 2300 | 8.7 | 0.96 | |
| 12O | Inside | 370 | 4.4 | 1.48 | 4.8 | 76 | a,b,c | 21 515 ± 1 521 | 1452 ± 4 | 2277 ± 82 | 8.3 ± 0.0 | 0.96 ± 0.00 |
| 13A | Outside | 0.37 | 6.1 | 0.16 | 2.2 | 0.1 | 22 966 | 1145 | 1755 | 6.9 | 0.97 | |
| 13O | Outside | 1.5 | 4.3 | 0.12 | 2.7 | 0.3 | a,b,c | 17 761 ± 1626 | 639 ± 10 | 977 ± 60 | 5.8 ± 0.1 | 0.98 ± 0.00 |
| 14A | Outside | 0.91 | 6.1 | 0.1 | 2.5 | 0.4 | 22 291 | 969 | 1355 | 7.2 | 0.98 | |
| 14O | Outside | 0.79 | 4.8 | 0.34 | 4.1 | 0.2 | a,b,c | 20 118 ± 1308 | 1167 ± 16 | 1684 ± 36 | 7.8 ± 0.1 | 0.97 ± 0.00 |
| 20A | Outside | 0.35 | 5.4 | 1.99 | 4.7 | 0.3 | 23 354 | 1424 | 2108 | 8.2 | 0.97 | |
| 20O | Outside | 0.54 | 4.5 | 1.96 | 5.1 | 0.1 | a,b,c | 19 364 ± 1469 | 1427 ± 55 | 2160 ± 144 | 8.0 ± 0.1 | 0.97 ± 0.01 |
| Soil | Sampling | 137Cs | Corg(NaOH) | WC | TADR | Number of | Observed | Shannon | Good's estimator | |||
| sample | location | (Bq g−1) | pH | (g kg−1) | (%) | h−1) | Subsample | high-quality reads | OTUs | Chao1 | diversity index | of coverage |
| 1A | Inside | 420 | 5.1 | 2.56 | 5.8 | 83 | 21 279 | 1806 | 2702 | 8.6 | 0.96 | |
| 1O | Inside | 420 | 4.8 | 2.61 | 7.3 | 86 | a,b,c | 19 818 ± 1094 | 2087 ± 42 | 3030 ± 78 | 9.3 ± 0.1 | 0.95 ± 0.00 |
| 3A | Inside | 300 | 5.6 | 1.48 | 5.7 | 28 | 18 822 | 1628 | 2568 | 8.4 | 0.96 | |
| 3O | Inside | 750 | 5 | 1.77 | 5.1 | 153 | a,b,c | 20 827 ± 1469 | 1622 ± 36 | 2433 ± 29 | 8.0 ± 0.2 | 0.96 ± 0.00 |
| 4A | Inside | 61 | 5.4 | 0.49 | 3.5 | 9 | 23 083 | 1454 | 2197 | 8.3 | 0.96 | |
| 4O | Inside | 140 | 4.8 | 0.88 | 5.1 | 29 | a,b,c | 20 931 ± 489 | 1478 ± 47 | 2324 ± 51 | 8.4 ± 0.2 | 0.96 ± 0.00 |
| 8A | Inside | 640 | 5.2 | 4.12 | 6.2 | 137 | 22 159 | 1353 | 2102 | 8.1 | 0.97 | |
| 8O | Inside | 670 | 4.5 | 2.93 | 3.6 | 137 | a,b,c | 23 603 ± 90 | 1498 ± 24 | 2278 ± 54 | 8.4 ± 0.1 | 0.96 ± 0.00 |
| 10A | Inside | 290 | 4.7 | 2.15 | 4.9 | 90 | 23 613 | 1333 | 2054 | 7.9 | 0.97 | |
| 10O | Inside | 710 | 4.9 | 2.77 | 5.2 | 145 | a,b,c | 21 544 ± 2042 | 1524 ± 27 | 2275 ± 11 | 8.5 ± 0.1 | 0.96 ± 0.00 |
| 12A | Inside | 260 | 5.4 | 1.16 | 3.7 | 47 | 19 690 | 1585 | 2300 | 8.7 | 0.96 | |
| 12O | Inside | 370 | 4.4 | 1.48 | 4.8 | 76 | a,b,c | 21 515 ± 1 521 | 1452 ± 4 | 2277 ± 82 | 8.3 ± 0.0 | 0.96 ± 0.00 |
| 13A | Outside | 0.37 | 6.1 | 0.16 | 2.2 | 0.1 | 22 966 | 1145 | 1755 | 6.9 | 0.97 | |
| 13O | Outside | 1.5 | 4.3 | 0.12 | 2.7 | 0.3 | a,b,c | 17 761 ± 1626 | 639 ± 10 | 977 ± 60 | 5.8 ± 0.1 | 0.98 ± 0.00 |
| 14A | Outside | 0.91 | 6.1 | 0.1 | 2.5 | 0.4 | 22 291 | 969 | 1355 | 7.2 | 0.98 | |
| 14O | Outside | 0.79 | 4.8 | 0.34 | 4.1 | 0.2 | a,b,c | 20 118 ± 1308 | 1167 ± 16 | 1684 ± 36 | 7.8 ± 0.1 | 0.97 ± 0.00 |
| 20A | Outside | 0.35 | 5.4 | 1.99 | 4.7 | 0.3 | 23 354 | 1424 | 2108 | 8.2 | 0.97 | |
| 20O | Outside | 0.54 | 4.5 | 1.96 | 5.1 | 0.1 | a,b,c | 19 364 ± 1469 | 1427 ± 55 | 2160 ± 144 | 8.0 ± 0.1 | 0.97 ± 0.01 |
The table comprises soil physico-chemical parameters (from Chapon et al.2012), TADR by cell, OTU counts and estimators of alpha-diversity. In the soil sample name, ‘A’ stands for April and ‘O’ for October.
Of the reads, 95.7% were affiliated with Bacteria and 4.3% were affiliated with Archaea. The most consistently detected bacterial phyla (relative abundance >1%) across all samples were Proteobacteria, Acidobacteria, AD3, Planctomycetes, Chloroflexi, Verrucomicrobia and Actinobacteria, with an uneven distribution across the samples (Fig. 1). Combined, these seven groups accounted for 83.3% of the sequences. Twenty-seven additional bacterial phyla were detected as well as three archaeal phyla.
Trench T22 hosts specific microbiota
The rarefaction curves calculated using different estimators revealed a higher apparent species richness in almost all samples collected in the trench, as compared to the samples collected outside of the trench (Table 1). Accordingly, mean Chao1 richness and Shannon diversity indices were significantly higher in samples collected inside as opposed to outside the trench (P< 0.05; Fig. S2).
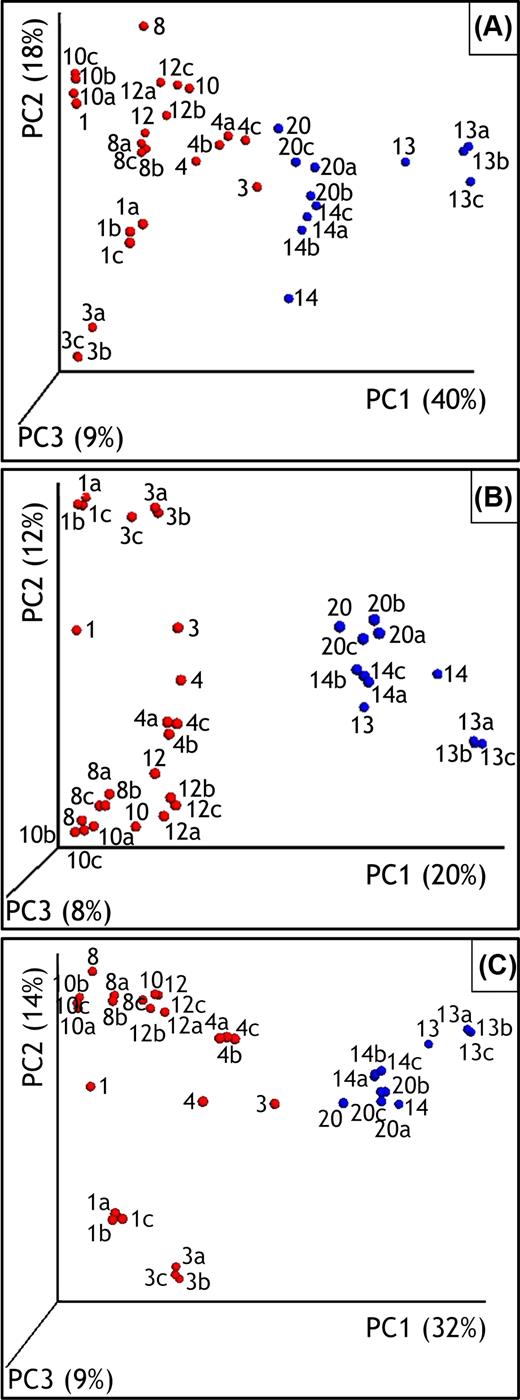
Principal coordinates analysis (PCoA) based on bacterial and archaeal OTUs and performed with Bray-Curtis, Unweighted and Weighted Unifrac methods all showed a clear separation between the communities inside and outside the trench (Fig. 2). The first principal components of the PCoA accounted for 20%–40% of the variation between the samples, depending on the method used. Some divergence between samples taken at the same location in April and in October could be observed, particularly for samples collected within the trench. By contrast, high consistency between replicates (labeled as ‘a’, ‘b’ or ‘c’) was observed for each soil sample collected in October 2009, indicating high reproducibility of the experimental procedure.
Three-dimensional PCoA plots of weighted UniFrac (A), unweighted UniFrac (B) and Bray−Curtis (C) distances between the soil samples collected in the trench (red) and in the surrounding soils (blue). Samples are identified with a number corresponding to the sampling position. To enhance readability, the samples collected in April 2009 are labeled with a single number, whereas the triplicates performed in October 2009 are labeled as Xa, Xb or Xc. The % variation explained by the PCs is indicated on the respective axes and refers to the fraction of the total variance.
Impact of long-term exposure to radioactive contaminated wastes on prokaryotic microbiota
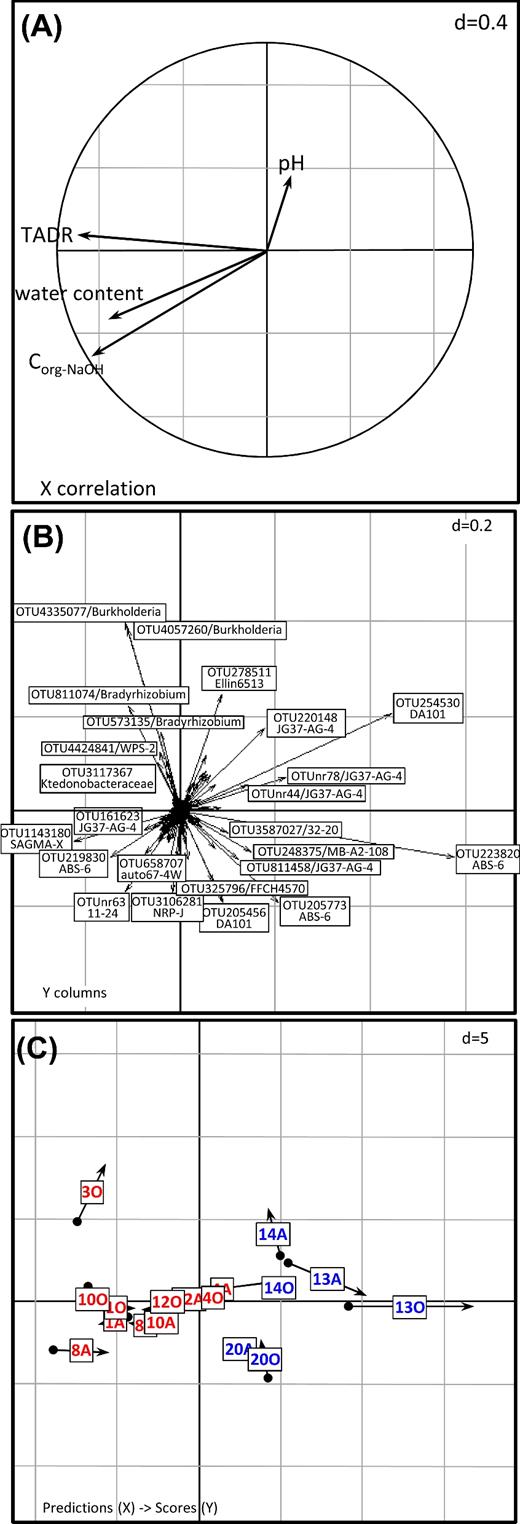
To examine how the prokaryotic microbiota was affected by radioactivity, the TADR was calculated for each soil sample. On average, estimated TADR by cell were two orders of magnitude higher in trench soils than in samples collected outside of the trench; the maximum TADR was ca. 150 mGy h−1 in sample 8 in October 2009, whereas the lowest value was 0.1 mGy h−1 in samples 13 and 20 in April and October 2009, respectively (Table 1). Internal absorbed dose rates contributed to only 3% of the estimated TADR by cell (Table S3A and B) which is mainly due to the small size of prokaryotic cells. Actinides were the primary contributors to the estimated TADR by cell, and 239Pu and 240Pu were responsible for 80%–96% of the estimated TADR by cell (Table S3A and B). Since the PCoA analysis was able to confirm the reproducibility of the experiment, the triplicates performed in October were averaged in order to create a single dataset and to facilitate depiction of the data. A RDA coupled to a permutation test (randtest function) was used to test whether the community composition at the different sampling sites was related to the physico-chemical properties of the soil samples (Ramette 2007). The four physico-chemical variables included in the analysis (TADR, Corg(NaOH), WC and pH) significantly explained 43% of the variation of the bacterial and archaeal communities (r2 = 0.43, P= 0.001 based on 999 permutations). The first two axes of the RDA explain 85% of the variance due to the soil physico-chemical properties, 71% of which is accounted for by the first axis, which correlated primarily with TADR (r = –0.90), Corg(NaOH) (r = –0.84) and WC (r = –0.76) (Fig. 3). Using the RDA function in the ‘vegan’ package to separately analyze the effect of each variable indicates that TADR was the principal variable to influence the prokaryotic communities at the out level (P< 0.05).
Multivariate RDA diagrams illustrating the relationship between the OTU-level community structure from soil samples collected in April (labeled with A) and October (labeled with O) 2009 and environmental parameters (TADR, Corg(NaOH), pH and WC). (A) Correlation of physicochemical explanatory variables with the first two axes of the PCAIV; (B) OTU scores. Only OTUs with the highest loadings are labeled with their ID number and lowest taxonomic rank. (C) Joint display of the position of the sampling sites by averaging (points) and by regression (arrowheads). Red: samples from the trench; blue: samples from the surrounding area.
The same analysis at the phylum level confirmed the effect of the environmental variables on the bacterial communities since 52% of the variation of the prokaryotic communities at the phylum level were explained by the environmental variables (r2 = 0.523, P= 0.001 based on 999 permutations). The first two axes of the RDA explained 92% of the variance, among which 74% is explained by the first axis which correlated mainly with TADR (r = –0.89) and to a lesser extent with Corg(NaOH) (r = –0.69) and WC (r = –0.66) (data not shown). The analysis of the effect of each variable separately showed that the principal variables influencing the prokaryotic communities at the phylum level were TADR (P< 0.05) and Corg(NaOH) (P< 0.1).
Primary differences between prokaryotic microbiota of the trench and surrounding soils
Non-parametric tests (Kruskal-Wallis) were used to determine the significance of differences in prokaryotic diversity between samples and to identify the taxa representative of either the trench or the surrounding soils. This analysis revealed that among the 7663 OTUs identified, 1781 OTUs displayed significant changes in abundance between the highly contaminated trench soils and the low-contamination surrounding soils (P< 0.05). Specifically, 973 OTUs were significantly more abundant in the trench soils than in the surrounding soils, whereas 808 OTUs were significantly more abundant in the surrounding soils than in the trench soils. Furthermore, these 1781 OTUs accounted for 50% of the total prokaryotic microbiota in the trench and for 67% of the total prokaryotic microbiota in the surrounding soils, and could be classified into 26 bacterial phyla and three archaeal phyla.
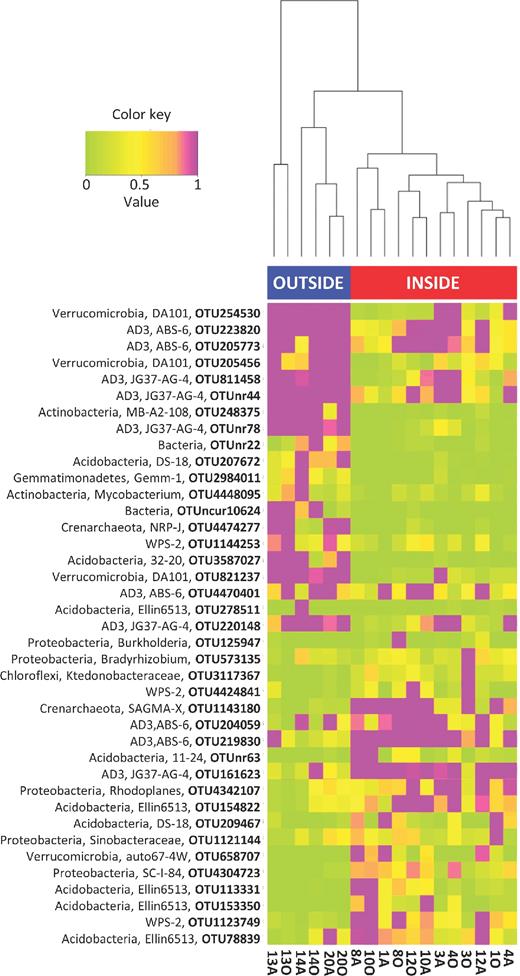
To further explore the primary differences between microbiota of the trench soils and surrounding soils, a heatmap was constructed with the most abundant OTUs (>1% relative abundance in at least one sample; see Table S4 for details) differing significantly between the trench and the surrounding soil microbiota (Fig. 4). The surrounding soils were characterized by 20 abundant OTUs affiliated with the phyla Crenarchaeota (NRP-J order), Acidobacteria (Ellin6513, 32–20, DS-18 orders), Actinobacteria (Mycobacterium genus and MB-A2–108 class), AD3 (ABS-6 and JG37-AG-4 classes), Gemmatimonadetes (Gemm-1 class), Verrucomicrobia (DA101 genus) and WPS-2 (Fig. 4). On the other hand, the trench prokaryotic community was characterized by 19 abundant OTUs. These OTUs were affiliated with the phyla Crenarchaeota (SAGMA-X family), Acidobacteria (11–24, Ellin6513, DS-18 orders), AD3 (ABS-6 and JG37-AG-4 classes), Chloroflexi (Ktedonobacteraceae family), Proteobacteria (Bradyrhizobium, Rhodoplanes and Burkholderia genera, SC-I-84 order and Sinobacteraceae family), Verrucomicrobia (auto67–4W family) and WPS-2 (Fig. 4). Within the Archaea, the representative sequence of OTU 1143180 (Crenarchaeota (SAGMA-X)) exhibited 100% sequence similarity with Candidatus Nitrosotalea devanaterra, an acidophilic ammonia oxidizer (Lehtovirta-Morley et al.2011) as well as sequences detected in acid water from a gold mine (AB050208; Takai et al.2001). The OTUs specific to the trench did not have any cultured representatives within the Bacteria, with the exception of Bradyrhyzobium, Rhodoplanes, Burkholderia and Sinobacteraceae.
Cluster analysis of the soil samples and the corresponding heat map constructed with abundant OTUs (>1% relative abundance in at least one soil sample) differing significantly between the trench and the surrounding soil microbiota. Red: samples from the trench; blue: samples from the surrounding area. The highest and lowest taxonomic affiliations are indicated for each OTU.
Relationships with other environments were investigated by performing a BLAST search of the non-redundant database, using representative sequences of the 19 OTUs specific to the trench soils. Interestingly, among the closest neighbors (98%–100% sequence similarity), we found sequences derived not only from a wide range of forest soils (data not shown) but also from cold environments (alpine tundra, boreal wetlands and forest, Finland, Himalayan mountains, Alaska, Iceland, Greenland, Antarctic habitats, the Arctic), acidic environments (acid water, acid mine drainage, acidic fen soil, acidic subalpine forest, acidic wetland), volcanic soils (Kasatochi Island in Alaska, a lava forest soil in Korea, Hawaiian volcanic deposit), RN- and heavy metal-contaminated soils, sediment and water samples (neptunium-contaminated sediments, soil samples of uranium mining waste pile, acid water from gold and potassium mines, lead-contaminated forest soil) as shown in Table S5. The representative sequence of OTU 573135, which is affiliated with Bradyrhizobium, was reported to have a 100% sequence similarity with many sequences, including a bacterium cultured from dust samples from the International Space Station (LT617088; Mora et al.2016).
Representation of cultured isolates in the prokaryotic community
We previously characterized a collection of 303 isolates recovered from Chernobyl soil samples by 16S rDNA partial sequencing (Chapon et al.2012). Here, we analyzed whether the 284 isolates with available appropriate sequence data (i.e. the V4 domain of the 16S rDNA) were detected in the pyrosequencing dataset, and if they represent an abundant taxon in the prokaryotic community. In total, 206 sequences derived from isolates exhibited at least 97% sequence similarity with a pyrotag, falling within 47 OTUs of the pyrosequencing dataset. A total of 183 isolates fall within 42 OTUs that represented a very minor fraction of the community across all samples, since altogether they accounted for <0.2% of the total. By contrast, 23 isolates fall within five well-represented OTUs (maximum relative abundance ranging from 0.71% to 6.97%): OTUs 125947, 156722, 196652 and 4057260, all affiliated to Burkholderia spp. and OTU 573135 affiliated to Bradyrhizobium spp. (Table S6). The relative abundance of OTUs 4057260 and 573135 reached 6.97% and 2.90%, respectively, in the most contaminated soil (750 Bq/g of 137Cs), which was collected in October 2009 at sampling point 3 (sample 3O; Tables 1 and S6). Furthermore, OTUs 125947 and 573135 are characteristic of the trench prokaryotic community.
DISCUSSION
The current work explored the diversity of soil prokaryotes that have been evolving since more than two decades in RN-contaminated soils inside and outside of the trench T22, located within the Chernobyl exclusion zone. The presence of complex prokaryotic communities in contaminated soil and control samples from this site was revealed by a genetic fingerprint method (DGGE) in our previous study (Chapon et al.2012). We observed that apparent diversity in the trench soils was similar to that observed in the surrounding soils on DGGE. However, this approach did not uncover any dominant community fingerprint clearly related to RN content, nor did it provide any direct taxonomic information. Here, we revisited this previous work to address these issues using 454 pyrosequencing of 16S rRNA genes. We surveyed the prokaryotic diversity of this understudied environment, and we assessed the effects of RN contamination and Corg(NaOH) content on soil microbiota using a comparative analysis with low-contamination soils collected outside of the trench in the same area. The 137Cs content in the soil samples collected outside of the trench is consistent with worldwide upper background levels of 137Cs due to Chernobyl fallout (e.g. in France: from 0.01 to 0.1 Bq g−1 in upper soils, max value in hotspots: 10 Bq g−1; IRSN 2016). In addition, Michel et al. (2015) reported several values for Ukraine (from 0.3 mBq g−1 to 6.6 Bq g−1) outside of the Chernobyl exclusion zone.
The seven dominant bacterial phyla in the Chernobyl soils were Proteobacteria, Acidobacteria, AD3, Planctomycetes, Chloroflexi, Verrucomicrobia and Actinobacteria. These phyla are commonly encountered as the dominant taxa in soils (Janssen 2006; Rastogi et al.2009). Here, the abundance of Chloroflexi (mean relative abundance 9.4%, ranging from 2.4% to 16.1%) and AD3 (mean relative abundance 14.8%, ranging from 3.3% to 39.0%) was high, both inside and outside the trench. On a global scale, this is the most noticeable feature of Chernobyl soil prokaryotic communities. This high abundance is not an artifact linked to DNA extraction or PCR amplification, since other soil samples processed in the same experimental run did not exhibit the same profile (data not shown). High abundance of Chloroflexi/AD3 has been described in cold environments (Costello and Schmidt 2006; Taş et al.2014; Hultman et al.2015; Frey et al.2016). Members of Chloroflexi/AD3 are reported to predominate following acute gamma irradiation of soils (McNamara et al.2007; El-Sayed and Ghanem 2009) and have been detected in natural uranium ores (Mondani et al.2011) and in uranium-contaminated sites (Selenska-Pobell 2002; Barns et al.2007; Hug et al.2013). It has been proposed that degradation of plant polymers is widespread across this phylum (Hug et al.2013). Thus, members of this phylum are well adapted to cold environments; they are potentially able to degrade lignocellulosic material; and they can tolerate uranium and/or its associated radioactivity. The high abundance of Chloroflexi/AD3 members both inside and outside trench T22 is coherent, taking into account the facts that Chernobyl upper soils are partly frozen during winter (Bixio et al.2002), that wood debris were buried in the trench, and that bacteria and archaea are exposed to different levels of irradiation and contamination whether they are inside or outside of the trench. Nevertheless, when examining the results in greater detail, we noted that specific phylotypes within Chloroflexi/AD3 were selected by the soil trench. Indeed, the abundance of seven OTUs affiliated with classes ABS-6 and JG37-AG-4 was dramatically reduced in the trench, whereas three OTUs from the same classes were strongly enriched in the trench, suggesting that these phylotypes may have distinct ecological niches and physiological characteristics such as tolerance to RNs exposure.
Our detailed comparative analysis of soils collected inside and outside the trench has revealed several specific patterns of soil prokaryotic assemblages. First, the Chao1 richness and Shannon diversity indices were significantly higher inside the trench as compared to the samples collected outside of the trench. On average, 1589 observed OTUs were detected in the trench soils, whereas 1103 OTUs were detected in the surrounding soils, indicating a positive effect of trench conditions on diversity. This had not been evidenced in our previous study, probably because of the limitation of DGGE to resolve complex profiles. Second, the PCoA ordination demonstrated significant differences between the prokaryotic communities from the trench and the surrounding area. When the trenches were dug, large amounts of highly irradiated and contaminated wood debris were buried, providing a considerable input of RNs as well as organic matter into this oligotrophic environment. The introduction of organic matter is expected to enhance diversity either by stimulating heterotrophic microorganisms which were already present as minor species or by introducing new species from outside of the site. For instance, Field et al. (2010) showed that bacterial diversity was significantly enhanced upon cellulosic waste burial in the soil, with a corresponding shift in OTU number from approximately 750 to 1600. By contrast, RN contamination is expected to have a negative impact on microbial communities due to radiotoxicity. According to Field et al. (2010), the impact of organic matter in the trench should have been larger than that actually observed in this study. We thus hypothesized that the presence of RNs counterbalanced the positive effect of lignocellulosic material on prokaryotic diversity in the trench, which would suggest that the RNs remaining in the trench still exert a negative impact on the prokaryotic communities. This hypothesis is supported by the statistical analysis of the data, since the RDA coupled to a permutation test revealed that the principal variable, among all measured variables, influencing the prokaryotic communities at the OTU and phylum level was TADR and to a lesser extent Corg(NaOH) when the analysis was performed at the phylum level. However, we could not exclude that other confounding factors not monitored during our study might also have influenced the prokaryotic communities composition.
A total of 808 OTUs were significantly enriched in the prokaryotic community outside of the trench, including 20 abundant OTUs. These abundant OTUs were affiliated to different phyla and do not have any cultured representatives, with the exception of Mycobacterium. Members of these phylotypes probably correspond to RN-sensitive species and/or oligotrophic microorganisms. We also identified specific OTUs that distinguished the trench microbiota from surrounding microbiota, indicating that specific phylotypes were selected by the trench soils. Close relationships with a wide range of cold environments and acidic soils were revealed, as well as volcanic soils or soils that have been subjected to fire, showing that traces of past events are still detectable in the trench prokaryotic community. In addition, relationships with radioactive environments were shown for five OTUs specific to the trench soils. Thus, members of the phylotypes that were selected by the trench conditions, in particular by high TADR, may have the capacity to survive under stressful conditions. These phylotypes are distributed among several phyla, illustrating that adaptation to life in radioactive environment is widespread in prokaryotes.
A limited number of reads from the 454 dataset identified as Deinococcus were detected in the soil samples, representing a very minor fraction of the total abundance (0.004%–0.084%; mean relative abundance 0.017%). However, we failed to confirm the presence of this bacterium in Chernobyl soils using specific primers (Theodorakopoulos et al.2013), suggesting that the very low relative abundance is the consequence of sequencing errors or PCR bias. Furthermore, Deinococcus-related species were not identified among the cultured bacteria retrieved from the soil samples (Chapon et al.2012). Taken together, these results strongly suggest that Deinococcus are either absent or are in extremely low abundance in this environment. This may be related to the high moisture content of Chernobyl soils, whereas Deinococcus members have been shown to be well adapted to dry or low-moisture soils (Fredrickson et al.2004; Chanal et al.2006; Theodorakopoulos et al.2013; Li et al.2015).
Finally, the identification of representatives of two abundant OTUs typical of trench microbiota (OTU 125947 and 573135) in our culture collection has yielded some interesting results. These isolates, related to the genera Bradyrhizobium and Burkholderia, represent appropriate models for assessing the impact of long-term RN exposure on bacteria. Future studies could focus on these isolates to help determine the mechanisms of adaptation to this very unusual ecological niche.
CONCLUSION
In this study, we demonstrated that prokaryotic communities from Chernobyl disposal trench T22, as well as samples collected in the proximity, share common traits with prokaryotic assemblages from cold environments on a global scale although a significant difference in diversity and community composition could be observed between the trench and its proximal environment. Our results indicate that the introduction of contaminated lignocellulosic material within trench T22 has shaped prokaryotic communities by modifying species composition and abundance. In this regard, TADR was the principal variable to influence the prokaryotic communities while still exerting a negative effect on prokaryotic diversity. We identified specialized phylotypes that have successfully colonized the trench, two of which have cultured representatives in our culture collection. Further work, such as shotgun metagenome sequencing and experiments on these bacterial isolates, may be helpful in determining the function of bacterial communities in this ecosystem.
SUPPLEMENTARY DATA
Supplementary data are available at FEMSEC online.
Acknowledgements
We thank Brandon Loveall of Improvence for English proofreading of the manuscript. We thank the anonymous reviewers and the editor for their constructive comments which significantly contributed to improve the manuscript.
FUNDING
This work was supported by the National Center for Scientific Research and the Institute for Radioprotection and Nuclear Safety through the TRASSE GNR (Research Action Number 2009–1A). Nicolas Theodorakopoulos was the recipient of a PhD grant co-funded by the Institute for Radioprotection and Nuclear Safety and the Provence Alpes Côtes d’Azur regional council.
Conflict of interest. None declared.