-
PDF
- Split View
-
Views
-
Cite
Cite
Tobin J Hammer, Jon G Sanders, Noah Fierer, Not all animals need a microbiome, FEMS Microbiology Letters, Volume 366, Issue 10, May 2019, fnz117, https://doi.org/10.1093/femsle/fnz117
Close - Share Icon Share
ABSTRACT
It is often taken for granted that all animals host and depend upon a microbiome, yet this has only been shown for a small proportion of species. We propose that animals span a continuum of reliance on microbial symbionts. At one end are the famously symbiont-dependent species such as aphids, humans, corals and cows, in which microbes are abundant and important to host fitness. In the middle are species that may tolerate some microbial colonization but are only minimally or facultatively dependent. At the other end are species that lack beneficial symbionts altogether. While their existence may seem improbable, animals are capable of limiting microbial growth in and on their bodies, and a microbially independent lifestyle may be favored by selection under some circumstances. There is already evidence for several ‘microbiome-free’ lineages that represent distantly related branches in the animal phylogeny. We discuss why these animals have received such little attention, highlighting the potential for contaminants, transients, and parasites to masquerade as beneficial symbionts. We also suggest ways to explore microbiomes that address the limitations of DNA sequencing. We call for further research on microbiome-free taxa to provide a more complete understanding of the ecology and evolution of macrobe-microbe interactions.
INTRODUCTION
Beginning with Antonie van Leeuwenhoek and continuing into the early 20th century, microbiologists and zoologists discovered a seemingly ubiquitous world of microbial life in and on the bodies of animals. From minute beetles to humans, microorganisms were repeatedly observed in intimate association with animal tissues (Buchner 1965 and references therein). Such findings led to a widespread opinion among these early pioneers that microbial symbiosis must be ‘an elementary principle of all organisms’ (Buchner 1965, p. 69). But the study of symbiosis then fell out of fashion and laid relatively dormant until methodological advances, particularly cultivation-independent DNA sequencing, spurred a second wave of microbial exploration (Moran 2006).
There has been a recent explosion of studies finding microbes on and in a wide range of animals, reviving the view that all macroscopic organisms are hosts to microbial symbionts or microbiomes (Table S1, Supporting Information). Furthermore, in a number of host groups, symbionts have been shown to play critical and often surprising roles in development, physiology, behavior, defense from enemies and a variety of other traits (e.g. Ezenwa et al. 2012; McFall-Ngai et al. 2013; Sommer and Bäckhed 2013; Douglas 2014; Gerardo and Parker 2014). The apparent ubiquity of microbiomes, combined with evidence that symbionts mediate certain traits in certain animals, gives an impression that the microbiome has far-reaching influences on the biology of all animals (Table S1, Supporting Information).
But is the paradigm that all animals have and depend upon microbial symbionts actually supported by firm evidence? We argue that the field has overcorrected from the days when symbiont roles in animal biology were underappreciated. Now, reinforced by methodological issues (discussed below) and particularly exciting case studies, researchers often assume microbiomes where there may be none. Many authors have claimed that all (or ‘virtually all’) animals host a microbiome (Table S1,Supporting Information), but in fact, next to nothing is known about microbial associations for the majority of animal species on the planet. We suggest that the presence of a resident and functionally relevant microbiome is not the appropriate null hypothesis for explorations into these uncharted waters. It is important to not just ask ‘who are the microbes?’ and ‘what are they doing?’, but also whether there is even a microbiome in the first place.
Moreover, by generalizing all animals as microbe-rich holobionts, this paradigm obscures an interesting aspect of animal diversity—that animals vary strongly in the degree to which they host and depend upon microbial symbionts. This more nuanced perspective encourages us to rethink the approaches we use to study animal microbiomes, and prompts new questions about how and why animals evolve along the spectrum of microbiome dependence.
What is a microbiome anyway?
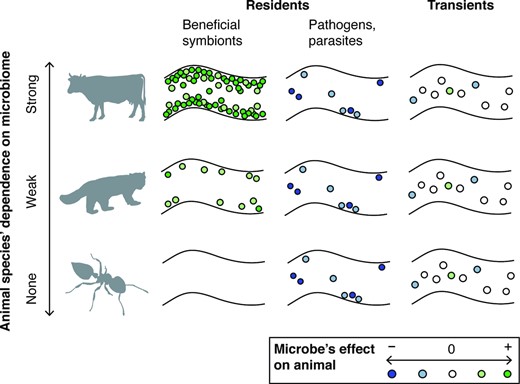
Whether all animals do or do not have microbiomes, microbiota or symbionts depends upon one's definition of these terms. It is almost certainly true that every animal interacts with microbes in some way, for at least part of its life cycle. But terms such as microbiome (Lederberg and McCray 2001) are so all-encompassing that the same word lumps very different kinds of microbial associations into a single concept, glossing over fundamental differences across animal species (also see Harris 1993). To highlight these differences, we focus on two axes of variation—residency and function—that are of particular significance in characterizing microbiomes and their constituent members (Fig. 1). Both categories can be fluid and highly context-dependent (e.g. Weeks et al. 2007; Mason et al. 2011; Chamberland et al. 2017; Douglas 2018). Nevertheless, they are a useful way to roughly categorize both microbes and microbiomes by their average or typical effects, as shown by the mainstream use of related terms like ‘pathogen’, ‘transient’ and ‘beneficial microbiome’.
A schematic of microbial associations for three animal species across the microbial dependency spectrum, exemplified by a cow, red panda, and Crematogaster ant (see text for more information on these species). All species contain some transient and parasitic/pathogenic microbes, but differ in the degree to which they are colonized by beneficial symbionts. Shown is a section of the gut, but the same principles could apply to non-gut symbioses as well. Inset: classification of individual microbes by their effect on host fitness, from negative ( − ) to neutral ( 0 ) to positive ( + ). Note that microbial residency and function categories can be fluid and context-dependent (not shown). For clarity, we do not depict the case of an animal highly dependent on transient microbes as food.
By residency, we refer to the degree to which a microbial population remains stably associated with a host. Resident microbes, also known as symbionts [‘living together’ (Douglas 1994)], differ from transient microbes in that they replicate inside a host at a rate exceeding loss due to death or excretion. Microbiologists and ecologists have long placed an important distinction between these two categories (e.g. Janzen 1977; Harris 1993; Berg 1996; Snell Taylor et al. 2018), and an increasing number of animal microbiome studies are making efforts to differentiate resident from transient taxa as well (David et al. 2014; Lee et al. 2016; Hammer et al. 2017; Auchtung et al. 2018).
By ‘function’, we refer to the types and degree of microbial effects on host fitness. Transient microbes could have positive, negative, or negligible effects on hosts. Due to their growth and metabolic activity, resident microbes are likely to have some effect on their host, but it could be negative—in which case they would traditionally be described as parasites or pathogens. Alternatively, resident microbes could increase host fitness; in these cases, the benefits to hosts can take many forms, and can vary in their importance depending on the microbial and host taxa involved and on environmental conditions.
These axes can also apply at the level of whole assemblages of microbes (Fig. 1). For example, some animals such as termites and humans host microbiomes that are predominantly composed of resident, beneficial microbes (Walter and Ley 2011; Brune and Dietrich 2015). Noting that a range of microbial associations can occur (Fig. 1), the focus of our discussion here is on the opposite end of the spectrum: animals that typically host few or no living microbial cells. When microbes are present in these species, they may be entirely transient and irrelevant to host fitness, or resident yet detrimental to host fitness. For clarity and emphasis, we refer to these species as lacking microbiomes.
Microbiome presence and function vary within hosts
To appreciate how microbiomes can vary across animal species, it is helpful to consider the enormous variation in symbiont abundance and function that exists within individual hosts. For example, humans host trillions of microbes in the gut and on skin (Sender, Fuchs and Milo 2016), but the brain, blood, placenta, eye surface and many other tissues are effectively sterile under normal conditions (Glassing et al. 2016; Perez-Muñoz et al. 2017; Wan et al. 2018). Many arthropods that host localized, intracellular endosymbionts in the body cavity lack resident gut microbes altogether (Engel and Moran 2013; Jing et al. 2014; Ross et al. 2018). Large variation can occur even within a single organ. For example, bacteria in the human small intestine are several orders of magnitude lower in abundance than in the large intestine, and may compete with the host for nutrients, rather than contribute nutrients (Walter and Ley 2011; Donaldson, Lee and Mazmanian 2016). Insects reliant on a resident gut microbiome also typically confine their symbionts to specific regions within the digestive tract (Sudakaran et al. 2012; Ohbayashi et al. 2015; Kwong and Moran 2016; Lanan et al. 2016; Salem et al.2017)
In invertebrates, symbionts are often only abundant and active during those phases of the host life cycle in which they contribute specific functions, such as nutrient provisioning of newly emerged adults (Vigneron et al. 2014) or pathogen defense of eggs (Flórez et al. 2017). In honey bees and bumble bees, larvae contain very few, if any microbes (Martinson, Moy and Moran 2012), while adult workers host a dense and specific gut microbiome important for digestion and immune defense (Kwong and Moran 2016). The marine snail Gigantopelta chessoia provides another example. Young adults lack symbionts and graze on free-living microbes. As they continue growing, they develop a unique organ to acquire and house chemosynthetic symbionts, upon which they then depend entirely for nutrition (Chen et al. 2018). Symbionts may even be costly when present in the ‘wrong’ life stage, as shown in the coral Orbicella faveolata (Hartmann et al. 2019).
Likewise, not all microbial lineages are abundant and active within a given microbiome, despite statements that all animals form symbioses with a diverse array of microorganisms, including bacteria, archaea, fungi and other eukaryotes (Table S1, Supporting Information). For example, unlike bacteria, fungi have been shown to be only transiently present in the healthy human gut and typically derived from food or the mouth (Auchtung et al. 2018). In invertebrates, resident microbiomes are often dominated by only one or a few species (e.g. Haynes et al. 2003; Dubilier, Bergin and Lott 2008; Salem et al. 2017; Matsuura et al. 2018), not a diverse community spanning multiple domains (but see Brune and Dietrich 2015). Even species ingesting or exposed to microbially diverse substrates, such as soil or seawater, can actively filter environmental microbes to permit only a single species to colonize (Nyholm and McFall-Ngai 2004; Ohbayashi et al. 2015).
These examples clearly illustrate how animals are capable of controlling where, when and which microbes colonize their bodies. Likewise, they show how animals may rely on only some microbes, for only some functions, at only some stages in their life cycle. Given such diversity in microbiome abundance and function within individual animals, it is reasonable to expect analogous variation extending across animal lineages. In other words, while some animals are strongly dependent on symbionts for one or more functions, others have evolved lifestyles with little or no direct contributions by symbionts.
How and why animals may exist without symbionts
We know of no strong reasons to expect a priori that all animals must engage in symbioses with beneficial microbes. The often-repeated statement that microbes are everywhere or that animals inhabit a ‘bacterial world’ (McFall-Ngai et al. 2013) may be taken to suggest that symbiotic associations are inescapable. Yet many animals inhabit and/or feed on relatively sterile microenvironments (such as internal tissues of plants or other animals), and it is well known that animals can control or prevent the growth of environmentally derived microbes through chemical, mechanical or immunological means. If it is possible for an animal to avoid colonization by all but one species of microbe, why not by all microbes?
Furthermore, even if a specific trait is microbially mediated in one animal species, it may not be in another. For example, bobtail squid are a now-classic model system in which bioluminescent symbionts provide hosts with camouflage (McFall-Ngai 2008), but most other cephalopods (and most bioluminescent animals in general) produce light on their own (Haddock, Moline and Case 2010). Egg production in the wasp Asobara tabida depends on the presence of the endosymbiont Wolbachia, but not in a closely related species that lacks Wolbachia (Dedeine et al. 2001). Likewise, leaves and wood are difficult-to-digest, nitrogen-poor diets that may bring to mind symbiont-dependent animals, such as leafcutter ants and termites (Aylward, Currie and Suen 2012; Brune and Dietrich 2015). However, a variety of other animals use host-encoded mechanisms to subsist on leaves or wood without direct microbial assistance (German and Bittong 2009; Shelomi, Watanabe and Arakawa 2014; Hammer et al. 2017; Besser et al. 2018). (These capabilities are sometimes conferred by genes horizontally acquired from microbes, but we do not consider such genes, nor mitochondria, to be part of an animal's microbiome).
Microbiome-free animals would miss out on the well-documented benefits that symbionts are capable of providing, and excluding microbes is likely neither cheap nor easy—particularly for species that encounter large numbers of potential colonists in their food or environment. On the other hand, they would avoid paying the widely acknowledged costs of hosting and relying on symbionts (Vorburger and Gouskov 2011; Bennett and Moran 2015; Hammer et al. 2017; Rodrigo et al. 2017). As examples of these costs, symbionts can limit host dispersal (Simonsen et al. 2017), compete with hosts for nutrients (Plante, Jumars and Baross 1990; Gaskins, Collier and Anderson 2002), and become parasitic or pathogenic (Sachs and Wilcox 2006; Young et al. 2017; Vonaesch et al. 2018). Symbionts are often more sensitive than their hosts to stressors such as heat, salinity or chemicals (Wernegreen 2012; Nougué et al. 2015; Motta, Raymann and Moran 2018), limiting their host's ability to tolerate or adapt to these factors. The genomes and functions of heritable symbionts may degenerate over time, requiring hosts to continually evolve compensatory adaptations (e.g. Campbell et al. 2018) and potentially increasing the extinction risk of host lineages (Bennett and Moran 2015).
In general, the idiosyncratic constellation of selective pressures acting on a given animal may tip the cost-benefit balance toward a strategy of avoiding, rather than investing in, a microbiome. A non-trivial, but important challenge for future research is to systematically investigate the tradeoffs experienced by organisms adopting either strategy. To that end, a more comprehensive documentation of microbiome-free animal diversity is needed.
Evidence for no- and low-microbiome animals
There is already evidence that a microbiome-free lifestyle not only exists, but is widely distributed across the animal tree of life (Table 1). Based on molecular, culturing, and/or microscopy-based methods, comparatively few or no microbes were detected in the gut or entire body of these animals; in a couple cases, experiments also support a lack of dependence on microbes. Although the prevalence of microbiome-free animals cannot currently be estimated, the extreme diversity, abundance, and ecological importance of some of these groups, such as caterpillars and ants, suggests that independence from microbes may be a highly successful strategy.
Fourteen animal groups harboring few or no resident microbes.
| Common name . | Classification . | Diet . | Tissue type . | Evidence* . | References . | |||
|---|---|---|---|---|---|---|---|---|
| . | . | . | . | 1 . | 2 . | 3 . | 4 . | . |
| Arthropoda | ||||||||
| Ants | Insecta, Hymenoptera, Formicidae (various species) | Herbivorous, carnivorous or omnivorous | Gut, whole body | ✓ | ✓ | ✓ | (Russell, Sanders and Moreau 2016; Hu et al. 2017; Sanders et al. 2017) | |
| Solitary bees | Insecta, Hymenoptera, Anthophila (various species) | Herbivorous | Gut | ✓ | ✓ | ✓ | (Dobson and Peng 1997; Martinson et al. 2011, Kwong et al. 2017) | |
| Caterpillars | Insecta, Lepidoptera (various species) | Herbivorous or carnivorous | Gut, whole body | ✓ | ✓ | ✓ | ✓ | (Staudacher et al. 2016; Whitaker et al. 2016; Hammer et al. 2017; Phalnikar et al. 2019; Vilanova et al. 2016) |
| Herbivorous beetles | Insecta, Coleoptera (various species) | Herbivorous | Gut, whole body | ✓ | ✓ | ✓ | (Taylor 1985; Peterson and Schalk 1994; Kelley and Dobler 2011) | |
| Dragonflies | Insecta, Odonata, Libellulidae (various species) | Carnivorous | Gut | ✓ | ✓ | (Deb, Nair and Agashe 2018) | ||
| Stick insects | Insecta, Phasmatodea (various species) | Herbivorous | Gut, whole body | ✓ | ✓ | (Shelomi et al. 2013, 2015) | ||
| Spider mites | Trombidiformes, Tetranychidae, Tetranychus urticae | Herbivorous | Whole body | ✓ | (Santos-Matos et al. 2017) | |||
| Gribble worms | Malacostraca, Isopoda, Limnoriidae (various species) | Xylophagous | Gut | ✓ | (Boyle and Mitchell 1978; King et al. 2010) | |||
| Opossum shrimp | Malacostraca, Mysida, Mysidae, Mysis stenolepis | Detritivorous | Gut | ✓ | (Friesen, Mann and Novitsky 1986) | |||
| Mud shrimp | Malacostraca, Decapoda, Axiidae, Calocaris macandreae | Detritivorous and necrophagous | Gut | ✓ | (Pinn et al. 1999) | |||
| Annelida | ||||||||
| Potworms | Clitellata, Haplotaxida, Enchytraeidae, Cognettia sphagnetorum | Soil | Gut | ✓ | (Latter 1977) | |||
| Ctenophora | ||||||||
| Comb jellies | Tentaculata, Lobata, Bolinopsidae, Mnemiopsis leidyi | Carnivorous | Whole body | ✓ | ✓ | (Hammann, Moss and Zimmer 2015) | ||
| Mollusca | ||||||||
| Oysters | Bivalvia, Ostreoida, Ostreidae, Magallana gigas | Planktivorous | Whole body | ✓ | ✓ | (Garland, Nash and McMeekin 1982) | ||
| Nematomorpha | ||||||||
| Horsehair worms | (various species) | Endoparasitic | Whole body | ✓ | (Duron and Gavotte 2007; Hudson and Floate 2009) | |||
| Common name . | Classification . | Diet . | Tissue type . | Evidence* . | References . | |||
|---|---|---|---|---|---|---|---|---|
| . | . | . | . | 1 . | 2 . | 3 . | 4 . | . |
| Arthropoda | ||||||||
| Ants | Insecta, Hymenoptera, Formicidae (various species) | Herbivorous, carnivorous or omnivorous | Gut, whole body | ✓ | ✓ | ✓ | (Russell, Sanders and Moreau 2016; Hu et al. 2017; Sanders et al. 2017) | |
| Solitary bees | Insecta, Hymenoptera, Anthophila (various species) | Herbivorous | Gut | ✓ | ✓ | ✓ | (Dobson and Peng 1997; Martinson et al. 2011, Kwong et al. 2017) | |
| Caterpillars | Insecta, Lepidoptera (various species) | Herbivorous or carnivorous | Gut, whole body | ✓ | ✓ | ✓ | ✓ | (Staudacher et al. 2016; Whitaker et al. 2016; Hammer et al. 2017; Phalnikar et al. 2019; Vilanova et al. 2016) |
| Herbivorous beetles | Insecta, Coleoptera (various species) | Herbivorous | Gut, whole body | ✓ | ✓ | ✓ | (Taylor 1985; Peterson and Schalk 1994; Kelley and Dobler 2011) | |
| Dragonflies | Insecta, Odonata, Libellulidae (various species) | Carnivorous | Gut | ✓ | ✓ | (Deb, Nair and Agashe 2018) | ||
| Stick insects | Insecta, Phasmatodea (various species) | Herbivorous | Gut, whole body | ✓ | ✓ | (Shelomi et al. 2013, 2015) | ||
| Spider mites | Trombidiformes, Tetranychidae, Tetranychus urticae | Herbivorous | Whole body | ✓ | (Santos-Matos et al. 2017) | |||
| Gribble worms | Malacostraca, Isopoda, Limnoriidae (various species) | Xylophagous | Gut | ✓ | (Boyle and Mitchell 1978; King et al. 2010) | |||
| Opossum shrimp | Malacostraca, Mysida, Mysidae, Mysis stenolepis | Detritivorous | Gut | ✓ | (Friesen, Mann and Novitsky 1986) | |||
| Mud shrimp | Malacostraca, Decapoda, Axiidae, Calocaris macandreae | Detritivorous and necrophagous | Gut | ✓ | (Pinn et al. 1999) | |||
| Annelida | ||||||||
| Potworms | Clitellata, Haplotaxida, Enchytraeidae, Cognettia sphagnetorum | Soil | Gut | ✓ | (Latter 1977) | |||
| Ctenophora | ||||||||
| Comb jellies | Tentaculata, Lobata, Bolinopsidae, Mnemiopsis leidyi | Carnivorous | Whole body | ✓ | ✓ | (Hammann, Moss and Zimmer 2015) | ||
| Mollusca | ||||||||
| Oysters | Bivalvia, Ostreoida, Ostreidae, Magallana gigas | Planktivorous | Whole body | ✓ | ✓ | (Garland, Nash and McMeekin 1982) | ||
| Nematomorpha | ||||||||
| Horsehair worms | (various species) | Endoparasitic | Whole body | ✓ | (Duron and Gavotte 2007; Hudson and Floate 2009) | |||
Fourteen animal groups harboring few or no resident microbes.
| Common name . | Classification . | Diet . | Tissue type . | Evidence* . | References . | |||
|---|---|---|---|---|---|---|---|---|
| . | . | . | . | 1 . | 2 . | 3 . | 4 . | . |
| Arthropoda | ||||||||
| Ants | Insecta, Hymenoptera, Formicidae (various species) | Herbivorous, carnivorous or omnivorous | Gut, whole body | ✓ | ✓ | ✓ | (Russell, Sanders and Moreau 2016; Hu et al. 2017; Sanders et al. 2017) | |
| Solitary bees | Insecta, Hymenoptera, Anthophila (various species) | Herbivorous | Gut | ✓ | ✓ | ✓ | (Dobson and Peng 1997; Martinson et al. 2011, Kwong et al. 2017) | |
| Caterpillars | Insecta, Lepidoptera (various species) | Herbivorous or carnivorous | Gut, whole body | ✓ | ✓ | ✓ | ✓ | (Staudacher et al. 2016; Whitaker et al. 2016; Hammer et al. 2017; Phalnikar et al. 2019; Vilanova et al. 2016) |
| Herbivorous beetles | Insecta, Coleoptera (various species) | Herbivorous | Gut, whole body | ✓ | ✓ | ✓ | (Taylor 1985; Peterson and Schalk 1994; Kelley and Dobler 2011) | |
| Dragonflies | Insecta, Odonata, Libellulidae (various species) | Carnivorous | Gut | ✓ | ✓ | (Deb, Nair and Agashe 2018) | ||
| Stick insects | Insecta, Phasmatodea (various species) | Herbivorous | Gut, whole body | ✓ | ✓ | (Shelomi et al. 2013, 2015) | ||
| Spider mites | Trombidiformes, Tetranychidae, Tetranychus urticae | Herbivorous | Whole body | ✓ | (Santos-Matos et al. 2017) | |||
| Gribble worms | Malacostraca, Isopoda, Limnoriidae (various species) | Xylophagous | Gut | ✓ | (Boyle and Mitchell 1978; King et al. 2010) | |||
| Opossum shrimp | Malacostraca, Mysida, Mysidae, Mysis stenolepis | Detritivorous | Gut | ✓ | (Friesen, Mann and Novitsky 1986) | |||
| Mud shrimp | Malacostraca, Decapoda, Axiidae, Calocaris macandreae | Detritivorous and necrophagous | Gut | ✓ | (Pinn et al. 1999) | |||
| Annelida | ||||||||
| Potworms | Clitellata, Haplotaxida, Enchytraeidae, Cognettia sphagnetorum | Soil | Gut | ✓ | (Latter 1977) | |||
| Ctenophora | ||||||||
| Comb jellies | Tentaculata, Lobata, Bolinopsidae, Mnemiopsis leidyi | Carnivorous | Whole body | ✓ | ✓ | (Hammann, Moss and Zimmer 2015) | ||
| Mollusca | ||||||||
| Oysters | Bivalvia, Ostreoida, Ostreidae, Magallana gigas | Planktivorous | Whole body | ✓ | ✓ | (Garland, Nash and McMeekin 1982) | ||
| Nematomorpha | ||||||||
| Horsehair worms | (various species) | Endoparasitic | Whole body | ✓ | (Duron and Gavotte 2007; Hudson and Floate 2009) | |||
| Common name . | Classification . | Diet . | Tissue type . | Evidence* . | References . | |||
|---|---|---|---|---|---|---|---|---|
| . | . | . | . | 1 . | 2 . | 3 . | 4 . | . |
| Arthropoda | ||||||||
| Ants | Insecta, Hymenoptera, Formicidae (various species) | Herbivorous, carnivorous or omnivorous | Gut, whole body | ✓ | ✓ | ✓ | (Russell, Sanders and Moreau 2016; Hu et al. 2017; Sanders et al. 2017) | |
| Solitary bees | Insecta, Hymenoptera, Anthophila (various species) | Herbivorous | Gut | ✓ | ✓ | ✓ | (Dobson and Peng 1997; Martinson et al. 2011, Kwong et al. 2017) | |
| Caterpillars | Insecta, Lepidoptera (various species) | Herbivorous or carnivorous | Gut, whole body | ✓ | ✓ | ✓ | ✓ | (Staudacher et al. 2016; Whitaker et al. 2016; Hammer et al. 2017; Phalnikar et al. 2019; Vilanova et al. 2016) |
| Herbivorous beetles | Insecta, Coleoptera (various species) | Herbivorous | Gut, whole body | ✓ | ✓ | ✓ | (Taylor 1985; Peterson and Schalk 1994; Kelley and Dobler 2011) | |
| Dragonflies | Insecta, Odonata, Libellulidae (various species) | Carnivorous | Gut | ✓ | ✓ | (Deb, Nair and Agashe 2018) | ||
| Stick insects | Insecta, Phasmatodea (various species) | Herbivorous | Gut, whole body | ✓ | ✓ | (Shelomi et al. 2013, 2015) | ||
| Spider mites | Trombidiformes, Tetranychidae, Tetranychus urticae | Herbivorous | Whole body | ✓ | (Santos-Matos et al. 2017) | |||
| Gribble worms | Malacostraca, Isopoda, Limnoriidae (various species) | Xylophagous | Gut | ✓ | (Boyle and Mitchell 1978; King et al. 2010) | |||
| Opossum shrimp | Malacostraca, Mysida, Mysidae, Mysis stenolepis | Detritivorous | Gut | ✓ | (Friesen, Mann and Novitsky 1986) | |||
| Mud shrimp | Malacostraca, Decapoda, Axiidae, Calocaris macandreae | Detritivorous and necrophagous | Gut | ✓ | (Pinn et al. 1999) | |||
| Annelida | ||||||||
| Potworms | Clitellata, Haplotaxida, Enchytraeidae, Cognettia sphagnetorum | Soil | Gut | ✓ | (Latter 1977) | |||
| Ctenophora | ||||||||
| Comb jellies | Tentaculata, Lobata, Bolinopsidae, Mnemiopsis leidyi | Carnivorous | Whole body | ✓ | ✓ | (Hammann, Moss and Zimmer 2015) | ||
| Mollusca | ||||||||
| Oysters | Bivalvia, Ostreoida, Ostreidae, Magallana gigas | Planktivorous | Whole body | ✓ | ✓ | (Garland, Nash and McMeekin 1982) | ||
| Nematomorpha | ||||||||
| Horsehair worms | (various species) | Endoparasitic | Whole body | ✓ | (Duron and Gavotte 2007; Hudson and Floate 2009) | |||
In addition to the animals listed in Table 1, an apparent lack of symbionts has been mentioned for parenchyma-feeding microleafhoppers (Buchner 1965; Bennett and Moran 2015), predatory mantids (Nalepa, Bignell and Bandi 2001), water bugs and water striders (Ohbayashi et al. 2015) and several other lineages (Buchner 1965). Reports of negligible microbial contributions to digestion in wood-eating catfish (German and Bittong 2009), some herbivorous land crabs (Linton and Greenaway 2007), and detritivorous flies (Šustr, Stingl and Brune 2014) are also noteworthy. While it is impossible to disprove the existence of undetected microbes or microbial functions in any of the animals mentioned above, we argue that a microbiome-free state is the appropriate null hypothesis, and that evidence must be acquired to reject it.
Associations among sets of interacting species often form a continuum from mutualism to antagonism (e.g. Johnson, Graham and Smith 1997; Pierce et al. 2002; Gómez, Schupp and Jordano 2018). Analogously, though we have highlighted the extreme case of species which do not appear to host, nor depend on an appreciable assemblage of resident microbes (Table 1), animals are likely to span a continuum of dependency on microbiomes. Many animals may have an intermediate lifestyle where resident microbes are only occasionally present and/or at low abundances. In such cases, microbes may be largely commensal or parasitic, and it is costlier for hosts to suppress them entirely than to tolerate some level of growth. Alternatively, resident microbes may be only modestly beneficial to their hosts, or only beneficial in specific contexts.
Certain birds, bats and pandas appear to match this ‘low-microbiome’ scenario. Some species have comparatively few gut microbes (Hammer et al. 2017; Contijoch et al. 2019), and unlike humans and other strongly microbe-dependent mammals (Ley et al. 2008), many bird and bat gut microbiomes are dominated by classically ‘weedy’ Bacilli (particularly Staphylococcus and Streptococcus) and Proteobacteria (Phillips et al. 2012; Waite, Deines and Taylor 2012; Carrillo-Araujo et al. 2015; Hird et al. 2015; Ingala et al. 2018). Gut microbiomes of terrestrial mammalian carnivores, as well as herbivorous pandas, also frequently have a high proportion of Streptococcus and Proteobacteria (Ley et al. 2008; Xue et al. 2015). As discussed further below, some species within these groups have relatively simple guts with short retention times, traits that also suggest a lower reliance on microbes for digestion and nutrition.
Why microbiome-free animals have gone largely unnoticed
Why has the paradigm of universal animal microbiomes been so broadly accepted (Table S1, Supporting Information)? One answer is that the tools widely used to characterize the identities, dynamics and functional potential of microbial communities have become cheaper and easier to use than experimental techniques for testing microbial impacts on host physiology and fitness. Sequencing marker genes (such as the 16S rRNA gene) of previously uncharacterized microbiomes is relatively fast and inexpensive, and does not require laboratory rearing or specialized knowledge of the animal at hand. These surveys are therefore an attractive first step to take before embarking on slower and more expensive experiments. But sequencing-based methods—including shotgun metagenomics—come with important caveats, as we discuss below. Viewed through DNA sequencing glasses, a motley assortment of contaminants, transients and parasites can look a lot like the beneficial microbiome that we have increasingly come to expect.
Contaminants
Sequence data from samples with low microbial biomass are especially likely to be overwhelmed by contaminants introduced during sample handling and processing (Salter et al. 2014; de Goffau et al. 2018; Eisenhofer et al. 2019). It is difficult to know the true extent of contamination among published animal microbiome studies as a large number do not report information on negative controls. Furthermore, well-known contaminant taxa (Salter et al. 2014; Eisenhofer et al. 2019) are often related to widespread plant, water or soil-associated taxa that, in theory, could be animal symbionts. However, it is notable that characteristic human skin bacteria such as Corynebacterium and Propionibacterium (Byrd, Belkaid and Segre 2018) have been reported from a range of animals, such as caterpillars (Pinto-Tomás et al. 2011), a parasitoid wasp (Brucker and Bordenstein 2013), the nematode Caenorhabditis elegans (Berg et al. 2016), sandpipers (Risely et al. 2018) and orca skin (Hooper et al. 2019). As these samples were all processed by humans, a parsimonious explanation for the presence of such taxa is contamination.
Ignoring the potential for contamination to mask low-density or sterile environments can lead to spurious conclusions (de Goffau et al. 2018; Eisenhofer et al. 2019). For example, microbial DNA was sequenced from human placenta samples, supporting the idea that maternal bacterial symbionts colonize fetuses in utero (Funkhouser and Bordenstein 2013). The ‘placenta microbiome’ is now well-understood to be an artifact of contamination (Lauder et al. 2016; Perez-Muñoz et al. 2017; Leiby et al. 2018). Similarly, we predict that some studies purporting to characterize animal microbiomes will also turn out to have been inadvertent surveys of human, lab and reagent contaminants. Going forward, including appropriate negative controls and critically evaluating sequence data for the presence of contaminants are crucial steps toward recognizing no- or low-microbiome animals.
Transients
A trickier, but also important challenge for microbiome studies is to distinguish transient from resident microbes, as animals will often contain a variety of transient microbes from food or other environmental sources. For example, gut microbiomes of wild caterpillars are dominated by transient, diet-associated bacteria (Whitaker et al. 2016; Hammer et al. 2017). Diet-derived bacteria and fungi are also detectable—though proportionally much rarer—in the human gut (David et al. 2014). Even if these transient cells are dead, they could be interpreted as symbionts in microbiome datasets if they are sufficiently common and abundant. Most sequencing-based surveys do not discriminate between metabolically active, viable but inactive and dead cells (Emerson et al. 2017). Distinguishing among these categories is crucial, as a microbiome dominated by transient taxa may be functionally irrelevant as a whole. Aquatic filter-feeders and animals feeding on decaying organic material, soil or dung might rely on ingested and digested microbes as food, though these microbes may be more akin to prey than symbionts.
When unrecognized, transient microbes can generate specious correlations between microbiomes and host traits or environmental factors. Much of the recent microbiome literature has focused on how microbiomes vary across host development, diets, environments, taxonomy or phylogeny. Such findings are likely to be biologically relevant when the microbiome is indeed important to host health. Yet variation in microbial abundances or composition across such factors can be byproducts of pre-existing variation in microbial loads or profiles in environmental inputs. For example, two caterpillars may appear to have different microbiomes simply because they consume leaves with different types and amounts of microbes (Hammer et al. 2017)—even if these microbes only pass through the gut. Analogously but at a broader scale, patterns such as ‘phylosymbiosis’ can occur when phylogenetically conserved host traits differentially filter environmental microbes (Moran and Sloan 2015; Mazel et al. 2018); if these microbes are only transient, such patterns may not signify a meaningful host-microbiome association.
The presence of transient microbes also means that inferences of microbial functional potential—whether assessed via marker gene-based predictions (Langille et al. 2013), metagenomic sequencing, or in vitro tests of microbes isolated from an animal—are not necessarily realized in vivo. For example, the detection of cellulase genes and cellulolytic bacterial isolates from panda guts may suggest that pandas rely on gut microbes to digest cellulose (Zhu et al. 2011; Fan et al. 2012). However, these genes and bacterial taxa are of low abundance in the gut (Xue et al. 2015; Guo et al. 2018; Contijoch et al. 2019), and physiological studies have shown that little fiber degradation actually occurs during digestion (Wei et al. 1999; Senshu et al. 2014; Dierenfeld et al. 2018). Instead, pandas primarily rely on plant cell contents for nutrition (Dierenfeld et al. 2018), which they may access with endogenous enzymes. In this case, the occurrence of cellulase genes and cellulolytic activity likely tells us more about what these microbes do in bamboo or panda feces than in the gut itself.
Parasites and pathogens
Even apparently healthy animals are often colonized by microbial parasites or latent pathogens, and with observational data alone, they may be easily confused with beneficial symbionts. Neither prevalence nor abundance is a guarantee that a microbe is beneficial. Parasites may be highly prevalent in a population; for example, many healthy adult humans host Demodex mites on their skin (Thoemmes et al. 2014) and the protist Blastocystis in their gut (Lepczyńska et al. 2017). In the absence of beneficial symbionts, parasites and pathogens are likely to make up a high proportion of sequence libraries (Fig. 1). Stress, altered diets and other effects of animal sampling or husbandry may also cause normally low-level or transient microbes to become abundant residents. Laboratory rearing, a common precursor to microbiome sampling, can increase the absolute abundance of parasitic or commensal microbes (Lighthart 1988; Turelli 1994) as well as change microbial community structure (e.g. Chandler et al. 2011; Hammer, McMillan and Fierer 2014; Staudacher et al. 2016). In the laboratory, normally pathogenic microbes can also evolve into beneficial symbionts (Tso et al. 2018). As a result, captive animals may host microbiomes that are unrepresentative not just in their composition, but also in their effects on host fitness.
How to recognize microbiome-free animals
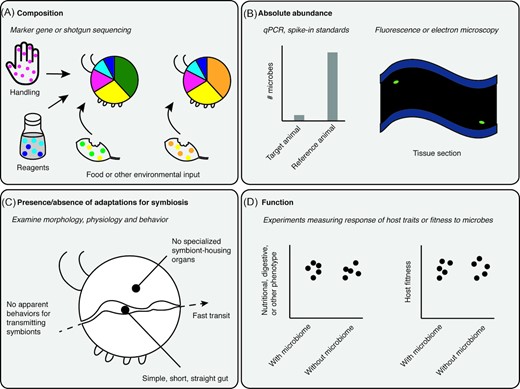
The near-ubiquity of laboratory contaminants, transients, parasitic and pathogenic microbes, in tandem with biases in research efforts and publications, has likely concealed cases of animals lacking resident and beneficial microbiomes. How can we better recognize them? Awareness that such animals are possible could lead researchers to re-evaluate their data or motivate exploration of as-yet unstudied animal lineages. A useful starting point is to incorporate information on animal nutrition, physiology, morphology and natural history to predict whether an animal is likely to host, and need, microbial symbionts. Perhaps most importantly, it will be necessary to both interpret sequencing-based data more critically, and to broaden the approaches we use to study microbiomes (Fig. 2).
Signs that an animal may lack a resident and beneficial microbiome, as shown with different types of data, listed in bold text (A–D). Some commonly used methods are listed in italicized text. (A), High inter-individual variability, proportions of environmental or diet-derived taxa, and/or contamination. (B), Low absolute abundance (numbers of microbial cells or genomes relative to a reference animal known to host dense, functional symbionts), visualized by microscopy showing host tissues lacking microbial colonization. (C), No obvious adaptations for acquiring, transmitting, or hosting symbionts. (D), Suppressing or eliminating putative microbial symbionts does not alter host traits nor decrease host fitness.
Reporting microbial population sizes should be standard practice in microbiome studies, as it is in surveys of plant and animal communities. Information on the absolute abundance of microbes can help differentiate contaminants and transients from residents, and reveal ecological and evolutionary patterns that would be difficult to discern with compositional data alone (Fig. 3). Quantitative PCR is a useful method that is becoming more widely adopted (e.g. Hammer et al. 2017; Kwong et al. 2017; Sanders et al. 2017); flow cytometry (Props et al. 2016; Vandeputte et al. 2017) and the inclusion of spike-in DNA standards (Smets et al. 2016; Tourlousse et al. 2017) are other options. However, it should be kept in mind that a high-density microbiome could simply reflect consumption of microbially rich food (Hammer et al. 2017). Thus, counting and characterizing microbial communities in an animal's diet or other environmental inputs can lend further insight into whether an animal has a resident microbiome. Finally, microscopy is a complementary approach for assessing microbial colonization (e.g. Boyle and Mitchell 1978; Martinson, Moy and Moran 2012; Sanders et al. 2017) that avoids some of the pitfalls inherent in DNA-based methods.
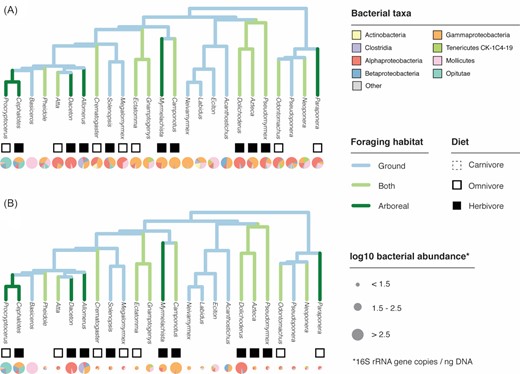
Incorporating absolute abundances aids biological interpretation of microbiome data. In the case of a survey of gut microbiomes from rainforest ants (Sanders et al. 2017), simply plotting bacterial taxonomic composition on the ant phylogeny (A) shows little if any pattern across ant lineages. However, incorporating absolute microbial abundance information (B) reveals a link between bacterial abundance (indicated by pie chart size) and arboreal-foraging, herbivorous ants. Both panels: bacterial composition indicated by pie charts in lower row; inferred diet indicated by squares in upper row (filled = herbivorous; open = omnivorous; none = carnivorous); foraging habitat indicated by branch color on tree. For details on ant traits, ancestral state reconstruction and phylogenetic relationships, see (Blanchard and Moreau 2017). Normalized bacterial abundances are log10-transformed and binned into three levels, and ant diet categories are inferred from ∂15 N measurements from (Sanders et al. 2017).
To distinguish harmful from beneficial residents and to test for specific microbial services, we also recommend complementing observational studies with experiments when possible. A useful experimental design is to manipulate the presence or activity of a microbiome and examine whether, and if so how and to what degree, host fitness is affected. For example, rearing gnotobiotic animals or administering antibiotics have been used to test microbiome function in a wide range of animals (e.g. Ridley et al. 2012; Hammer et al. 2017; Zheng et al. 2017; Hu et al. 2018; Kohl et al. 2018). However, interpreting results from these experiments is not always straightforward. Germ-free or antibiotic-treated individuals may show elevated growth even in species normally reliant on microbes (Wostmann 1981; Gaskins, Collier and Anderson 2002). Furthermore, potential symbiont functions can be overlooked in experiments lacking field-realistic conditions such as environmental stressors, food availability and quality, or the presence of natural enemies.
Lastly, we can hearken back to the physiological, morphological and natural history-driven approaches that were commonly used in the pre-sequencing era. For example, it is worth asking whether an animal appears to invest in acquiring and maintaining symbionts—such as through exudates, excretions and secretions, conspicuous transmission behaviors or specialized organs such as bacteriomes, trophosomes or expanded gut compartments (e.g. Hosokawa et al. 2008; McFall-Ngai 2008; Brune and Dietrich 2015; Chen et al. 2018; Li et al. 2018). Existing literature on relationships between microbes and gut structure (Langer and Snipes 1991; Stevens and Hume 1995; Hackstein and van Alen 1996) could also help us predict the degree to which an animal depends on gut microbes. For example, a relatively low reliance on microbially mediated digestion, detoxification and nutrient synthesis could be expected in short and simple guts with rapid transit times—as are common among nectivorous and frugivorous birds (Karasov and Levey 1990; del Rio and Karasov 2002), bats (Tedman and Hall 1985; Roswag, Becker and Encarnação 2012), pandas (Wei et al. 1999), small insectivorous mammals such as moles and shrews (Stevens and Hume 1995; Langer 2002), and some insects such as caterpillars (Engel and Moran 2013). The possibility that a given animal does not directly rely on microbial symbionts can be further evaluated by measuring characteristic microbial byproducts (e.g. methane or short-chain fatty acids), enzyme activities or levels of a substrate of interest (e.g. fiber or dietary toxins) in vivo.
OUTLOOK
Much as microbes themselves span a continuum in their reliance on hosts (Moran 2001; Sachs, Skophammer and Regus 2011), animals vary in the degree to which they rely on microbes—from those that cannot survive without symbionts to those that are completely independent. Such variation has been obscured by the increasingly popular assumption that the biology of all animals is mediated by microbiomes (Table S1, Supporting Information). Awareness that not all animal lineages host resident and beneficial microbiomes brings into focus fundamental questions about the macroevolution of microbiomes. For example, what factors are associated with the gain and loss of microbial dependence? How do evolutionary transitions in symbiotic states occur? Do these transitions influence animal diversification? Answering such questions will require a more complete picture of microbiome dependence across the animal tree of life than we can currently assemble. Future explorations will benefit not only from using a broader set of methods, but also from keeping an open mind to the diverse ways that animals do, or do not, interact with microbes.
ACKNOWLEDGEMENTS
We thank the reviewers for comments that improved the manuscript. TJH was supported by a Graduate Student Research Award from the Cooperative Institute for Research in Environmental Sciences. JGS was supported by a postdoctoral fellowship from the Cornell Institute for Host–Microbe Interaction and Disease. NF was funded by the U.S. National Science Foundation (DEB 1542653). We acknowledge Alex Wild for the photograph upon which the Crematogaster silhouette is based. Cow and caterpillar silhouettes are from phylopic.org. The authors have no conflict of interest to declare.
Conflicts of interest. None declared.