-
PDF
- Split View
-
Views
-
Cite
Cite
Belén Adiego-Pérez, Paola Randazzo, Jean Marc Daran, René Verwaal, Johannes A Roubos, Pascale Daran-Lapujade, John van der Oost, Multiplex genome editing of microorganisms using CRISPR-Cas, FEMS Microbiology Letters, Volume 366, Issue 8, April 2019, fnz086, https://doi.org/10.1093/femsle/fnz086
Close - Share Icon Share
ABSTRACT
Microbial production of chemical compounds often requires highly engineered microbial cell factories. During the last years, CRISPR-Cas nucleases have been repurposed as powerful tools for genome editing. Here, we briefly review the most frequently used CRISPR-Cas tools and describe some of their applications. We describe the progress made with respect to CRISPR-based multiplex genome editing of industrial bacteria and eukaryotic microorganisms. We also review the state of the art in terms of gene expression regulation using CRISPRi and CRISPRa. Finally, we summarize the pillars for efficient multiplexed genome editing and present our view on future developments and applications of CRISPR-Cas tools for multiplex genome editing.
INTRODUCTION
Industrial microbiology plays a key role in the transition towards a more sustainable industry to produce food and feed ingredients, bio-based materials, biofuels and direct synthesis of cosmetic and pharmaceutical compounds (Lee et al.2019). Oil-based production processes are gradually being substituted by bio-based processes, in which genetically engineered microorganisms are generally crucial to achieve cost-effective productivities and yields (Hong and Nielsen 2012; Dai and Nielsen 2015). The implementation of CRISPR-Cas tools has revolutionized genome editing and mitigated the investment in the metabolic engineering programs required to generate highly engineered microbial cell factories (Donohoue, Barrangou and May 2018; Choi et al. 2019).
CRISPR-Cas systems (Clustered Regularly Interspaced Short Palindromic Repeats and CRISPR-associated proteins) are bacterial and archaeal adaptive immune defence systems, which can be repurposed as versatile genetic editing or regulation tools in a broad range of organisms. The effector endonucleases of these systems are guided by short RNA molecules encoded by CRISPR arrays. Native CRISPR arrays consist of a succession of spacers originating from invader organisms separated by direct repeats (Mojica et al. 2005; Barrangou et al. 2007). Transcription of the CRISPR array results in a long precursor-crRNA transcript (pre-crRNA), that is subsequently being processed to short functional CRISPR RNA (crRNA) guides (Brouns et al. 2008). To date, six different types of CRISPR-Cas systems (I–VI) have been described that are divided into two major classes (Class 1 and Class 2) (Makarova and Koonin 2015; Makarova et al. 2015; Shmakov et al. 2015, 2017; Koonin, Makarova and Zhang 2017). This review focuses on the application of DNA-targeting class 2 CRISPR systems (included in types II and V), that all consist of large multi-domain effector proteins able to use crRNA guides to target complementary DNA. Recent reviews have covered applications of CRISPR-Cas editing in bacteria (Choi and Lee 2016), in Streptomyces (Alberti and Corre 2019), in filamentous fungi (Shi et al. 2017), in yeast (Stovicek, Holkenbrink and Borodina 2017; Raschmanová et al. 2018), in microalgae and cyanobacteria (Naduthodi, Barbosa and Van der Oost 2018), and in general industrial microorganisms (Ferreira, David and Nielsen 2018).
After an introduction of single target genome editing tools, we focus on the spectacular development of multiplexed genome editing by Cas9 (type II) and Cas12a (type V) in industrial microorganisms. Both bacterial and eukaryotic examples are described, although more attention is given to yeast and filamentous fungi, since the diversity of strategies using Cas endonucleases for genome editing applications is more extensive in this group of organisms.
Single target genome editing and regulation
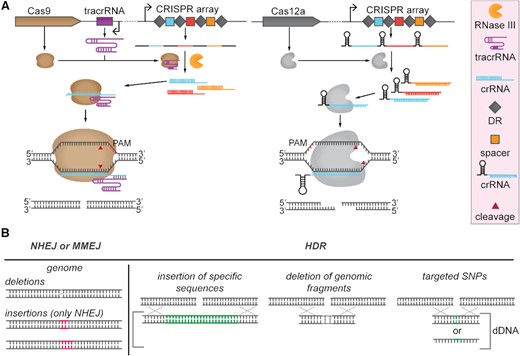
Since its establishment as a genome editing tool (Gasiunas et al. 2012; Jinek et al. 2012), Cas9 from Streptococcus pyogenes (SpCas9) has become the most widely used RNA-guided endonuclease for genome editing and transcription regulation purposes (Table 1). Expression of this type II Cas nuclease together with a guide RNA (gRNA) is sufficient for generating targeted blunt double-stranded breaks (DSBs). The gRNA bound by SpCas9 consists of two small RNA molecules: a CRISPR RNA (crRNA) and a trans-activating CRISPR RNA (tracrRNA). To simplify gRNA expression, a synthetic chimeric construct named single guide RNA (sgRNA) can be synthesized by fusing the tracrRNA and the crRNA (Jinek et al. 2012). Targeting of complementary DNA sequences (protospacers) by the Cas9:gRNA complex requires a protospacer adjacent motif (PAM), in case of Cas9 positioned downstream of the target sequence (Deveau et al. 2008). Correct PAM identification and base-pairing will trigger cleavage of the non-target and target DNA strands by the RuvC and HNH nuclease domains, respectively (Gasiunas et al. 2012; Jinek et al. 2012) (Fig. 1).
(A) Cas9 and Cas12a expression and cleavage schemes. Left panel: Cas9 requires tracrRNA transcription and RNase III expression for CRISPR array transcript processing. Cas9 forms a complex with crRNA and tracrRNA and cleaves target DNA generating blunt ends. Right panel: Cas12a processes its own CRISPR array transcript to obtain individual crRNAs without the requirement of any tracrRNA or RNAse III co-expression. Cas12a stays in complex with crRNA and cleaves target DNA generating staggered ends. (B) Double strand break (DSB) repair mechanisms. DSBs can be repaired via non-homologous end joining (NHEJ), alternative non-homologous end joining repair pathways such as microhomology-mediated end joining (MMEJ), or via homologous direct recombination. NHEJ and MMEJ repair pathways can lead to the incorporation of deletions or insertions (only in case of NHEJ) in the targeted region. HDR is combined with the supplementation of donor DNA (dDNA), which can be double stranded or single stranded. dDNA can be used for insertion of long DNA sequences, deletion of genomic fragments, or introduction of single point mutations (SNPs).
Characteristics of the most commonly used Cas orthologues for genome editing.
| . | Cas9 . | Cas12a . | ||
|---|---|---|---|---|
| Ortholog . | SpCas9 . | FnCas12a . | AsCas12a . | LbCas12a . |
| Subtype | II-A | V-A | ||
| Organism of origin | Streptococcus pyogenes | Francisella novicida | Acidaminococcus sp. | Lachnospiraceae bacterium |
| Nuclease domain | HNH, RuvC | RuvC | ||
| tracrRNA | Yes | No | ||
| PAM (5′–3′) | NGG | TTTV | TTTV | TTTV |
| Size (amino acids) | 1368 | 1302 | 1307 | 1228 |
| RNA processing | No/RNaseIII | Yes/WED III | Yes/WED III | Yes/WED III |
| Minimum guide length (mature) | ∼100 nt | ∼44 nt | ||
| Reference(s) | (Deltcheva et al. 2011) | (Zetsche, Heidenreich and Mohanraju et al. 2017) | ||
| . | Cas9 . | Cas12a . | ||
|---|---|---|---|---|
| Ortholog . | SpCas9 . | FnCas12a . | AsCas12a . | LbCas12a . |
| Subtype | II-A | V-A | ||
| Organism of origin | Streptococcus pyogenes | Francisella novicida | Acidaminococcus sp. | Lachnospiraceae bacterium |
| Nuclease domain | HNH, RuvC | RuvC | ||
| tracrRNA | Yes | No | ||
| PAM (5′–3′) | NGG | TTTV | TTTV | TTTV |
| Size (amino acids) | 1368 | 1302 | 1307 | 1228 |
| RNA processing | No/RNaseIII | Yes/WED III | Yes/WED III | Yes/WED III |
| Minimum guide length (mature) | ∼100 nt | ∼44 nt | ||
| Reference(s) | (Deltcheva et al. 2011) | (Zetsche, Heidenreich and Mohanraju et al. 2017) | ||
Characteristics of the most commonly used Cas orthologues for genome editing.
| . | Cas9 . | Cas12a . | ||
|---|---|---|---|---|
| Ortholog . | SpCas9 . | FnCas12a . | AsCas12a . | LbCas12a . |
| Subtype | II-A | V-A | ||
| Organism of origin | Streptococcus pyogenes | Francisella novicida | Acidaminococcus sp. | Lachnospiraceae bacterium |
| Nuclease domain | HNH, RuvC | RuvC | ||
| tracrRNA | Yes | No | ||
| PAM (5′–3′) | NGG | TTTV | TTTV | TTTV |
| Size (amino acids) | 1368 | 1302 | 1307 | 1228 |
| RNA processing | No/RNaseIII | Yes/WED III | Yes/WED III | Yes/WED III |
| Minimum guide length (mature) | ∼100 nt | ∼44 nt | ||
| Reference(s) | (Deltcheva et al. 2011) | (Zetsche, Heidenreich and Mohanraju et al. 2017) | ||
| . | Cas9 . | Cas12a . | ||
|---|---|---|---|---|
| Ortholog . | SpCas9 . | FnCas12a . | AsCas12a . | LbCas12a . |
| Subtype | II-A | V-A | ||
| Organism of origin | Streptococcus pyogenes | Francisella novicida | Acidaminococcus sp. | Lachnospiraceae bacterium |
| Nuclease domain | HNH, RuvC | RuvC | ||
| tracrRNA | Yes | No | ||
| PAM (5′–3′) | NGG | TTTV | TTTV | TTTV |
| Size (amino acids) | 1368 | 1302 | 1307 | 1228 |
| RNA processing | No/RNaseIII | Yes/WED III | Yes/WED III | Yes/WED III |
| Minimum guide length (mature) | ∼100 nt | ∼44 nt | ||
| Reference(s) | (Deltcheva et al. 2011) | (Zetsche, Heidenreich and Mohanraju et al. 2017) | ||
The more recently characterized endonuclease Cas12a (formerly called Cpf1) (type V) can cleave dsDNA directed by a crRNA, hence without the requirement of a tracrRNA (Zetsche et al. 2015) (Table 1). Cas12a does not possess an HNH domain, and its RuvC domain has been demonstrated to cleave both the non-target and the target DNA strands (Swarts, Van der Oost and Jinek 2017; Swarts and Jinek 2018). Moreover, Cas12a is able to process its crRNA guide autonomously (Fonfara et al. 2016; Swarts, van der Oost and Jinek 2017; Zetsche et al. 2017), while Cas9 relies on the activity of an additional non-Cas, dsRNA (crRNA/tracrRNA) targeting ribonuclease (RNaseIII) (Deltcheva et al. 2011). Both Cas9 and Cas12a can use multiple crRNA guides for creating simultaneous DSBs at different target loci in the genome (Fig. 1). Recently, two distinct Cas12 subtypes (Cas12b, CasX/Cas12e) were shown to also edit genomes of bacteria and mammalian cells (Liu et al. 2019; Strecker et al. 2019).
Genomic DSBs can be repaired by homology-directed repair (HDR), non-homologous end joining repair (NHEJ) or alternative non-homologous end joining systems such as microhomology-mediated end joining (MMEJ) (Chayot et al. 2010; Sfeir and Symington 2015; Yao et al. 2017). The error-prone NHEJ repair system is often most prevalent in eukaryotes (Pawelczak et al. 2018) (Fig. 1), whereas it has been predicted to be encoded by only ∼26% of publicly available prokaryotic genomes (Bowater and Doherty 2006; McGovern et al. 2016; Nayak and Metcalf 2017). The less studied alternative MMEJ repair system has been reported to also be present in bacteria and fungi (Sfeir and Symington 2015). This repair system has been proven to be active together with other repair mechanisms in the fungi Aspergillus niger and Yarrowia lipolytica (Shi et al. 2018) or in NHEJ-free bacteria (Chayot et al. 2010). HDR can be used in a targeted way: (i) to insert DNA fragments in targeted genomic locations; (ii) to delete small and large DNA fragments; or (iii) to introduce point mutations. HDR requires the introduction of a single or double-stranded DNA repair fragment into the cell, called donor DNA (dDNA), encoding the desired novel property or designed nucleotide change. To avoid targeting after the designed change, the recombinant sequence generally contains one or more silent mutations in the protospacer or PAM recognition sequence or partial deletion thereof. On the other hand, in organisms with highly active NHEJ or MMEJ repair systems, the introduction of non-specific insertions (only in case of NHEJ) and/or deletions (indels) in a certain target sequences can lead to gene disruption (Cong et al. 2013).
Inactive or deactivated versions of both Cas9 and Cas12a (named dCas9 and dCas12a) have been designed by substituting one or more of the catalytic amino acids in the nuclease domains (Gasiunas et al. 2012; Jinek et al. 2012; Zetsche et al. 2015; Swarts and Jinek 2018). These variants have been used to regulate gene expression in many organisms since they retain the target-binding ability (Berlec et al. 2018). By directing dCas9 or dCas12a to the promoter or coding sequence of a target gene, transcriptional repression (silencing) can be achieved by steric hindrance of the RNA polymerase and/or of transcription factors required for transcription of the target gene. This CRISPR interference (CRISPRi) technique has initially been established in Escherichia coli, resulting in significant transcriptional repression when targeting either the promoter or the non-template DNA strand of an open reading frame (Bikard et al. 2013; Qi et al. 2013). In eukaryotic microorganisms, gene repression is normally achieved by fusing repressor domains such as the mammalian transcriptional repressor domain Mxi1 or the Krüppel-associated box (KRAB domain) to the C-terminus of dCas9 or dCas12a (Jensen et al. 2017; Jensen 2018). Recently, native repression domains have been characterized in the yeast Saccharomyces cerevisiae with multiple Cas9 orthologs (Lian et al. 2017). This practice is more common in eukaryotic organisms since the use of only a dCas9:gRNA-complex seems not to be sufficient to significantly block transcription (Gilbert et al. 2013). Moreover, fusions of these deactivated variants to transcription activation domains are used to achieve gene activation (CRISPRa). In eukaryotes, VP16, VP64, Gal4AD or the synthetic VPR activator domains have been used successfully (Chavez et al. 2015; Jensen et al. 2017; Schwartz et al. 2018), while the omega (ω) subunit of the RNA polymerase has been used in bacteria (Bikard et al. 2013; La Russa and Qi 2015).
Multiplex genome editing
Editing of multiple loci is often required to introduce multiple heterologous genes and to fine-tune metabolic networks of microbial cell factories. In the pre-CRISPR era, iterative rounds of genome editing making use of selection markers were necessary to build strains expressing multiple-gene expression pathways. Establishment of marker-free CRISPR-Cas tools brought powerful nuclease-mediated multiplex genome engineering capabilities, considerably saving time and resources in strain construction programs. The multiplexing capabilities of CRISPR-Cas systems as genome editing tools have been widely exploited with Cas9 and more recently with Cas12a. One of the first microbial applications demonstrating the multiplexing capabilities of Cas9 was performed with the native Cas9 system of Streptococcus pneumoniae and two spacers expressed from a synthetic array integrated into the genome (Jiang et al. 2013). After this proof of principle, multiple studies explored the multiplexing capabilities of the endonuclease Cas9 in mammalian cells (Cong et al. 2013; Mali et al. 2013), as well as in industrially relevant prokaryotic and eukaryotic microorganisms (Table 2).
Multiplexed genome editing events in industrial microorganisms using CRISPR-Cas systems.
| Specie [strain(poidy)] . | Cas nuclease tool (expression), Plasmid (replication origin)/genome integrated . | Strategy for multiplexed gRNA expression/delivery (expression), plasmid (replication origin)/genome integrated . | Type of donor DNA: HFs;amount/concentration . | Type of modification: Number of target, editing efficiency . | Reference . |
|---|---|---|---|---|---|
| PROKARYOTES | |||||
| Escherichia coli[MG1655] | SpCas9 (inducible),Plasmid expression (repA101 ori) | Several sgRNA expression cassettes (constitutive),Plasmid expression | Circular dsDNA (pMB1 ori):∼300 bp | Knockouts: 2, 100%; 3, 88.3%; 4, >30% | (Feng et al. 2018) |
| Escherichia coli[MG1655] | SpCas9 (constitutive),Plasmid expression (repA101 (Ts) ori) | Several sgRNA expression cassettes (constitutive),Plasmid expression (pMB1 ori) | Circular dsDNA (pMB1 ori): 250–550 bp | Knockouts: 2, 97% ± 4%; 3, 47% ± 8% | (Jiang et al. 2015) |
| Escherichia coli[MG1655] | SpCas9 (inducible),Plasmid expression (ColE1 ori) | Several sgRNA expression cassettes (inducible),Plasmid expression (pMB1 ori) | Linear ssDNA: 70 bp; 5 pmol | Short insertions: 2, ∼70% | (Ronda et al. 2016) |
| Escherichia coli[MG1655] | SpCas9 (constitutive),Plasmid expression (p15A ori) | Several sgRNA expression cassettes (constitutive),Plasmid expression (ColE1 ori) | Linear ssDNA: ∼89 bp; 50 pmol | Point mutations: 2, 83%; 3, 23% | (Li et al. 2015) |
| Streptococcus pneumoniae[crR6c] | SpCas9 (constitutive),Genome integrated | Native-like CRISPR array (constitutive),Genome integrated | Linear dsDNA: not mentioned ;0.7 ng/µl to 2.5 µg/µl | Deletions: 2, 75% | (Jiang et al. 2013) |
| Streptomyces lividans | SpCas9 (constitutive),Plasmid expression (pSG5rep) | Several sgRNA expression cassettes (constitutive),Plasmid expression (pSG5rep) | Circular dsDNA (oriT): 1 kB | Short deletions (20–34 bp): 2, 100% (4/4) | (Cobb, Wang and Zhao 2015) |
| Streptomyces coelicolor[M145] | SpCas9 (constitutive),Plasmid expression (pSG5rep) | Several sgRNA expression cassettes (constitutive),Plasmid expression (pSG5rep) | Circular dsDNA (pSG5): ∼1 kB | Deletions (768–1053 bp): 2, 29–54% | (Huang et al. 2015) |
| Escherichia coli[MG1655] | FnCas12a (inducible),Plasmid expression (repA 101) | Native-like CRISPR array (constitutive),Plasmid expression (pSC101 ori) | Circular dsDNA (oriE): 500 bp | Gene insertions: 3, ∼20% | (Ao et al. 2018) |
| Streptomyces coelicolor[M145] | FnCas12a (constitutive),Plasmid expression (pSG5rep) | Native-like CRISPR array (constitutive),Plasmid expression (pSG5) | Circular dsDNA (pSG5): ∼1 kB | Knockouts: 2, 75% | (Li et al. 2018) |
| EUKARYOTES | |||||
| Saccharomyces cerevisiae[CEN.PK113–7D (n), CEN.PK2–1c (n), CEN.PK122 (2n)] | SpCas9 (constitutive),Genome integrated | Several sgRNA expression cassettes (RNA pol III promoter, constitutive),Plasmid expression (multicopy) | Linear dsDNA: 60 bp; 12 pmols | Knockouts: 2, 100%; 6, 65% | (Mans et al. 2015) |
| Saccharomyces cerevisiae[CEN.PK2–1c (n)] | SpCas9 (constitutive),Genome integrated | Several sgRNA expression cassettes with homology flanks to a linearized plasmid backbonePlasmid expression (multicopy) | Linear dsDNA: 500 bp; 0.6–1.54 pmols | Knockouts: 3, 64% | (Horwitz et al. 2015) |
| Saccharomyces cerevisiae[CEN.PK113–5D (n)] | FnCpf1 (constitutive),Genome integrated | Native-like CRISPR array (RNA pol III promoter, constitutive)Plasmid expression (multicopy) | Linear dsDNA: 60 bp; 12 pmols | Knockouts: 2, 100%; 4, 85% | (Swiat et al. 2017) |
| Saccharomyces cerevisiae[CEN.PK113–7D (n), Ethanol Red (2n)] | SpCas9 (constitutive),Plasmid expression (multicopy) | Several sgRNA expression cassettes (RNA pol III promoter, constitutive),Plasmid expression (multicopy) | Linear ssDNA: 40 bp; 300pmols | Knockouts: 2, 91–98% | (Generoso et al. 2016) |
| Saccharomyces cerevisiae[BY4741 (n)] | FnCpf1 (constitutive),Plasmid expression (centromeric) | Native-like CRISPR array (RNA pol III promoter, constitutive),Plasmid expression (multicopy) | Linear dsDNA: 50 bp | Multi-gene integrations: 2, 52%; 3, 43% | (Li, Wang and Wei 2018) |
| Saccharomyces cerevisiae[204 508; ATCC (mated) (2n)] | SpCas9 (constitutive), Plasmid expression (multicopy) | Synthetic array of ribozyme-flanked sgRNA (RNA pol III promoter, constitutive),Plasmid expression (multicopy) | Linear dsDNA: 50 bp; 44.94 pmol | Deletions: 2, 43%; 3, 19% | (Ryan et al. 2014) |
| Saccharomyces cerevisiae[CEN.PK113–7D (n)] | LbCpf1, AsCpf1 or FnCpf1 (constitutive), Plasmid expression (centromeric) | Native-like crRNA-array with homology flanks to a linearized plasmid backbone (RNA pol III promoter, constitutive),Plasmid expression (multicopy) | Linear dsDNA: 50 bp; 90–120 fmols | Multi-gene integrations: 3, 91% | (Verwaal et al. 2018) |
| Saccharomyces cerevisiae[BY4741 (n), CEN.PK2–1c (n)] | iSpCas9 (constitutive),Plasmid expression (multicopy) | Native-like crRNA-array (RNA pol III promoter, constitutive), separate expression of tracrRNA,Plasmid expression (multicopy) | Circular dsDNA, at 5’ of each spacer sequence (multicopy): 50 bp; 142 fmols | Knockouts: 3, 27–87% | (Bao et al. 2015) |
| Saccharomyces cerevisiae[CEN.PK113–7D (n)] | SpCas9 (constitutive),Genome integrated | Synthetic crRNA-array (RNA pol III promoter), separate expression of PaCsy4, Plasmid expression (multicopy) | Linear dsDNA: 60 bp; 12 pmols | Knockouts: 2, 100%; 4, 96% | (Ferreira, et al. 2018) |
| Saccharomyces cerevisiae[BY4741 (n)] | SpCas9 (constitutive),Plasmid expression (centromeric) | Several HDV ribozyme-sgRNA expression cassettes (RNA pol III promoter, constitutive),Plasmid expression (centromeric) | Linear dsDNA, barcoded: 60 bp; 55 pmols | Knockouts: 2, 65–87.5%; 3, 57.5–75%; 4, 27.5–15% | (Lee et al. 2015) |
| Saccharomyces cerevisiae[CEN.PK2–1C (n)] | SpCas9 (constitutive),Plasmid expression (centromeric) | Several sgRNA expression cassettes (RNA pol III promoter, constitutive),Plasmid expression (multicopy) | Linear dsDNA: 500 bp; 700 fmols | Multi-gene integrations: 3, 84% | (Ronda et al. 2015) |
| Saccharomyces cerevisiae[CEN.PK111–27B (n)] | SpCas9 (constitutive),Plasmid expression (centromeric) | Several sgRNA expression cassettes (RNA pol III promoter, constitutive),Plasmid expression (multicopy) | Linear dsDNA: 50 bp; 4 pmols | Multi-gene integrations: 2, 58%; 3, 30.6% | (Jakočiūnas et al. 2015) |
| Saccharomyces cerevisiae[CEN.PK2–1C] | SpCas9 (constitutive),Genome integrated | Several sgRNA expression cassettes (RNA pol III, constitutive), some target more than one site,Plasmid expression (multicopy) | Linear dsDNA: 60 bp; 26.96 pmol | Deletions: 9, 50% (only 2 transformants on plate) | (Wijsman et al. 2019) |
| Saccharomyces cerevisiae[CEN.PK 113–5D] | SpCas9 (constitutive),Multicopy plasmid | Synthetic crRNA-array (with one RNA pol III promoter for the expression of four gRNAs), gRNAs between tRNAgly sequences, Plasmid expression (multicopy) | Linear dsDNA: 50 bp; 266.9 pmol | Deletions (8 bp): 8, 86.7% | (Zhang et al. 2019) |
| Ogataea parapolymorpha[CBS 11 895 (n)] | SpCas9 (constitutive),Plasmid expression (centromeric) | Synthetic array of ribozyme-flanked sgRNA (RNA pol II promoter, constitutive),Plasmid expression (centromeric) | Linear dsDNA: 480 bp, 1.6 pmols | Knockouts: 2, 2–5% | (Juergens et al. 2018) |
| Ogataea polymorpha[CGMCC7.89 (n)] | iSpCas9 (constitutive),Genome integrated | Several sgRNA expression cassettes expressed (RNA pol III promoter, constitutive),Genome integrated | Linear dsDNA: 1 kb, 1.7 pmols | Multi-gene integrations: 3, 30.56 ± 2.40% | (Wang et al. 2018a) |
| Yarrowia lipolytica[ATCC 201 249 (n), ATCC MYA-2613 (n)] | SpCas9 (constitutive),Plasmid expression (centromeric) | Synthetic array of ribozyme-flanked sgRNAs (RNA pol II promoter, constitutive),Plasmid expression (centromeric) | On multicopy plasmid: ∼450bp | Knockouts: 2, 36.7 ± 8.5%; 3, 19.3 ± 9.2% | (Gao et al. 2016) |
| Penicillium chrysogenum[DS68530] | SpCas9 (transient), Delivered as a RNP | In vitro synthetized sgRNA in RNP, protoplast-mediated transformation, Transient expression | Linear dsDNA: ≥ 1 kb, 1–11 µg | Cassette integration: 2, 50% | (Pohl et al. 2016) |
| Aspergillus nidulans[IBT27263 (n)] | SpCas9 (constitutive), Plasmid expression (centromeric) | Synthetic array of tRNA-flanked sgRNAs (RNA pol III promoter, constitutive),Plasmid expression (centromeric) | Linear ssDNA: 45 bp, 1 µmol | Multi-purpose: 3, 90% | (Nodvig et al. 2018) |
| Trichoderma reesei[ATCC 13 631, (n); ATCC 56 765 (n)] | SpCas9 (inducible), Genome integrated | In vitro synthetized sgRNA delivery by protoplasts transformation, Transient expression | Linear dsDNA: 200 bp, 296 pmols | Knockouts: 2, 16–45%; 3, 4.2% | (Liu et al. 2015) |
| Scheffersomyces stipitis[UC7, (n)] | SpCas9 (constitutive), Plasmid expression (centromeric) | Several sgRNA expression cassettes (RNA pol III promoter, constitutive),Plasmid expression (centromeric) | Linear dsDNA: 500 bp | Knockouts: 2, 40% | (Cao et al. 2018) |
| Kluyveromyces lactis[ATCC8585 (n)] | SpCas9 (constitutive),Genome integrated | Several sgRNA expression cassettes with homology flanks to a linearized plasmid backbone,Plasmid expression (multicopy) | Linear dsDNA: 1 kb, 0.6–1.54 pmols | Multi-gene integration: 3, 2.1% | (Horwitz et al. 2015) |
| Myceliophthora thermophila[ATCC 42 464, (n)] | SpCas9 (constitutive),Plasmid expression (centromeric) | Several sgRNA expression cassettes (RNA pol III promoter, constitutive),Transient expression | Linear dsDNA: 600 bp; ∼12pmols | Knockouts: 2, 61–70%; 3, 30%; 4, 22% | (Liu et al. 2017) |
| Saccharomyces pastorianus[CBS1483, (n#)] | SpCas9 (constitutive),Plasmid expression (centromeric) | Synthetic array of ribozyme-flanked sgRNA (RNA pol II promoter, constitutive),Plasmid expression (centromeric) | Linear dsDNA: 60 bp; 12pmols | Knockouts: 2, 100% | (Gorter de Vries et al. 2017) |
| Komagataella phaffi[CBS7435 (n)] | SpCas9 (constitutive),Plasmid expression (centromeric) | Several sgRNA (ribozyme-flanked) expression cassettes (RNA pol II promoter, constitutive),Plasmid expression (centromeric) | Linear dsDNA: 1 kb; ∼400–770fmols | Knockouts: 2, 69 ± 13% | (Weninger et al. 2016) |
| Phaeodactylum tricornutum(2n) | SpCas9 (transient),Delivered as a RNP | In vitro synthetized sgRNA in RNP, biolystic delivery, Transient expression | – | Knockouts: 2, 65–100%; 3, 15.4% | (Serif et al. 2018) |
| Magnaporthe oryzae(2n) | SpCas9 (transient),Delivered as a RNP | In vitro synthetized sgRNA in RNP, protoplast-mediated transformation, Transient expression | Linear dsDNA: ∼40 bp | Knockouts by SNP: 2, 3.4–12.3% | (Foster et al. 2018) |
| Fusarium fujikuroi[NJtech 02, CCTCC M2015614] | SpCas9 (constitutive),Plasmid expression (centromeric) | Several sgRNA expression cassettes (RNA pol III promoter, constitutive),Plasmid expression (centromeric) | – | Knockouts by disruption: 2, 20.8%; 3, 4.2% | (Shi et al. 2019) |
| Specie [strain(poidy)] . | Cas nuclease tool (expression), Plasmid (replication origin)/genome integrated . | Strategy for multiplexed gRNA expression/delivery (expression), plasmid (replication origin)/genome integrated . | Type of donor DNA: HFs;amount/concentration . | Type of modification: Number of target, editing efficiency . | Reference . |
|---|---|---|---|---|---|
| PROKARYOTES | |||||
| Escherichia coli[MG1655] | SpCas9 (inducible),Plasmid expression (repA101 ori) | Several sgRNA expression cassettes (constitutive),Plasmid expression | Circular dsDNA (pMB1 ori):∼300 bp | Knockouts: 2, 100%; 3, 88.3%; 4, >30% | (Feng et al. 2018) |
| Escherichia coli[MG1655] | SpCas9 (constitutive),Plasmid expression (repA101 (Ts) ori) | Several sgRNA expression cassettes (constitutive),Plasmid expression (pMB1 ori) | Circular dsDNA (pMB1 ori): 250–550 bp | Knockouts: 2, 97% ± 4%; 3, 47% ± 8% | (Jiang et al. 2015) |
| Escherichia coli[MG1655] | SpCas9 (inducible),Plasmid expression (ColE1 ori) | Several sgRNA expression cassettes (inducible),Plasmid expression (pMB1 ori) | Linear ssDNA: 70 bp; 5 pmol | Short insertions: 2, ∼70% | (Ronda et al. 2016) |
| Escherichia coli[MG1655] | SpCas9 (constitutive),Plasmid expression (p15A ori) | Several sgRNA expression cassettes (constitutive),Plasmid expression (ColE1 ori) | Linear ssDNA: ∼89 bp; 50 pmol | Point mutations: 2, 83%; 3, 23% | (Li et al. 2015) |
| Streptococcus pneumoniae[crR6c] | SpCas9 (constitutive),Genome integrated | Native-like CRISPR array (constitutive),Genome integrated | Linear dsDNA: not mentioned ;0.7 ng/µl to 2.5 µg/µl | Deletions: 2, 75% | (Jiang et al. 2013) |
| Streptomyces lividans | SpCas9 (constitutive),Plasmid expression (pSG5rep) | Several sgRNA expression cassettes (constitutive),Plasmid expression (pSG5rep) | Circular dsDNA (oriT): 1 kB | Short deletions (20–34 bp): 2, 100% (4/4) | (Cobb, Wang and Zhao 2015) |
| Streptomyces coelicolor[M145] | SpCas9 (constitutive),Plasmid expression (pSG5rep) | Several sgRNA expression cassettes (constitutive),Plasmid expression (pSG5rep) | Circular dsDNA (pSG5): ∼1 kB | Deletions (768–1053 bp): 2, 29–54% | (Huang et al. 2015) |
| Escherichia coli[MG1655] | FnCas12a (inducible),Plasmid expression (repA 101) | Native-like CRISPR array (constitutive),Plasmid expression (pSC101 ori) | Circular dsDNA (oriE): 500 bp | Gene insertions: 3, ∼20% | (Ao et al. 2018) |
| Streptomyces coelicolor[M145] | FnCas12a (constitutive),Plasmid expression (pSG5rep) | Native-like CRISPR array (constitutive),Plasmid expression (pSG5) | Circular dsDNA (pSG5): ∼1 kB | Knockouts: 2, 75% | (Li et al. 2018) |
| EUKARYOTES | |||||
| Saccharomyces cerevisiae[CEN.PK113–7D (n), CEN.PK2–1c (n), CEN.PK122 (2n)] | SpCas9 (constitutive),Genome integrated | Several sgRNA expression cassettes (RNA pol III promoter, constitutive),Plasmid expression (multicopy) | Linear dsDNA: 60 bp; 12 pmols | Knockouts: 2, 100%; 6, 65% | (Mans et al. 2015) |
| Saccharomyces cerevisiae[CEN.PK2–1c (n)] | SpCas9 (constitutive),Genome integrated | Several sgRNA expression cassettes with homology flanks to a linearized plasmid backbonePlasmid expression (multicopy) | Linear dsDNA: 500 bp; 0.6–1.54 pmols | Knockouts: 3, 64% | (Horwitz et al. 2015) |
| Saccharomyces cerevisiae[CEN.PK113–5D (n)] | FnCpf1 (constitutive),Genome integrated | Native-like CRISPR array (RNA pol III promoter, constitutive)Plasmid expression (multicopy) | Linear dsDNA: 60 bp; 12 pmols | Knockouts: 2, 100%; 4, 85% | (Swiat et al. 2017) |
| Saccharomyces cerevisiae[CEN.PK113–7D (n), Ethanol Red (2n)] | SpCas9 (constitutive),Plasmid expression (multicopy) | Several sgRNA expression cassettes (RNA pol III promoter, constitutive),Plasmid expression (multicopy) | Linear ssDNA: 40 bp; 300pmols | Knockouts: 2, 91–98% | (Generoso et al. 2016) |
| Saccharomyces cerevisiae[BY4741 (n)] | FnCpf1 (constitutive),Plasmid expression (centromeric) | Native-like CRISPR array (RNA pol III promoter, constitutive),Plasmid expression (multicopy) | Linear dsDNA: 50 bp | Multi-gene integrations: 2, 52%; 3, 43% | (Li, Wang and Wei 2018) |
| Saccharomyces cerevisiae[204 508; ATCC (mated) (2n)] | SpCas9 (constitutive), Plasmid expression (multicopy) | Synthetic array of ribozyme-flanked sgRNA (RNA pol III promoter, constitutive),Plasmid expression (multicopy) | Linear dsDNA: 50 bp; 44.94 pmol | Deletions: 2, 43%; 3, 19% | (Ryan et al. 2014) |
| Saccharomyces cerevisiae[CEN.PK113–7D (n)] | LbCpf1, AsCpf1 or FnCpf1 (constitutive), Plasmid expression (centromeric) | Native-like crRNA-array with homology flanks to a linearized plasmid backbone (RNA pol III promoter, constitutive),Plasmid expression (multicopy) | Linear dsDNA: 50 bp; 90–120 fmols | Multi-gene integrations: 3, 91% | (Verwaal et al. 2018) |
| Saccharomyces cerevisiae[BY4741 (n), CEN.PK2–1c (n)] | iSpCas9 (constitutive),Plasmid expression (multicopy) | Native-like crRNA-array (RNA pol III promoter, constitutive), separate expression of tracrRNA,Plasmid expression (multicopy) | Circular dsDNA, at 5’ of each spacer sequence (multicopy): 50 bp; 142 fmols | Knockouts: 3, 27–87% | (Bao et al. 2015) |
| Saccharomyces cerevisiae[CEN.PK113–7D (n)] | SpCas9 (constitutive),Genome integrated | Synthetic crRNA-array (RNA pol III promoter), separate expression of PaCsy4, Plasmid expression (multicopy) | Linear dsDNA: 60 bp; 12 pmols | Knockouts: 2, 100%; 4, 96% | (Ferreira, et al. 2018) |
| Saccharomyces cerevisiae[BY4741 (n)] | SpCas9 (constitutive),Plasmid expression (centromeric) | Several HDV ribozyme-sgRNA expression cassettes (RNA pol III promoter, constitutive),Plasmid expression (centromeric) | Linear dsDNA, barcoded: 60 bp; 55 pmols | Knockouts: 2, 65–87.5%; 3, 57.5–75%; 4, 27.5–15% | (Lee et al. 2015) |
| Saccharomyces cerevisiae[CEN.PK2–1C (n)] | SpCas9 (constitutive),Plasmid expression (centromeric) | Several sgRNA expression cassettes (RNA pol III promoter, constitutive),Plasmid expression (multicopy) | Linear dsDNA: 500 bp; 700 fmols | Multi-gene integrations: 3, 84% | (Ronda et al. 2015) |
| Saccharomyces cerevisiae[CEN.PK111–27B (n)] | SpCas9 (constitutive),Plasmid expression (centromeric) | Several sgRNA expression cassettes (RNA pol III promoter, constitutive),Plasmid expression (multicopy) | Linear dsDNA: 50 bp; 4 pmols | Multi-gene integrations: 2, 58%; 3, 30.6% | (Jakočiūnas et al. 2015) |
| Saccharomyces cerevisiae[CEN.PK2–1C] | SpCas9 (constitutive),Genome integrated | Several sgRNA expression cassettes (RNA pol III, constitutive), some target more than one site,Plasmid expression (multicopy) | Linear dsDNA: 60 bp; 26.96 pmol | Deletions: 9, 50% (only 2 transformants on plate) | (Wijsman et al. 2019) |
| Saccharomyces cerevisiae[CEN.PK 113–5D] | SpCas9 (constitutive),Multicopy plasmid | Synthetic crRNA-array (with one RNA pol III promoter for the expression of four gRNAs), gRNAs between tRNAgly sequences, Plasmid expression (multicopy) | Linear dsDNA: 50 bp; 266.9 pmol | Deletions (8 bp): 8, 86.7% | (Zhang et al. 2019) |
| Ogataea parapolymorpha[CBS 11 895 (n)] | SpCas9 (constitutive),Plasmid expression (centromeric) | Synthetic array of ribozyme-flanked sgRNA (RNA pol II promoter, constitutive),Plasmid expression (centromeric) | Linear dsDNA: 480 bp, 1.6 pmols | Knockouts: 2, 2–5% | (Juergens et al. 2018) |
| Ogataea polymorpha[CGMCC7.89 (n)] | iSpCas9 (constitutive),Genome integrated | Several sgRNA expression cassettes expressed (RNA pol III promoter, constitutive),Genome integrated | Linear dsDNA: 1 kb, 1.7 pmols | Multi-gene integrations: 3, 30.56 ± 2.40% | (Wang et al. 2018a) |
| Yarrowia lipolytica[ATCC 201 249 (n), ATCC MYA-2613 (n)] | SpCas9 (constitutive),Plasmid expression (centromeric) | Synthetic array of ribozyme-flanked sgRNAs (RNA pol II promoter, constitutive),Plasmid expression (centromeric) | On multicopy plasmid: ∼450bp | Knockouts: 2, 36.7 ± 8.5%; 3, 19.3 ± 9.2% | (Gao et al. 2016) |
| Penicillium chrysogenum[DS68530] | SpCas9 (transient), Delivered as a RNP | In vitro synthetized sgRNA in RNP, protoplast-mediated transformation, Transient expression | Linear dsDNA: ≥ 1 kb, 1–11 µg | Cassette integration: 2, 50% | (Pohl et al. 2016) |
| Aspergillus nidulans[IBT27263 (n)] | SpCas9 (constitutive), Plasmid expression (centromeric) | Synthetic array of tRNA-flanked sgRNAs (RNA pol III promoter, constitutive),Plasmid expression (centromeric) | Linear ssDNA: 45 bp, 1 µmol | Multi-purpose: 3, 90% | (Nodvig et al. 2018) |
| Trichoderma reesei[ATCC 13 631, (n); ATCC 56 765 (n)] | SpCas9 (inducible), Genome integrated | In vitro synthetized sgRNA delivery by protoplasts transformation, Transient expression | Linear dsDNA: 200 bp, 296 pmols | Knockouts: 2, 16–45%; 3, 4.2% | (Liu et al. 2015) |
| Scheffersomyces stipitis[UC7, (n)] | SpCas9 (constitutive), Plasmid expression (centromeric) | Several sgRNA expression cassettes (RNA pol III promoter, constitutive),Plasmid expression (centromeric) | Linear dsDNA: 500 bp | Knockouts: 2, 40% | (Cao et al. 2018) |
| Kluyveromyces lactis[ATCC8585 (n)] | SpCas9 (constitutive),Genome integrated | Several sgRNA expression cassettes with homology flanks to a linearized plasmid backbone,Plasmid expression (multicopy) | Linear dsDNA: 1 kb, 0.6–1.54 pmols | Multi-gene integration: 3, 2.1% | (Horwitz et al. 2015) |
| Myceliophthora thermophila[ATCC 42 464, (n)] | SpCas9 (constitutive),Plasmid expression (centromeric) | Several sgRNA expression cassettes (RNA pol III promoter, constitutive),Transient expression | Linear dsDNA: 600 bp; ∼12pmols | Knockouts: 2, 61–70%; 3, 30%; 4, 22% | (Liu et al. 2017) |
| Saccharomyces pastorianus[CBS1483, (n#)] | SpCas9 (constitutive),Plasmid expression (centromeric) | Synthetic array of ribozyme-flanked sgRNA (RNA pol II promoter, constitutive),Plasmid expression (centromeric) | Linear dsDNA: 60 bp; 12pmols | Knockouts: 2, 100% | (Gorter de Vries et al. 2017) |
| Komagataella phaffi[CBS7435 (n)] | SpCas9 (constitutive),Plasmid expression (centromeric) | Several sgRNA (ribozyme-flanked) expression cassettes (RNA pol II promoter, constitutive),Plasmid expression (centromeric) | Linear dsDNA: 1 kb; ∼400–770fmols | Knockouts: 2, 69 ± 13% | (Weninger et al. 2016) |
| Phaeodactylum tricornutum(2n) | SpCas9 (transient),Delivered as a RNP | In vitro synthetized sgRNA in RNP, biolystic delivery, Transient expression | – | Knockouts: 2, 65–100%; 3, 15.4% | (Serif et al. 2018) |
| Magnaporthe oryzae(2n) | SpCas9 (transient),Delivered as a RNP | In vitro synthetized sgRNA in RNP, protoplast-mediated transformation, Transient expression | Linear dsDNA: ∼40 bp | Knockouts by SNP: 2, 3.4–12.3% | (Foster et al. 2018) |
| Fusarium fujikuroi[NJtech 02, CCTCC M2015614] | SpCas9 (constitutive),Plasmid expression (centromeric) | Several sgRNA expression cassettes (RNA pol III promoter, constitutive),Plasmid expression (centromeric) | – | Knockouts by disruption: 2, 20.8%; 3, 4.2% | (Shi et al. 2019) |
#Anaeuploid.
Multiplexed genome editing events in industrial microorganisms using CRISPR-Cas systems.
| Specie [strain(poidy)] . | Cas nuclease tool (expression), Plasmid (replication origin)/genome integrated . | Strategy for multiplexed gRNA expression/delivery (expression), plasmid (replication origin)/genome integrated . | Type of donor DNA: HFs;amount/concentration . | Type of modification: Number of target, editing efficiency . | Reference . |
|---|---|---|---|---|---|
| PROKARYOTES | |||||
| Escherichia coli[MG1655] | SpCas9 (inducible),Plasmid expression (repA101 ori) | Several sgRNA expression cassettes (constitutive),Plasmid expression | Circular dsDNA (pMB1 ori):∼300 bp | Knockouts: 2, 100%; 3, 88.3%; 4, >30% | (Feng et al. 2018) |
| Escherichia coli[MG1655] | SpCas9 (constitutive),Plasmid expression (repA101 (Ts) ori) | Several sgRNA expression cassettes (constitutive),Plasmid expression (pMB1 ori) | Circular dsDNA (pMB1 ori): 250–550 bp | Knockouts: 2, 97% ± 4%; 3, 47% ± 8% | (Jiang et al. 2015) |
| Escherichia coli[MG1655] | SpCas9 (inducible),Plasmid expression (ColE1 ori) | Several sgRNA expression cassettes (inducible),Plasmid expression (pMB1 ori) | Linear ssDNA: 70 bp; 5 pmol | Short insertions: 2, ∼70% | (Ronda et al. 2016) |
| Escherichia coli[MG1655] | SpCas9 (constitutive),Plasmid expression (p15A ori) | Several sgRNA expression cassettes (constitutive),Plasmid expression (ColE1 ori) | Linear ssDNA: ∼89 bp; 50 pmol | Point mutations: 2, 83%; 3, 23% | (Li et al. 2015) |
| Streptococcus pneumoniae[crR6c] | SpCas9 (constitutive),Genome integrated | Native-like CRISPR array (constitutive),Genome integrated | Linear dsDNA: not mentioned ;0.7 ng/µl to 2.5 µg/µl | Deletions: 2, 75% | (Jiang et al. 2013) |
| Streptomyces lividans | SpCas9 (constitutive),Plasmid expression (pSG5rep) | Several sgRNA expression cassettes (constitutive),Plasmid expression (pSG5rep) | Circular dsDNA (oriT): 1 kB | Short deletions (20–34 bp): 2, 100% (4/4) | (Cobb, Wang and Zhao 2015) |
| Streptomyces coelicolor[M145] | SpCas9 (constitutive),Plasmid expression (pSG5rep) | Several sgRNA expression cassettes (constitutive),Plasmid expression (pSG5rep) | Circular dsDNA (pSG5): ∼1 kB | Deletions (768–1053 bp): 2, 29–54% | (Huang et al. 2015) |
| Escherichia coli[MG1655] | FnCas12a (inducible),Plasmid expression (repA 101) | Native-like CRISPR array (constitutive),Plasmid expression (pSC101 ori) | Circular dsDNA (oriE): 500 bp | Gene insertions: 3, ∼20% | (Ao et al. 2018) |
| Streptomyces coelicolor[M145] | FnCas12a (constitutive),Plasmid expression (pSG5rep) | Native-like CRISPR array (constitutive),Plasmid expression (pSG5) | Circular dsDNA (pSG5): ∼1 kB | Knockouts: 2, 75% | (Li et al. 2018) |
| EUKARYOTES | |||||
| Saccharomyces cerevisiae[CEN.PK113–7D (n), CEN.PK2–1c (n), CEN.PK122 (2n)] | SpCas9 (constitutive),Genome integrated | Several sgRNA expression cassettes (RNA pol III promoter, constitutive),Plasmid expression (multicopy) | Linear dsDNA: 60 bp; 12 pmols | Knockouts: 2, 100%; 6, 65% | (Mans et al. 2015) |
| Saccharomyces cerevisiae[CEN.PK2–1c (n)] | SpCas9 (constitutive),Genome integrated | Several sgRNA expression cassettes with homology flanks to a linearized plasmid backbonePlasmid expression (multicopy) | Linear dsDNA: 500 bp; 0.6–1.54 pmols | Knockouts: 3, 64% | (Horwitz et al. 2015) |
| Saccharomyces cerevisiae[CEN.PK113–5D (n)] | FnCpf1 (constitutive),Genome integrated | Native-like CRISPR array (RNA pol III promoter, constitutive)Plasmid expression (multicopy) | Linear dsDNA: 60 bp; 12 pmols | Knockouts: 2, 100%; 4, 85% | (Swiat et al. 2017) |
| Saccharomyces cerevisiae[CEN.PK113–7D (n), Ethanol Red (2n)] | SpCas9 (constitutive),Plasmid expression (multicopy) | Several sgRNA expression cassettes (RNA pol III promoter, constitutive),Plasmid expression (multicopy) | Linear ssDNA: 40 bp; 300pmols | Knockouts: 2, 91–98% | (Generoso et al. 2016) |
| Saccharomyces cerevisiae[BY4741 (n)] | FnCpf1 (constitutive),Plasmid expression (centromeric) | Native-like CRISPR array (RNA pol III promoter, constitutive),Plasmid expression (multicopy) | Linear dsDNA: 50 bp | Multi-gene integrations: 2, 52%; 3, 43% | (Li, Wang and Wei 2018) |
| Saccharomyces cerevisiae[204 508; ATCC (mated) (2n)] | SpCas9 (constitutive), Plasmid expression (multicopy) | Synthetic array of ribozyme-flanked sgRNA (RNA pol III promoter, constitutive),Plasmid expression (multicopy) | Linear dsDNA: 50 bp; 44.94 pmol | Deletions: 2, 43%; 3, 19% | (Ryan et al. 2014) |
| Saccharomyces cerevisiae[CEN.PK113–7D (n)] | LbCpf1, AsCpf1 or FnCpf1 (constitutive), Plasmid expression (centromeric) | Native-like crRNA-array with homology flanks to a linearized plasmid backbone (RNA pol III promoter, constitutive),Plasmid expression (multicopy) | Linear dsDNA: 50 bp; 90–120 fmols | Multi-gene integrations: 3, 91% | (Verwaal et al. 2018) |
| Saccharomyces cerevisiae[BY4741 (n), CEN.PK2–1c (n)] | iSpCas9 (constitutive),Plasmid expression (multicopy) | Native-like crRNA-array (RNA pol III promoter, constitutive), separate expression of tracrRNA,Plasmid expression (multicopy) | Circular dsDNA, at 5’ of each spacer sequence (multicopy): 50 bp; 142 fmols | Knockouts: 3, 27–87% | (Bao et al. 2015) |
| Saccharomyces cerevisiae[CEN.PK113–7D (n)] | SpCas9 (constitutive),Genome integrated | Synthetic crRNA-array (RNA pol III promoter), separate expression of PaCsy4, Plasmid expression (multicopy) | Linear dsDNA: 60 bp; 12 pmols | Knockouts: 2, 100%; 4, 96% | (Ferreira, et al. 2018) |
| Saccharomyces cerevisiae[BY4741 (n)] | SpCas9 (constitutive),Plasmid expression (centromeric) | Several HDV ribozyme-sgRNA expression cassettes (RNA pol III promoter, constitutive),Plasmid expression (centromeric) | Linear dsDNA, barcoded: 60 bp; 55 pmols | Knockouts: 2, 65–87.5%; 3, 57.5–75%; 4, 27.5–15% | (Lee et al. 2015) |
| Saccharomyces cerevisiae[CEN.PK2–1C (n)] | SpCas9 (constitutive),Plasmid expression (centromeric) | Several sgRNA expression cassettes (RNA pol III promoter, constitutive),Plasmid expression (multicopy) | Linear dsDNA: 500 bp; 700 fmols | Multi-gene integrations: 3, 84% | (Ronda et al. 2015) |
| Saccharomyces cerevisiae[CEN.PK111–27B (n)] | SpCas9 (constitutive),Plasmid expression (centromeric) | Several sgRNA expression cassettes (RNA pol III promoter, constitutive),Plasmid expression (multicopy) | Linear dsDNA: 50 bp; 4 pmols | Multi-gene integrations: 2, 58%; 3, 30.6% | (Jakočiūnas et al. 2015) |
| Saccharomyces cerevisiae[CEN.PK2–1C] | SpCas9 (constitutive),Genome integrated | Several sgRNA expression cassettes (RNA pol III, constitutive), some target more than one site,Plasmid expression (multicopy) | Linear dsDNA: 60 bp; 26.96 pmol | Deletions: 9, 50% (only 2 transformants on plate) | (Wijsman et al. 2019) |
| Saccharomyces cerevisiae[CEN.PK 113–5D] | SpCas9 (constitutive),Multicopy plasmid | Synthetic crRNA-array (with one RNA pol III promoter for the expression of four gRNAs), gRNAs between tRNAgly sequences, Plasmid expression (multicopy) | Linear dsDNA: 50 bp; 266.9 pmol | Deletions (8 bp): 8, 86.7% | (Zhang et al. 2019) |
| Ogataea parapolymorpha[CBS 11 895 (n)] | SpCas9 (constitutive),Plasmid expression (centromeric) | Synthetic array of ribozyme-flanked sgRNA (RNA pol II promoter, constitutive),Plasmid expression (centromeric) | Linear dsDNA: 480 bp, 1.6 pmols | Knockouts: 2, 2–5% | (Juergens et al. 2018) |
| Ogataea polymorpha[CGMCC7.89 (n)] | iSpCas9 (constitutive),Genome integrated | Several sgRNA expression cassettes expressed (RNA pol III promoter, constitutive),Genome integrated | Linear dsDNA: 1 kb, 1.7 pmols | Multi-gene integrations: 3, 30.56 ± 2.40% | (Wang et al. 2018a) |
| Yarrowia lipolytica[ATCC 201 249 (n), ATCC MYA-2613 (n)] | SpCas9 (constitutive),Plasmid expression (centromeric) | Synthetic array of ribozyme-flanked sgRNAs (RNA pol II promoter, constitutive),Plasmid expression (centromeric) | On multicopy plasmid: ∼450bp | Knockouts: 2, 36.7 ± 8.5%; 3, 19.3 ± 9.2% | (Gao et al. 2016) |
| Penicillium chrysogenum[DS68530] | SpCas9 (transient), Delivered as a RNP | In vitro synthetized sgRNA in RNP, protoplast-mediated transformation, Transient expression | Linear dsDNA: ≥ 1 kb, 1–11 µg | Cassette integration: 2, 50% | (Pohl et al. 2016) |
| Aspergillus nidulans[IBT27263 (n)] | SpCas9 (constitutive), Plasmid expression (centromeric) | Synthetic array of tRNA-flanked sgRNAs (RNA pol III promoter, constitutive),Plasmid expression (centromeric) | Linear ssDNA: 45 bp, 1 µmol | Multi-purpose: 3, 90% | (Nodvig et al. 2018) |
| Trichoderma reesei[ATCC 13 631, (n); ATCC 56 765 (n)] | SpCas9 (inducible), Genome integrated | In vitro synthetized sgRNA delivery by protoplasts transformation, Transient expression | Linear dsDNA: 200 bp, 296 pmols | Knockouts: 2, 16–45%; 3, 4.2% | (Liu et al. 2015) |
| Scheffersomyces stipitis[UC7, (n)] | SpCas9 (constitutive), Plasmid expression (centromeric) | Several sgRNA expression cassettes (RNA pol III promoter, constitutive),Plasmid expression (centromeric) | Linear dsDNA: 500 bp | Knockouts: 2, 40% | (Cao et al. 2018) |
| Kluyveromyces lactis[ATCC8585 (n)] | SpCas9 (constitutive),Genome integrated | Several sgRNA expression cassettes with homology flanks to a linearized plasmid backbone,Plasmid expression (multicopy) | Linear dsDNA: 1 kb, 0.6–1.54 pmols | Multi-gene integration: 3, 2.1% | (Horwitz et al. 2015) |
| Myceliophthora thermophila[ATCC 42 464, (n)] | SpCas9 (constitutive),Plasmid expression (centromeric) | Several sgRNA expression cassettes (RNA pol III promoter, constitutive),Transient expression | Linear dsDNA: 600 bp; ∼12pmols | Knockouts: 2, 61–70%; 3, 30%; 4, 22% | (Liu et al. 2017) |
| Saccharomyces pastorianus[CBS1483, (n#)] | SpCas9 (constitutive),Plasmid expression (centromeric) | Synthetic array of ribozyme-flanked sgRNA (RNA pol II promoter, constitutive),Plasmid expression (centromeric) | Linear dsDNA: 60 bp; 12pmols | Knockouts: 2, 100% | (Gorter de Vries et al. 2017) |
| Komagataella phaffi[CBS7435 (n)] | SpCas9 (constitutive),Plasmid expression (centromeric) | Several sgRNA (ribozyme-flanked) expression cassettes (RNA pol II promoter, constitutive),Plasmid expression (centromeric) | Linear dsDNA: 1 kb; ∼400–770fmols | Knockouts: 2, 69 ± 13% | (Weninger et al. 2016) |
| Phaeodactylum tricornutum(2n) | SpCas9 (transient),Delivered as a RNP | In vitro synthetized sgRNA in RNP, biolystic delivery, Transient expression | – | Knockouts: 2, 65–100%; 3, 15.4% | (Serif et al. 2018) |
| Magnaporthe oryzae(2n) | SpCas9 (transient),Delivered as a RNP | In vitro synthetized sgRNA in RNP, protoplast-mediated transformation, Transient expression | Linear dsDNA: ∼40 bp | Knockouts by SNP: 2, 3.4–12.3% | (Foster et al. 2018) |
| Fusarium fujikuroi[NJtech 02, CCTCC M2015614] | SpCas9 (constitutive),Plasmid expression (centromeric) | Several sgRNA expression cassettes (RNA pol III promoter, constitutive),Plasmid expression (centromeric) | – | Knockouts by disruption: 2, 20.8%; 3, 4.2% | (Shi et al. 2019) |
| Specie [strain(poidy)] . | Cas nuclease tool (expression), Plasmid (replication origin)/genome integrated . | Strategy for multiplexed gRNA expression/delivery (expression), plasmid (replication origin)/genome integrated . | Type of donor DNA: HFs;amount/concentration . | Type of modification: Number of target, editing efficiency . | Reference . |
|---|---|---|---|---|---|
| PROKARYOTES | |||||
| Escherichia coli[MG1655] | SpCas9 (inducible),Plasmid expression (repA101 ori) | Several sgRNA expression cassettes (constitutive),Plasmid expression | Circular dsDNA (pMB1 ori):∼300 bp | Knockouts: 2, 100%; 3, 88.3%; 4, >30% | (Feng et al. 2018) |
| Escherichia coli[MG1655] | SpCas9 (constitutive),Plasmid expression (repA101 (Ts) ori) | Several sgRNA expression cassettes (constitutive),Plasmid expression (pMB1 ori) | Circular dsDNA (pMB1 ori): 250–550 bp | Knockouts: 2, 97% ± 4%; 3, 47% ± 8% | (Jiang et al. 2015) |
| Escherichia coli[MG1655] | SpCas9 (inducible),Plasmid expression (ColE1 ori) | Several sgRNA expression cassettes (inducible),Plasmid expression (pMB1 ori) | Linear ssDNA: 70 bp; 5 pmol | Short insertions: 2, ∼70% | (Ronda et al. 2016) |
| Escherichia coli[MG1655] | SpCas9 (constitutive),Plasmid expression (p15A ori) | Several sgRNA expression cassettes (constitutive),Plasmid expression (ColE1 ori) | Linear ssDNA: ∼89 bp; 50 pmol | Point mutations: 2, 83%; 3, 23% | (Li et al. 2015) |
| Streptococcus pneumoniae[crR6c] | SpCas9 (constitutive),Genome integrated | Native-like CRISPR array (constitutive),Genome integrated | Linear dsDNA: not mentioned ;0.7 ng/µl to 2.5 µg/µl | Deletions: 2, 75% | (Jiang et al. 2013) |
| Streptomyces lividans | SpCas9 (constitutive),Plasmid expression (pSG5rep) | Several sgRNA expression cassettes (constitutive),Plasmid expression (pSG5rep) | Circular dsDNA (oriT): 1 kB | Short deletions (20–34 bp): 2, 100% (4/4) | (Cobb, Wang and Zhao 2015) |
| Streptomyces coelicolor[M145] | SpCas9 (constitutive),Plasmid expression (pSG5rep) | Several sgRNA expression cassettes (constitutive),Plasmid expression (pSG5rep) | Circular dsDNA (pSG5): ∼1 kB | Deletions (768–1053 bp): 2, 29–54% | (Huang et al. 2015) |
| Escherichia coli[MG1655] | FnCas12a (inducible),Plasmid expression (repA 101) | Native-like CRISPR array (constitutive),Plasmid expression (pSC101 ori) | Circular dsDNA (oriE): 500 bp | Gene insertions: 3, ∼20% | (Ao et al. 2018) |
| Streptomyces coelicolor[M145] | FnCas12a (constitutive),Plasmid expression (pSG5rep) | Native-like CRISPR array (constitutive),Plasmid expression (pSG5) | Circular dsDNA (pSG5): ∼1 kB | Knockouts: 2, 75% | (Li et al. 2018) |
| EUKARYOTES | |||||
| Saccharomyces cerevisiae[CEN.PK113–7D (n), CEN.PK2–1c (n), CEN.PK122 (2n)] | SpCas9 (constitutive),Genome integrated | Several sgRNA expression cassettes (RNA pol III promoter, constitutive),Plasmid expression (multicopy) | Linear dsDNA: 60 bp; 12 pmols | Knockouts: 2, 100%; 6, 65% | (Mans et al. 2015) |
| Saccharomyces cerevisiae[CEN.PK2–1c (n)] | SpCas9 (constitutive),Genome integrated | Several sgRNA expression cassettes with homology flanks to a linearized plasmid backbonePlasmid expression (multicopy) | Linear dsDNA: 500 bp; 0.6–1.54 pmols | Knockouts: 3, 64% | (Horwitz et al. 2015) |
| Saccharomyces cerevisiae[CEN.PK113–5D (n)] | FnCpf1 (constitutive),Genome integrated | Native-like CRISPR array (RNA pol III promoter, constitutive)Plasmid expression (multicopy) | Linear dsDNA: 60 bp; 12 pmols | Knockouts: 2, 100%; 4, 85% | (Swiat et al. 2017) |
| Saccharomyces cerevisiae[CEN.PK113–7D (n), Ethanol Red (2n)] | SpCas9 (constitutive),Plasmid expression (multicopy) | Several sgRNA expression cassettes (RNA pol III promoter, constitutive),Plasmid expression (multicopy) | Linear ssDNA: 40 bp; 300pmols | Knockouts: 2, 91–98% | (Generoso et al. 2016) |
| Saccharomyces cerevisiae[BY4741 (n)] | FnCpf1 (constitutive),Plasmid expression (centromeric) | Native-like CRISPR array (RNA pol III promoter, constitutive),Plasmid expression (multicopy) | Linear dsDNA: 50 bp | Multi-gene integrations: 2, 52%; 3, 43% | (Li, Wang and Wei 2018) |
| Saccharomyces cerevisiae[204 508; ATCC (mated) (2n)] | SpCas9 (constitutive), Plasmid expression (multicopy) | Synthetic array of ribozyme-flanked sgRNA (RNA pol III promoter, constitutive),Plasmid expression (multicopy) | Linear dsDNA: 50 bp; 44.94 pmol | Deletions: 2, 43%; 3, 19% | (Ryan et al. 2014) |
| Saccharomyces cerevisiae[CEN.PK113–7D (n)] | LbCpf1, AsCpf1 or FnCpf1 (constitutive), Plasmid expression (centromeric) | Native-like crRNA-array with homology flanks to a linearized plasmid backbone (RNA pol III promoter, constitutive),Plasmid expression (multicopy) | Linear dsDNA: 50 bp; 90–120 fmols | Multi-gene integrations: 3, 91% | (Verwaal et al. 2018) |
| Saccharomyces cerevisiae[BY4741 (n), CEN.PK2–1c (n)] | iSpCas9 (constitutive),Plasmid expression (multicopy) | Native-like crRNA-array (RNA pol III promoter, constitutive), separate expression of tracrRNA,Plasmid expression (multicopy) | Circular dsDNA, at 5’ of each spacer sequence (multicopy): 50 bp; 142 fmols | Knockouts: 3, 27–87% | (Bao et al. 2015) |
| Saccharomyces cerevisiae[CEN.PK113–7D (n)] | SpCas9 (constitutive),Genome integrated | Synthetic crRNA-array (RNA pol III promoter), separate expression of PaCsy4, Plasmid expression (multicopy) | Linear dsDNA: 60 bp; 12 pmols | Knockouts: 2, 100%; 4, 96% | (Ferreira, et al. 2018) |
| Saccharomyces cerevisiae[BY4741 (n)] | SpCas9 (constitutive),Plasmid expression (centromeric) | Several HDV ribozyme-sgRNA expression cassettes (RNA pol III promoter, constitutive),Plasmid expression (centromeric) | Linear dsDNA, barcoded: 60 bp; 55 pmols | Knockouts: 2, 65–87.5%; 3, 57.5–75%; 4, 27.5–15% | (Lee et al. 2015) |
| Saccharomyces cerevisiae[CEN.PK2–1C (n)] | SpCas9 (constitutive),Plasmid expression (centromeric) | Several sgRNA expression cassettes (RNA pol III promoter, constitutive),Plasmid expression (multicopy) | Linear dsDNA: 500 bp; 700 fmols | Multi-gene integrations: 3, 84% | (Ronda et al. 2015) |
| Saccharomyces cerevisiae[CEN.PK111–27B (n)] | SpCas9 (constitutive),Plasmid expression (centromeric) | Several sgRNA expression cassettes (RNA pol III promoter, constitutive),Plasmid expression (multicopy) | Linear dsDNA: 50 bp; 4 pmols | Multi-gene integrations: 2, 58%; 3, 30.6% | (Jakočiūnas et al. 2015) |
| Saccharomyces cerevisiae[CEN.PK2–1C] | SpCas9 (constitutive),Genome integrated | Several sgRNA expression cassettes (RNA pol III, constitutive), some target more than one site,Plasmid expression (multicopy) | Linear dsDNA: 60 bp; 26.96 pmol | Deletions: 9, 50% (only 2 transformants on plate) | (Wijsman et al. 2019) |
| Saccharomyces cerevisiae[CEN.PK 113–5D] | SpCas9 (constitutive),Multicopy plasmid | Synthetic crRNA-array (with one RNA pol III promoter for the expression of four gRNAs), gRNAs between tRNAgly sequences, Plasmid expression (multicopy) | Linear dsDNA: 50 bp; 266.9 pmol | Deletions (8 bp): 8, 86.7% | (Zhang et al. 2019) |
| Ogataea parapolymorpha[CBS 11 895 (n)] | SpCas9 (constitutive),Plasmid expression (centromeric) | Synthetic array of ribozyme-flanked sgRNA (RNA pol II promoter, constitutive),Plasmid expression (centromeric) | Linear dsDNA: 480 bp, 1.6 pmols | Knockouts: 2, 2–5% | (Juergens et al. 2018) |
| Ogataea polymorpha[CGMCC7.89 (n)] | iSpCas9 (constitutive),Genome integrated | Several sgRNA expression cassettes expressed (RNA pol III promoter, constitutive),Genome integrated | Linear dsDNA: 1 kb, 1.7 pmols | Multi-gene integrations: 3, 30.56 ± 2.40% | (Wang et al. 2018a) |
| Yarrowia lipolytica[ATCC 201 249 (n), ATCC MYA-2613 (n)] | SpCas9 (constitutive),Plasmid expression (centromeric) | Synthetic array of ribozyme-flanked sgRNAs (RNA pol II promoter, constitutive),Plasmid expression (centromeric) | On multicopy plasmid: ∼450bp | Knockouts: 2, 36.7 ± 8.5%; 3, 19.3 ± 9.2% | (Gao et al. 2016) |
| Penicillium chrysogenum[DS68530] | SpCas9 (transient), Delivered as a RNP | In vitro synthetized sgRNA in RNP, protoplast-mediated transformation, Transient expression | Linear dsDNA: ≥ 1 kb, 1–11 µg | Cassette integration: 2, 50% | (Pohl et al. 2016) |
| Aspergillus nidulans[IBT27263 (n)] | SpCas9 (constitutive), Plasmid expression (centromeric) | Synthetic array of tRNA-flanked sgRNAs (RNA pol III promoter, constitutive),Plasmid expression (centromeric) | Linear ssDNA: 45 bp, 1 µmol | Multi-purpose: 3, 90% | (Nodvig et al. 2018) |
| Trichoderma reesei[ATCC 13 631, (n); ATCC 56 765 (n)] | SpCas9 (inducible), Genome integrated | In vitro synthetized sgRNA delivery by protoplasts transformation, Transient expression | Linear dsDNA: 200 bp, 296 pmols | Knockouts: 2, 16–45%; 3, 4.2% | (Liu et al. 2015) |
| Scheffersomyces stipitis[UC7, (n)] | SpCas9 (constitutive), Plasmid expression (centromeric) | Several sgRNA expression cassettes (RNA pol III promoter, constitutive),Plasmid expression (centromeric) | Linear dsDNA: 500 bp | Knockouts: 2, 40% | (Cao et al. 2018) |
| Kluyveromyces lactis[ATCC8585 (n)] | SpCas9 (constitutive),Genome integrated | Several sgRNA expression cassettes with homology flanks to a linearized plasmid backbone,Plasmid expression (multicopy) | Linear dsDNA: 1 kb, 0.6–1.54 pmols | Multi-gene integration: 3, 2.1% | (Horwitz et al. 2015) |
| Myceliophthora thermophila[ATCC 42 464, (n)] | SpCas9 (constitutive),Plasmid expression (centromeric) | Several sgRNA expression cassettes (RNA pol III promoter, constitutive),Transient expression | Linear dsDNA: 600 bp; ∼12pmols | Knockouts: 2, 61–70%; 3, 30%; 4, 22% | (Liu et al. 2017) |
| Saccharomyces pastorianus[CBS1483, (n#)] | SpCas9 (constitutive),Plasmid expression (centromeric) | Synthetic array of ribozyme-flanked sgRNA (RNA pol II promoter, constitutive),Plasmid expression (centromeric) | Linear dsDNA: 60 bp; 12pmols | Knockouts: 2, 100% | (Gorter de Vries et al. 2017) |
| Komagataella phaffi[CBS7435 (n)] | SpCas9 (constitutive),Plasmid expression (centromeric) | Several sgRNA (ribozyme-flanked) expression cassettes (RNA pol II promoter, constitutive),Plasmid expression (centromeric) | Linear dsDNA: 1 kb; ∼400–770fmols | Knockouts: 2, 69 ± 13% | (Weninger et al. 2016) |
| Phaeodactylum tricornutum(2n) | SpCas9 (transient),Delivered as a RNP | In vitro synthetized sgRNA in RNP, biolystic delivery, Transient expression | – | Knockouts: 2, 65–100%; 3, 15.4% | (Serif et al. 2018) |
| Magnaporthe oryzae(2n) | SpCas9 (transient),Delivered as a RNP | In vitro synthetized sgRNA in RNP, protoplast-mediated transformation, Transient expression | Linear dsDNA: ∼40 bp | Knockouts by SNP: 2, 3.4–12.3% | (Foster et al. 2018) |
| Fusarium fujikuroi[NJtech 02, CCTCC M2015614] | SpCas9 (constitutive),Plasmid expression (centromeric) | Several sgRNA expression cassettes (RNA pol III promoter, constitutive),Plasmid expression (centromeric) | – | Knockouts by disruption: 2, 20.8%; 3, 4.2% | (Shi et al. 2019) |
#Anaeuploid.
The crRNA processing activity of the recently characterized Cas12a increases the simplicity of multiplexing. By expressing a single CRISPR array containing multiple spacers under the transcriptional regulation of a single promoter and terminator, multiple loci can be targeted simultaneously (Zetsche et al. 2017). Therefore, there is no need of supplying multiple targeting expression constructs. Recent studies have demonstrated the multiplex editing potential of the Cas12a endonuclease in a wide range of microorganisms (Table 2).
Aneuploidy and polyploidy are common conditions among eukaryotic industrial microorganisms. The requirement for simultaneous targeting of multiple alleles in non-haploid strain results in a decrease of the CRISPR editing efficiencies (Mertens et al. 2019). The term ‘cis-multiplexing’ is used when targeting a single genomic locus found multiple times across the genome of a non-haploid organism. ‘Trans-multiplexing’ refers to the simultaneous introduction of modifications in multiple genes that occur in more than one copy in the genome (Ryan et al. 2014). Several examples of both cis- and trans- multiplexing have recently been described (Table 2).
gRNA expression systems for efficient multiplex genome editing
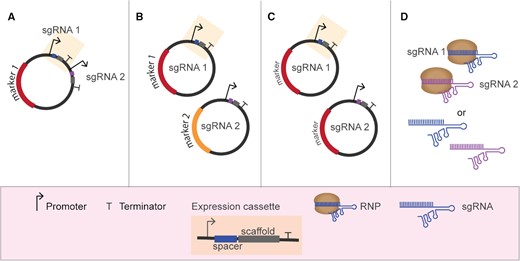
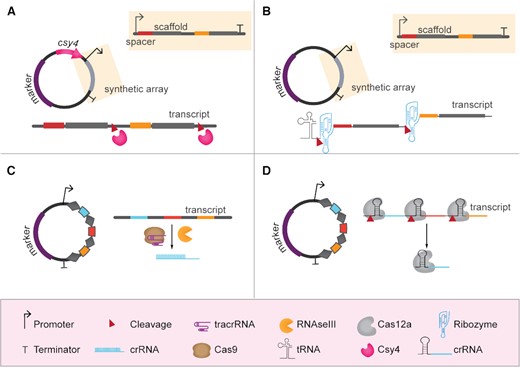
In most reported cases of genome editing of industrial microorganisms, and in all bacterial examples, gRNAs are generally expressed from multicopy plasmids that, after successful editing, can be removed through counter selection (Bao et al. 2015; Si et al. 2017) or growth on non-selective medium (Mans et al. 2015; Wijsman et al. 2019). In case of multiplex genome editing, two strategies have been described for the delivery of the different gRNAs: (i) multiple gRNA expression cassettes transcribing a sgRNA molecule from one or more plasmids (Fig. 2); (ii) polycistronic expression of gRNAs, either inspired by native CRISPR systems or by synthetically designed ones (Fig. 3). Individual gRNAs can also be in vitro transcribed and supplied directly to filamentous fungi or microalgae (Table 2).
Multiplexing using single gRNA expression cassettes. (A) Expression of several sgRNA cassettes from a single expression vector. (B) Expression of several sgRNA cassettes from multiple expression vectors (each harboring a different marker). (C) Expression of several sgRNA cassettes from multiple expression vectors (all harboring the same marker). (D) Transient supplementation with in vitro assembled RNPs or in vitro transcribed gRNAs.
Multiplexing using gRNA polycistronic cassettes. (A) Expression of gRNAs from synthetic array dependent on Csy4 processing. In this case, Csy4 has to be co-expressed. (B) Expression of gRNAs from synthetic array dependent on endoribonuclease splicing. In most of the reviewed examples, these synthetic arrays are expressed using tRNAs as RNA pol III promoters. (C) Expression of gRNAs from native-like CRISPR array dependent on Cas9, tracrRNA and RNAse III processing. (D) Expression of gRNAs from native-like CRISPR array dependent on Cas12a processing.
Multiplexing using multiple single gRNA expression cassettes
The initial attempts of multiplex genome editing using SpCas9 relied on combined expression of several individual gRNA expression cassettes. In bacteria, different strength natural or synthetic promoters (constitutive or inducible) are routinely used for gRNA expression (Table 2). In eukaryotes, efficient gRNA expression might be controlled by an RNA polymerase-III-dependent promoter. In a cell that expresses a nuclear-localized SpCas9, this gRNA can direct the nuclease to its target. In addition, fusions of gRNAs and tRNA auto-splicing sequences (used as promoters) have also been demonstrated to yield multiple functional gRNAs in eukaryotes (Schwartz et al. 2016; Song et al. 2018). Alternatively, gRNAs flanked with self-cleaving ribozymes on both ends have been expressed from RNA polymerase II-dependent promoters to provide transcripts with modified ends and increase transcript stability in the nucleus (Gao and Zhao 2014; Nødvig et al. 2015; Weninger et al. 2016; Wong et al. 2017). Moreover, expression of a bacterial T7 RNA polymerase in the yeasts Y. lipolytica and Kluyveromyces lactis has been used for guide expression (Morse et al. 2018).
In bacteria, the maximal number of reported simultaneous editing events using SpCas9 varies from organism to organism (Table 2). Expression of multiple gRNAs from multiple expression cassettes is done from plasmid-borne or genome-integrated constructs. Most reported examples of multiplex genome editing using gRNAs expressed from multiple expression cassettes have been performed in E. coli (Jiang et al. 2015; Li et al. 2015; Ronda et al. 2016), with a maximum of four genes targeted simultaneously and an editing efficiency of ∼30% (Feng et al. 2018). The authors of the study developed a CRISPR multiplex genome editing technique that uncouples transformation and editing. This separation is achieved by inducing Cas12a expression only after transformation and seems to be key, together with recombineering, for increased editing efficiencies (Reisch and Prather 2015; Feng et al. 2018).
In eukaryotes, the combination of several gRNA expression modules in a single amplicon or spread over several co-transformed plasmids resulted in successful editing in several organisms such as S. cerevisiae (Horwitz et al. 2015; Jakočiūnas et al. 2015; Lee et al. 2015; Mans et al. 2015; Ronda et al. 2015; Generoso et al. 2016; Deaner, Holzman and Alper 2018), Komagataella phaffi (Weninger et al. 2016), the xylose-utilizing yeast Scheffersomyces stipitis (Cao et al. 2018), the enzyme producer Myceliophthora thermophila (Liu et al. 2017), K. lactis (Horwitz et al. 2015), Fusarium fujikuroi (Shi et al. 2019), the methylotrophic yeast Ogataea polymorpha (Wang et al. 2018a) and the oleaginous yeast Y. lipolytica (Gao et al. 2016) (Table 2). In many organisms, high editing efficiencies allow for straightforward screening and selection of the desired mutant without introducing selectable markers in their genome. However, the reported efficiencies for targeting two or more sites simultaneously in eukaryotes vary significantly from one organism to another (i.e. from 2 to 100%). Using SpCas9, nine editing events in S. cerevisiae is the highest number of simultaneous modifications reported in microbes to date (Table 2) (Wijsman et al. 2019).
In some filamentous fungi, the gRNA molecules can be synthetized in vitro and co-transformed with Cas9-encoding plasmids. In microalgae, in vitro synthetized gRNAs can be delivered together with in vitro produced Cas proteins as ribonucleoprotein complexes (RNPs) as well (Liu et al. 2015; Pohl et al. 2016; Shi et al. 2017; Naduthodi, Barbosa and van der Oost 2018). Simultaneous double editing has been achieved by following this strategy in Penicillium chrysogenum (Pohl et al. 2016,2018), in Phaeodactylum tricornutum (Serif et al. 2018) and in the rice blast fungus Magnaporthe oryzae (Foster et al. 2018). Although transformation of in vitro synthetized gRNA simplifies gRNA cloning work and avoids the requirement of identifying effective RNA polymerase III promoters in eukaryotes, efficient selection of transformants may still require chromosomal integration of selectable markers located in the dDNA molecules (Liu et al. 2015; Pohl et al. 2016). These markers can be removed from the genome by using counter-selection markers or Cre-recombinase-based approaches.
Finally, although SpCas9 has been successfully implemented in microalgae, we found few examples for the use thereof for multiplexed genome editing (Behler et al. 2018; Naduthodi, Barbosa and van der Oost 2018). Recently, a triple knockout strain of the microalgae P. tricornutum was generated in a single step transformation using six different Cas9-based RNP complexes (Serif et al. 2018).
Multiplexing using gRNA polycistronic cassette
Cas9-dependent multiplex genome editing efforts reported to date are generally based on the combination of individual expression cassettes required for single targeting editing experiments. Hence, they commonly suffer from (i) repeated usage of equal sets of promoters and terminators for gRNA expression, (ii) requirement for multiple selection markers, and more importantly, (iii) labor-intensive expression cassettes and plasmid construction. By expressing multiple gRNAs under the control of a single promoter, polycistronic expression cassettes overcome these hurdles. To date, two polycistronic cassette-based approaches have been shown that enable multiplex genome editing (Fig. 3).
The first approach uses tandem arrays of chimeric sgRNAs that require a dedicated maturation mechanism to release the individual sgRNAs after transcription (Fig. 3A and B). The Csy4 ribonuclease cleaves synthetic precursor sgRNA arrays when interspaced by 28-nt sequences recognized by Csy4 (Qi et al. 2012). As such, efficient quadruplex editing by SpCas9 has been accomplished in S. cerevisiae (Ferreira et al.2018). Alternatively, cleavage of the crRNA array can be performed by self-cleaving RNA sequences. As such, efficient duplex editing was achieved in the non-conventional yeasts O. parapolymorpha and Saccharomyces pastorianus using sgRNAs flanked with Hammerhead and Hepatitis Delta Virus ribozymes (Gorter de Vries et al. 2017; Juergens et al. 2018). Triplex editing was achieved in a diploid S. cerevisiae strain with a synthetic array of sgRNAs flanked by ribozymes (Ryan et al. 2014). In addition, tandem fusion of multiple sgRNAs-tRNAs enabling sgRNAs processing by the native tRNA-maturation system was shown to promote SpCas9-assisted multiplexed genome editing in filamentous fungi (Nodvig et al. 2018).
The second approach for polycistronic gRNA expression resembles native CRISPR arrays (Fig. 3C and D). The expression of Cas9 is required together with the transcription of a CRISPR array with multiple crRNAs and a tracrRNA. With this approach, an array of two guides was expressed and processed in a self-targeting system in E. coli for plasmid removal. Processing of the CRISPR array transcript occurred via unknown native endoribonucleases (Ronda et al. 2016). Again, resembling native CRISPR systems, a three-spacers array and a tracrRNA were co-expressed in S. cerevisiae and up to three genes were deleted with efficiencies ranging from 27–87% (depending on the sequence of the targeting guides) (Bao et al. 2015). More recently, several orthologues from the class V endonuclease Cas12a (AsCas12a, LbCas12a and FnCas12a) (Zetsche et al. 2015) were shown to deliver efficient multiplex genome editing of E. coli (Ao et al. 2018), Streptomyces coelicolor (Li et al. 2018) and S. cerevisiae (Swiat et al. 2017; Verwaal et al. 2018; Li, Wang and Wei 2018).
Efficient delivery of donor DNA for repair of CRISPR-mediated DNA breaks
Current multiplexing strategies are based on two different procedures for the simultaneous delivery of dDNA fragments. On one hand, transient delivery of these homology repair templates is accomplished by co-transforming multiple linear dDNAs with a corresponding set of gRNAs. This approach was implemented for efficient introduction of multiple barcoded repairs fragments (short DNA fragments that contain a unique sequence tag) (Ryan and Cate 2014), and editing of heterologous metabolic pathways in both eukaryotic and prokaryotic genomes (Jiang et al. 2013; Jakočiūnas et al. 2015; Li et al. 2015; Mans et al. 2015; Ronda et al. 2015; Verwaal et al. 2018; Wang et al. 2018a). While dDNA is often delivered in double-stranded configuration, it should be noted that successful repair using single-stranded DNA (ssDNA) has also been achieved in S. cerevisiae (DiCarlo et al. 2013; Generoso et al. 2016), Aspergilli (Nodvig et al. 2018), in E. coli (Li et al. 2015; Ronda et al. 2016), and in mammalian cells (Richardson et al. 2016; Song and Stieger 2017). Alternatively, a stable source of dDNA sequences is provided when these are cloned in one multi-copy plasmid (Cobb, Wang and Zhao 2015; Huang et al. 2015; Jiang et al. 2015; Gao et al. 2016; Ao et al. 2018; Feng et al. 2018; Li et al. 2018). In a more elegant way, these sequences can be cloned in tandem to the corresponding gRNAs under control of a single promoter. These hybrids are long transcripts that include multiple repair template-crRNA sequences between CRISPR repeats to be further processed into shorter gRNAs. These gRNAs will still include the transcribed sequence of each dDNA template (Bao et al. 2015; Garst et al. 2017; Roy et al. 2018).
Repair efficiencies of the double-stranded DNA break at a targeted locus depend on the size of the homology flanks (HFs) of the dDNA. Commonly used dDNA sequences may vary from short-sized HFs (∼50 bp for S. cerevisiae or ∼50–100 bp for E. coli), to longer, PCR-based HFs for many bacteria, non-conventional yeasts and fungi (200–1000 bp) (Table 2). Long HFs have been shown to increase the efficiency of HDR for both single target and multiplex genome editing in bacteria and eukaryotic microorganisms (Ronda et al. 2015; Ao et al. 2018; Wang et al. 2018a). In S. cerevisiae, multiplex gene deletions can be obtained simultaneously by using oligo-sized dDNAs with short HFs (∼50 bp), either as single-stranded (Generoso et al. 2016) or as annealed double-stranded dDNAs (Mans et al. 2015; Swiat et al. 2017; Ferreira, David and Nielsen 2018) (Table 2). The amount of dDNA has previously been considered a key factor for enhancing CRISPR-Cas-mediated editing (Horwitz et al. 2015). Cells are generally transformed with a relatively high concentration of dDNAs, in the order of picomols (Horwitz et al. 2015; Mans et al. 2015). This concentration is higher than the one usually used for in vitro plasmid assembly procedures in S. cerevisiae (Kuijpers et al. 2013).
Methods for increasing HDR frequencies versus NHEJ and alternative NHEJ repair
The potential of CRISPR-Cas9/Cas12a for implementing precise HDR-based genome editing is often hindered by (i) the presence of NHEJ or alternative-NHEJ repair systems, (ii) the presence of inefficient HDR systems, or (iii) an unfavorable balance between NHEJ and HDR repair mechanisms.
In bacteria, Cas9 and Cas12a are mainly used as counter-selection tools: the endonucleases create DSBs causing cell death due to the absence or poor efficiency of NHEJ repair systems. Only those cells that successfully obtained and integrated appropriate repair templates into their genome avoid the occurrence of persistent chromosomal breaks (Mougiakos et al. 2016). The expression of recombineering systems based on the λ-Red recombinase is extensively used in organisms such as E. coli, Lactobacillus (Oh and van Pijkeren 2014) or Pseudomonas putida (Aparicio, de Lorenzo and Martínez-García 2018) to boost the efficiency of HDR and increase the number of recombinants (Mougiakos et al. 2018).
In the case of a multiplex engineering approach in yeasts, the desired genome editing (and the cell viability) relies on the stochastic allocation of dDNAs into the nucleus. In S. cerevisiae, the relatively high activity of the HDR machinery facilitates precise editing at multiple loci simultaneously (Mans et al. 2015; Swiat et al. 2017; Ferreira, David and Nielsen 2018). In contrast, the prevalent NHEJ system in non-conventional yeasts (or other difficult-to-engineer organisms, such as microalgae) might need a more extensive screening due to the high heterogeneity of transformants (Serif et al. 2018; Wang et al. 2018a). To improve the efficiency of HDR in some yeasts such as Y. lipolytica, the KU70 gene responsible for DSB repair in the NHEJ pathway was disrupted (Jang et al. 2018). A similar approach is often applied in non-conventional yeasts, e.g. in Naumovozyma castellii, where simultaneous deletion of the orthologues of KU70 and KU80 completely abolished NHEJ repair during CRISPR-based editing (Vyas et al. 2018). Alternatively, researchers have inhibited certain endogenous DNA repair components to favor HDR in CRISPR experiments (Vyas et al. 2018).
The balance between DSB repair pathways is further influenced by the cell cycle phase in which a cell is. By making use of hydroxyurea-mediated cell cycle arrest (S-phase), the frequency of targeted integration is significantly increased in multiple fungi (Tsakraklides et al. 2015) and demonstrated specifically for CRISPR-based genome editing of Y. lipolytica (Jang et al. 2018). Finally, an improvement of HDR efficiency in single target genome editing has recently been achieved by active recruitment of the dDNA to the DSB making use of the ability of some proteins to bind DSBs in S. cerevisiae (Roy et al. 2018). Similar methods could be considered for obtaining an increased HDR frequency in a multiplexing set-up for a broader variety of microbes.
Multiplex gene repression and activation
Multiplexing can also be exploited for CRISPRi/CRISPRa approaches (Table 3). Again, both dCas9 and dCas12a have been used for controlling gene expression. In many microorganisms, CRISPRi and CRISPRa have mainly been used to re-direct the carbon flux towards the production of the desired product. This usually requires fine-tuning the expression of multiple genes, which can be achieved by multiplexed gRNA expression for polygenic targeting. Multiplexing using CRISPRi has been explored in several prokaryotic industrial organisms such as E. coli (Zhang et al. 2017; Gao et al. 2018; Tian et al. 2019), S. coelicolor (Li et al. 2018; Zhao et al. 2018), P. putida (Tan, Reisch and Prather 2018), Bacillus subtilis (Wu et al2018b), Corynebacterium glutamicum (Cleto et al. 2016) and the cyanobacterium Synechocystis (Yao et al. 2016; Kaczmarzyk et al. 2018). Furthermore, different approaches have been explored in the yeasts S. cerevisiae (Jensen et al. 2017; Lian et al. 2017), Y. lipolytica (Schwartz et al. 2017; Zhang et al. 2018) and Kluyveromyces marxianus (Löbs et al. 2018). Independent studies on the implementation of Cas12a for CRISPRi purposes reported changes in repression strength after altering the order of the spacers in the CRISPR array (Liao et al. 2018). Other studies did not observe the same effect (Wang et al. 2017; Zetsche et al. 2017; Zhang et al. 2017). This suggests that the phenomenon could be gRNA-sequence-dependent and therefore most likely related to transcript secondary structure formation.
Multiplexed genome regulation events in industrial microorganisms using CRISPRi and CRISPRa.
| Specie [strain(poidy)] . | Cas nuclease tool (CRISPRi/CRISPRa), expression, promoter Plasmid (replication origin)/genome integrated . | Strategy for multiplexed gRNA expression/delivery (expression) Plasmid (replication origin)/genome integrated . | Goal . | Number of targets . | Reference . |
|---|---|---|---|---|---|
| PROKARYOTES | |||||
| E. coli[B0013] | dSpCas9 (CRISPRi)—constitutive expression, trc promoterPlasmid expression | Several sgRNA expression cassettes (constitutive)Plasmid expression (CloDF13ori) | Increase malate titer | 3 targets | (Gao et al. 2018) |
| E. coli[MG1655] | ddAsCas12a (CRISPRi)—constitutive expression, j23100 promoterPlasmid expression (p15A ori) | Several sgRNA expression cassettes (j23119-SpeI constitutive promoter)Plasmid expression (ColE1 ori) | Proof of principle | 3 targets | (Zhang et al. 2017) |
| E. coli[MG1655] | dSpCas9 (CRISPRI/CRISPRa)—constitutive expression, endogenous S. pyogenes promoterPlasmid expression | Several sgRNA expression cassettes (j23119 constitutive promoter) fused to protein binding sequences (scaffold RNAs)Plasmid expression | Proof of principle. Activation of ethanol biosynthesis | 2 targets | (Dong et al. 2018) |
| Bacillus subtilis[BNY] | dSpCas9 (CRISPRi)—- inducible expression, xylA promoterGenome integrated | Several sgRNA expression cassettes (Pveg constitutive promoter)Genome integrated | Increase N-acetylglucosamine titer | 3 targets | (Wu et al. 2018b) |
| Streptomyces coelicolor | dSpCas9 (CRISPRi)—constitutive expression, ermE*p promoterGenome integrated | Several sgRNA expression cassettes (j23119p constitutive promoter)Genome integrated | Proof of principle (knock-out 4 pigmented antibiotic synthesis) | 3 and 4 targets | (Zhao et al. 2018) |
| Pseudomonas putida | dSpaCas9 (CRISPRi)—inducible expression, LacI-Ptac promoterGenome integrated | Several sgRNA expression cassettes (Ptet promoter)Plasmid expression (oriV, Rep) | Proof of principle | 2 targets | (Tan et al. 2018) |
| Corynebacterium glutamicum | dSpCas9 (CRISPRi)—inducible expression, Ptac promoterPlasmid expression | Several sgRNA expression cassettes (Ptac inducible promoter)Plasmid expression | Increase aminoacid production | 3 targets | (Cleto et al. 2016) |
| Bacillus subtilis[BNY] | dSpCas9 (CRISPRi)—inducible expression, PxylA promoterGenome integrated | Several sgRNA expression cassettes (Pveg promoter)Genome integrated | Increase GlcNAc titer | 3 targets | (Wu et al. 2018) |
| Synechocystis | dSpCas9 (CRISPRi)—constitutive expression, PpsbA2 promoterGenome integrated | Several sgRNA expression cassettes (PL31 constitutive promoter)Genome integrated | Reduction of PHB and glycogen accumulation during nitrogen starvation | 4 targets | (Yao et al. 2016) |
| Synechocystis | dSpCas9 (CRISPRi)—inducible expression, PL22 promoterGenome integrated | Several sgRNA expression cassettes (PL22 constitutive promoter)Genome integrated | Carbon flux re-direction for production of fatty alcohols | 6 targets | (Kaczmarzyk et al. 2018) |
| Streptomyces coelicolor[M145] | ddFnCas12a (CRISPRi)—constitutive expression, ermEp* promoterGenome integrated | Native-like CRISPR array (kasOp* constitutive promoter)Genome integrated | Proof of principle (knock-out 3 pigmented antibiotic synthesis) | 3 targets | (Li et al. 2018) |
| EUKARYOTES | |||||
| Saccharomyces cerevisiae[CEN.PK2-a and Sigma 10 560–4A] | dCas9-VPR (CRISPRi and CRISPRa)Plasmid expression (centromeric) | TEF1p-tRNA-sgRNA-tRNA (constitutive)Plasmid expression (centromeric) | Increase 2,3-butanediol titer | 5 targets (4 interference, 1 activation) | (Deaner, Holzman and Alper 2018) |
| Saccharomyces cerevisiae | dSpCas9 (CRISPRi/CRISPRa)—inducible expression, pGal10 promoterGenome integrated | Several sgRNA (with RNA scaffolds) expression cassettes (SNR52p constitutive RNA pol III promoter)Plasmid expression (centromeric) | Proof of principle, obtention of different violacein biosynthetic pathway products | 3 targets | (Zalatan et al. 2015) |
| Saccharomyces cerevisiae | dLbCas12a-VP (CRISPRa), dSpCas9-RD1152 (CRISPRi), SaCas9 (CRISPRd)—constitutive expression, pTDH3 promoterGenome integrated | Several sgRNA and gRNA in synthetic array between Csy4 recognition sites (TEF1p RNA pol II constitutive promoter)Plasmid expression | Increase β-carotene production | 3 targets (1 activation, 1 interference and 1 deletion) | (Lian et al. 2017) |
| Kluyveromyces marxianus | dSpCas9 (CRISPRi)—constitutive expression, TEF1p promoterPlasmid expression (centromeric) | Several sgRNA expression cassettes (RPR1-tRNAgly,RNA pol III constitutive promoter)Plasmid expression (centromeric) | Increase ethyl acetate production | 6 targets (4 genes) | (Löbs et al. 2018) |
| Yarrowia lipolytica | dSpCas9-Mxi1 (CRISPRi)—constitutive expression, UAS1B8-TEFPlasmid expression | Several sgRNA expression cassettes (SCR1-tRNAgly RNA pol III constitutive promoter)Plasmid expression | Increase HR by repression of the NHEJ machinery | 3 targets (2 genes) | (Schwartz et al. 2017) |
| Saccharomyces cerevisiae[BY4741 (n)] | dSpCas9-VPR (CIRSPRi and CRISPRa)Plasmid expression (centromeric) | Synthetic array of ribozyme-flanked sgRNAs (Gal1p, RNA pol II inducible promoter)Plasmid expression (centromeric) | Proof of principle | 2 targets | (Deaner, Mejia and Alper 2017) |
| Saccharomyces cerevisiae[BY4741 (n)] | dSpCas9-VPR (CIRSPRi and CRISPRa)Plasmid expression (centromeric) | Synthetic array of ribozyme-flanked sgRNAs (TEF1p, RNA pol II constitutive promoter)Plasmid expression (centromeric) | Proof of principle | 4 targets | (Deaner, Mejia and Alper 2017) |
| Yarrowia lipolytica[ATCC 201 249] | dSpCas9 or dFnCas12a (CRISPRi)—constitutive expressionPlasmid expression (centromeric) | Several sgRNA expression cassettes (SCR1-tRNAgly RNA pol III constitutive promoter)Plasmid expression (centromeric) | Proof of principle, decrease protodeoxy-violaceinic acid | 3 targets | (Zhang et al. 2018) |
| Specie [strain(poidy)] . | Cas nuclease tool (CRISPRi/CRISPRa), expression, promoter Plasmid (replication origin)/genome integrated . | Strategy for multiplexed gRNA expression/delivery (expression) Plasmid (replication origin)/genome integrated . | Goal . | Number of targets . | Reference . |
|---|---|---|---|---|---|
| PROKARYOTES | |||||
| E. coli[B0013] | dSpCas9 (CRISPRi)—constitutive expression, trc promoterPlasmid expression | Several sgRNA expression cassettes (constitutive)Plasmid expression (CloDF13ori) | Increase malate titer | 3 targets | (Gao et al. 2018) |
| E. coli[MG1655] | ddAsCas12a (CRISPRi)—constitutive expression, j23100 promoterPlasmid expression (p15A ori) | Several sgRNA expression cassettes (j23119-SpeI constitutive promoter)Plasmid expression (ColE1 ori) | Proof of principle | 3 targets | (Zhang et al. 2017) |
| E. coli[MG1655] | dSpCas9 (CRISPRI/CRISPRa)—constitutive expression, endogenous S. pyogenes promoterPlasmid expression | Several sgRNA expression cassettes (j23119 constitutive promoter) fused to protein binding sequences (scaffold RNAs)Plasmid expression | Proof of principle. Activation of ethanol biosynthesis | 2 targets | (Dong et al. 2018) |
| Bacillus subtilis[BNY] | dSpCas9 (CRISPRi)—- inducible expression, xylA promoterGenome integrated | Several sgRNA expression cassettes (Pveg constitutive promoter)Genome integrated | Increase N-acetylglucosamine titer | 3 targets | (Wu et al. 2018b) |
| Streptomyces coelicolor | dSpCas9 (CRISPRi)—constitutive expression, ermE*p promoterGenome integrated | Several sgRNA expression cassettes (j23119p constitutive promoter)Genome integrated | Proof of principle (knock-out 4 pigmented antibiotic synthesis) | 3 and 4 targets | (Zhao et al. 2018) |
| Pseudomonas putida | dSpaCas9 (CRISPRi)—inducible expression, LacI-Ptac promoterGenome integrated | Several sgRNA expression cassettes (Ptet promoter)Plasmid expression (oriV, Rep) | Proof of principle | 2 targets | (Tan et al. 2018) |
| Corynebacterium glutamicum | dSpCas9 (CRISPRi)—inducible expression, Ptac promoterPlasmid expression | Several sgRNA expression cassettes (Ptac inducible promoter)Plasmid expression | Increase aminoacid production | 3 targets | (Cleto et al. 2016) |
| Bacillus subtilis[BNY] | dSpCas9 (CRISPRi)—inducible expression, PxylA promoterGenome integrated | Several sgRNA expression cassettes (Pveg promoter)Genome integrated | Increase GlcNAc titer | 3 targets | (Wu et al. 2018) |
| Synechocystis | dSpCas9 (CRISPRi)—constitutive expression, PpsbA2 promoterGenome integrated | Several sgRNA expression cassettes (PL31 constitutive promoter)Genome integrated | Reduction of PHB and glycogen accumulation during nitrogen starvation | 4 targets | (Yao et al. 2016) |
| Synechocystis | dSpCas9 (CRISPRi)—inducible expression, PL22 promoterGenome integrated | Several sgRNA expression cassettes (PL22 constitutive promoter)Genome integrated | Carbon flux re-direction for production of fatty alcohols | 6 targets | (Kaczmarzyk et al. 2018) |
| Streptomyces coelicolor[M145] | ddFnCas12a (CRISPRi)—constitutive expression, ermEp* promoterGenome integrated | Native-like CRISPR array (kasOp* constitutive promoter)Genome integrated | Proof of principle (knock-out 3 pigmented antibiotic synthesis) | 3 targets | (Li et al. 2018) |
| EUKARYOTES | |||||
| Saccharomyces cerevisiae[CEN.PK2-a and Sigma 10 560–4A] | dCas9-VPR (CRISPRi and CRISPRa)Plasmid expression (centromeric) | TEF1p-tRNA-sgRNA-tRNA (constitutive)Plasmid expression (centromeric) | Increase 2,3-butanediol titer | 5 targets (4 interference, 1 activation) | (Deaner, Holzman and Alper 2018) |
| Saccharomyces cerevisiae | dSpCas9 (CRISPRi/CRISPRa)—inducible expression, pGal10 promoterGenome integrated | Several sgRNA (with RNA scaffolds) expression cassettes (SNR52p constitutive RNA pol III promoter)Plasmid expression (centromeric) | Proof of principle, obtention of different violacein biosynthetic pathway products | 3 targets | (Zalatan et al. 2015) |
| Saccharomyces cerevisiae | dLbCas12a-VP (CRISPRa), dSpCas9-RD1152 (CRISPRi), SaCas9 (CRISPRd)—constitutive expression, pTDH3 promoterGenome integrated | Several sgRNA and gRNA in synthetic array between Csy4 recognition sites (TEF1p RNA pol II constitutive promoter)Plasmid expression | Increase β-carotene production | 3 targets (1 activation, 1 interference and 1 deletion) | (Lian et al. 2017) |
| Kluyveromyces marxianus | dSpCas9 (CRISPRi)—constitutive expression, TEF1p promoterPlasmid expression (centromeric) | Several sgRNA expression cassettes (RPR1-tRNAgly,RNA pol III constitutive promoter)Plasmid expression (centromeric) | Increase ethyl acetate production | 6 targets (4 genes) | (Löbs et al. 2018) |
| Yarrowia lipolytica | dSpCas9-Mxi1 (CRISPRi)—constitutive expression, UAS1B8-TEFPlasmid expression | Several sgRNA expression cassettes (SCR1-tRNAgly RNA pol III constitutive promoter)Plasmid expression | Increase HR by repression of the NHEJ machinery | 3 targets (2 genes) | (Schwartz et al. 2017) |
| Saccharomyces cerevisiae[BY4741 (n)] | dSpCas9-VPR (CIRSPRi and CRISPRa)Plasmid expression (centromeric) | Synthetic array of ribozyme-flanked sgRNAs (Gal1p, RNA pol II inducible promoter)Plasmid expression (centromeric) | Proof of principle | 2 targets | (Deaner, Mejia and Alper 2017) |
| Saccharomyces cerevisiae[BY4741 (n)] | dSpCas9-VPR (CIRSPRi and CRISPRa)Plasmid expression (centromeric) | Synthetic array of ribozyme-flanked sgRNAs (TEF1p, RNA pol II constitutive promoter)Plasmid expression (centromeric) | Proof of principle | 4 targets | (Deaner, Mejia and Alper 2017) |
| Yarrowia lipolytica[ATCC 201 249] | dSpCas9 or dFnCas12a (CRISPRi)—constitutive expressionPlasmid expression (centromeric) | Several sgRNA expression cassettes (SCR1-tRNAgly RNA pol III constitutive promoter)Plasmid expression (centromeric) | Proof of principle, decrease protodeoxy-violaceinic acid | 3 targets | (Zhang et al. 2018) |
Multiplexed genome regulation events in industrial microorganisms using CRISPRi and CRISPRa.
| Specie [strain(poidy)] . | Cas nuclease tool (CRISPRi/CRISPRa), expression, promoter Plasmid (replication origin)/genome integrated . | Strategy for multiplexed gRNA expression/delivery (expression) Plasmid (replication origin)/genome integrated . | Goal . | Number of targets . | Reference . |
|---|---|---|---|---|---|
| PROKARYOTES | |||||
| E. coli[B0013] | dSpCas9 (CRISPRi)—constitutive expression, trc promoterPlasmid expression | Several sgRNA expression cassettes (constitutive)Plasmid expression (CloDF13ori) | Increase malate titer | 3 targets | (Gao et al. 2018) |
| E. coli[MG1655] | ddAsCas12a (CRISPRi)—constitutive expression, j23100 promoterPlasmid expression (p15A ori) | Several sgRNA expression cassettes (j23119-SpeI constitutive promoter)Plasmid expression (ColE1 ori) | Proof of principle | 3 targets | (Zhang et al. 2017) |
| E. coli[MG1655] | dSpCas9 (CRISPRI/CRISPRa)—constitutive expression, endogenous S. pyogenes promoterPlasmid expression | Several sgRNA expression cassettes (j23119 constitutive promoter) fused to protein binding sequences (scaffold RNAs)Plasmid expression | Proof of principle. Activation of ethanol biosynthesis | 2 targets | (Dong et al. 2018) |
| Bacillus subtilis[BNY] | dSpCas9 (CRISPRi)—- inducible expression, xylA promoterGenome integrated | Several sgRNA expression cassettes (Pveg constitutive promoter)Genome integrated | Increase N-acetylglucosamine titer | 3 targets | (Wu et al. 2018b) |
| Streptomyces coelicolor | dSpCas9 (CRISPRi)—constitutive expression, ermE*p promoterGenome integrated | Several sgRNA expression cassettes (j23119p constitutive promoter)Genome integrated | Proof of principle (knock-out 4 pigmented antibiotic synthesis) | 3 and 4 targets | (Zhao et al. 2018) |
| Pseudomonas putida | dSpaCas9 (CRISPRi)—inducible expression, LacI-Ptac promoterGenome integrated | Several sgRNA expression cassettes (Ptet promoter)Plasmid expression (oriV, Rep) | Proof of principle | 2 targets | (Tan et al. 2018) |
| Corynebacterium glutamicum | dSpCas9 (CRISPRi)—inducible expression, Ptac promoterPlasmid expression | Several sgRNA expression cassettes (Ptac inducible promoter)Plasmid expression | Increase aminoacid production | 3 targets | (Cleto et al. 2016) |
| Bacillus subtilis[BNY] | dSpCas9 (CRISPRi)—inducible expression, PxylA promoterGenome integrated | Several sgRNA expression cassettes (Pveg promoter)Genome integrated | Increase GlcNAc titer | 3 targets | (Wu et al. 2018) |
| Synechocystis | dSpCas9 (CRISPRi)—constitutive expression, PpsbA2 promoterGenome integrated | Several sgRNA expression cassettes (PL31 constitutive promoter)Genome integrated | Reduction of PHB and glycogen accumulation during nitrogen starvation | 4 targets | (Yao et al. 2016) |
| Synechocystis | dSpCas9 (CRISPRi)—inducible expression, PL22 promoterGenome integrated | Several sgRNA expression cassettes (PL22 constitutive promoter)Genome integrated | Carbon flux re-direction for production of fatty alcohols | 6 targets | (Kaczmarzyk et al. 2018) |
| Streptomyces coelicolor[M145] | ddFnCas12a (CRISPRi)—constitutive expression, ermEp* promoterGenome integrated | Native-like CRISPR array (kasOp* constitutive promoter)Genome integrated | Proof of principle (knock-out 3 pigmented antibiotic synthesis) | 3 targets | (Li et al. 2018) |
| EUKARYOTES | |||||
| Saccharomyces cerevisiae[CEN.PK2-a and Sigma 10 560–4A] | dCas9-VPR (CRISPRi and CRISPRa)Plasmid expression (centromeric) | TEF1p-tRNA-sgRNA-tRNA (constitutive)Plasmid expression (centromeric) | Increase 2,3-butanediol titer | 5 targets (4 interference, 1 activation) | (Deaner, Holzman and Alper 2018) |
| Saccharomyces cerevisiae | dSpCas9 (CRISPRi/CRISPRa)—inducible expression, pGal10 promoterGenome integrated | Several sgRNA (with RNA scaffolds) expression cassettes (SNR52p constitutive RNA pol III promoter)Plasmid expression (centromeric) | Proof of principle, obtention of different violacein biosynthetic pathway products | 3 targets | (Zalatan et al. 2015) |
| Saccharomyces cerevisiae | dLbCas12a-VP (CRISPRa), dSpCas9-RD1152 (CRISPRi), SaCas9 (CRISPRd)—constitutive expression, pTDH3 promoterGenome integrated | Several sgRNA and gRNA in synthetic array between Csy4 recognition sites (TEF1p RNA pol II constitutive promoter)Plasmid expression | Increase β-carotene production | 3 targets (1 activation, 1 interference and 1 deletion) | (Lian et al. 2017) |
| Kluyveromyces marxianus | dSpCas9 (CRISPRi)—constitutive expression, TEF1p promoterPlasmid expression (centromeric) | Several sgRNA expression cassettes (RPR1-tRNAgly,RNA pol III constitutive promoter)Plasmid expression (centromeric) | Increase ethyl acetate production | 6 targets (4 genes) | (Löbs et al. 2018) |
| Yarrowia lipolytica | dSpCas9-Mxi1 (CRISPRi)—constitutive expression, UAS1B8-TEFPlasmid expression | Several sgRNA expression cassettes (SCR1-tRNAgly RNA pol III constitutive promoter)Plasmid expression | Increase HR by repression of the NHEJ machinery | 3 targets (2 genes) | (Schwartz et al. 2017) |
| Saccharomyces cerevisiae[BY4741 (n)] | dSpCas9-VPR (CIRSPRi and CRISPRa)Plasmid expression (centromeric) | Synthetic array of ribozyme-flanked sgRNAs (Gal1p, RNA pol II inducible promoter)Plasmid expression (centromeric) | Proof of principle | 2 targets | (Deaner, Mejia and Alper 2017) |
| Saccharomyces cerevisiae[BY4741 (n)] | dSpCas9-VPR (CIRSPRi and CRISPRa)Plasmid expression (centromeric) | Synthetic array of ribozyme-flanked sgRNAs (TEF1p, RNA pol II constitutive promoter)Plasmid expression (centromeric) | Proof of principle | 4 targets | (Deaner, Mejia and Alper 2017) |
| Yarrowia lipolytica[ATCC 201 249] | dSpCas9 or dFnCas12a (CRISPRi)—constitutive expressionPlasmid expression (centromeric) | Several sgRNA expression cassettes (SCR1-tRNAgly RNA pol III constitutive promoter)Plasmid expression (centromeric) | Proof of principle, decrease protodeoxy-violaceinic acid | 3 targets | (Zhang et al. 2018) |
| Specie [strain(poidy)] . | Cas nuclease tool (CRISPRi/CRISPRa), expression, promoter Plasmid (replication origin)/genome integrated . | Strategy for multiplexed gRNA expression/delivery (expression) Plasmid (replication origin)/genome integrated . | Goal . | Number of targets . | Reference . |
|---|---|---|---|---|---|
| PROKARYOTES | |||||
| E. coli[B0013] | dSpCas9 (CRISPRi)—constitutive expression, trc promoterPlasmid expression | Several sgRNA expression cassettes (constitutive)Plasmid expression (CloDF13ori) | Increase malate titer | 3 targets | (Gao et al. 2018) |
| E. coli[MG1655] | ddAsCas12a (CRISPRi)—constitutive expression, j23100 promoterPlasmid expression (p15A ori) | Several sgRNA expression cassettes (j23119-SpeI constitutive promoter)Plasmid expression (ColE1 ori) | Proof of principle | 3 targets | (Zhang et al. 2017) |
| E. coli[MG1655] | dSpCas9 (CRISPRI/CRISPRa)—constitutive expression, endogenous S. pyogenes promoterPlasmid expression | Several sgRNA expression cassettes (j23119 constitutive promoter) fused to protein binding sequences (scaffold RNAs)Plasmid expression | Proof of principle. Activation of ethanol biosynthesis | 2 targets | (Dong et al. 2018) |
| Bacillus subtilis[BNY] | dSpCas9 (CRISPRi)—- inducible expression, xylA promoterGenome integrated | Several sgRNA expression cassettes (Pveg constitutive promoter)Genome integrated | Increase N-acetylglucosamine titer | 3 targets | (Wu et al. 2018b) |
| Streptomyces coelicolor | dSpCas9 (CRISPRi)—constitutive expression, ermE*p promoterGenome integrated | Several sgRNA expression cassettes (j23119p constitutive promoter)Genome integrated | Proof of principle (knock-out 4 pigmented antibiotic synthesis) | 3 and 4 targets | (Zhao et al. 2018) |
| Pseudomonas putida | dSpaCas9 (CRISPRi)—inducible expression, LacI-Ptac promoterGenome integrated | Several sgRNA expression cassettes (Ptet promoter)Plasmid expression (oriV, Rep) | Proof of principle | 2 targets | (Tan et al. 2018) |
| Corynebacterium glutamicum | dSpCas9 (CRISPRi)—inducible expression, Ptac promoterPlasmid expression | Several sgRNA expression cassettes (Ptac inducible promoter)Plasmid expression | Increase aminoacid production | 3 targets | (Cleto et al. 2016) |
| Bacillus subtilis[BNY] | dSpCas9 (CRISPRi)—inducible expression, PxylA promoterGenome integrated | Several sgRNA expression cassettes (Pveg promoter)Genome integrated | Increase GlcNAc titer | 3 targets | (Wu et al. 2018) |
| Synechocystis | dSpCas9 (CRISPRi)—constitutive expression, PpsbA2 promoterGenome integrated | Several sgRNA expression cassettes (PL31 constitutive promoter)Genome integrated | Reduction of PHB and glycogen accumulation during nitrogen starvation | 4 targets | (Yao et al. 2016) |
| Synechocystis | dSpCas9 (CRISPRi)—inducible expression, PL22 promoterGenome integrated | Several sgRNA expression cassettes (PL22 constitutive promoter)Genome integrated | Carbon flux re-direction for production of fatty alcohols | 6 targets | (Kaczmarzyk et al. 2018) |
| Streptomyces coelicolor[M145] | ddFnCas12a (CRISPRi)—constitutive expression, ermEp* promoterGenome integrated | Native-like CRISPR array (kasOp* constitutive promoter)Genome integrated | Proof of principle (knock-out 3 pigmented antibiotic synthesis) | 3 targets | (Li et al. 2018) |
| EUKARYOTES | |||||
| Saccharomyces cerevisiae[CEN.PK2-a and Sigma 10 560–4A] | dCas9-VPR (CRISPRi and CRISPRa)Plasmid expression (centromeric) | TEF1p-tRNA-sgRNA-tRNA (constitutive)Plasmid expression (centromeric) | Increase 2,3-butanediol titer | 5 targets (4 interference, 1 activation) | (Deaner, Holzman and Alper 2018) |
| Saccharomyces cerevisiae | dSpCas9 (CRISPRi/CRISPRa)—inducible expression, pGal10 promoterGenome integrated | Several sgRNA (with RNA scaffolds) expression cassettes (SNR52p constitutive RNA pol III promoter)Plasmid expression (centromeric) | Proof of principle, obtention of different violacein biosynthetic pathway products | 3 targets | (Zalatan et al. 2015) |
| Saccharomyces cerevisiae | dLbCas12a-VP (CRISPRa), dSpCas9-RD1152 (CRISPRi), SaCas9 (CRISPRd)—constitutive expression, pTDH3 promoterGenome integrated | Several sgRNA and gRNA in synthetic array between Csy4 recognition sites (TEF1p RNA pol II constitutive promoter)Plasmid expression | Increase β-carotene production | 3 targets (1 activation, 1 interference and 1 deletion) | (Lian et al. 2017) |
| Kluyveromyces marxianus | dSpCas9 (CRISPRi)—constitutive expression, TEF1p promoterPlasmid expression (centromeric) | Several sgRNA expression cassettes (RPR1-tRNAgly,RNA pol III constitutive promoter)Plasmid expression (centromeric) | Increase ethyl acetate production | 6 targets (4 genes) | (Löbs et al. 2018) |
| Yarrowia lipolytica | dSpCas9-Mxi1 (CRISPRi)—constitutive expression, UAS1B8-TEFPlasmid expression | Several sgRNA expression cassettes (SCR1-tRNAgly RNA pol III constitutive promoter)Plasmid expression | Increase HR by repression of the NHEJ machinery | 3 targets (2 genes) | (Schwartz et al. 2017) |
| Saccharomyces cerevisiae[BY4741 (n)] | dSpCas9-VPR (CIRSPRi and CRISPRa)Plasmid expression (centromeric) | Synthetic array of ribozyme-flanked sgRNAs (Gal1p, RNA pol II inducible promoter)Plasmid expression (centromeric) | Proof of principle | 2 targets | (Deaner, Mejia and Alper 2017) |
| Saccharomyces cerevisiae[BY4741 (n)] | dSpCas9-VPR (CIRSPRi and CRISPRa)Plasmid expression (centromeric) | Synthetic array of ribozyme-flanked sgRNAs (TEF1p, RNA pol II constitutive promoter)Plasmid expression (centromeric) | Proof of principle | 4 targets | (Deaner, Mejia and Alper 2017) |
| Yarrowia lipolytica[ATCC 201 249] | dSpCas9 or dFnCas12a (CRISPRi)—constitutive expressionPlasmid expression (centromeric) | Several sgRNA expression cassettes (SCR1-tRNAgly RNA pol III constitutive promoter)Plasmid expression (centromeric) | Proof of principle, decrease protodeoxy-violaceinic acid | 3 targets | (Zhang et al. 2018) |
Fewer examples can be found of the use of CRISPRa because of the requirement of functional transcription activators. Frequently, CRISPRa applications are used in combination with CRISPRi approaches (Lian et al. 2017). Combination of CRISPRi and CRISPRa using RNA scaffolds was accomplished using similar designs in E. coli (Dong et al. 2018) and S. cerevisiae (Zalatan et al. 2015; Lian et al. 2017). In both cases, protein-binding RNA sequences were fused to the 3′ end of sgRNAs of Cas9. These protein-recruitment RNA sequences have a high affinity for certain proteins, such as the MCP (used both in E. coli and S. cerevisiae) or the PCP and Com proteins (tested only in S. cerevisiae). By fusing the transcriptional activation domain SoxS (in E. coli) or VP64 (in S. cerevisiae) to these RNA binding proteins, expression of a gene downstream a target promoter can be enhanced, and pathway fluxes sequentially directed (Zalatan et al. 2015; Dong et al. 2018).
Challenges and outlook of multiplexing
In recent years, remarkable progress has been achieved in the field of multiplexed genome editing. Besides broadening the spectrum of microorganisms that can be engineered using CRISPR-Cas endonucleases, novel approaches have recently been implemented in terms of gRNA and dDNA delivery for increasing multiplexing efficiency. Below, a summary is provided of the major challenges related to the development of efficient multiplexing CRISPR-Cas systems in microorganisms.
Guide design: availability of highly efficient guides (sgRNA, crRNA) obtained either through software prediction based on generalized well defined guide-design principles, preferential PAM domains and secondary structure prediction (Chari et al. 2015; Graham and Root 2015; Moreno-Mateos et al. 2015; Labuhn et al. 2018; Liao et al. 2018), or through pre-characterization of the functionality of individual guide performance in single target genome editing experiments.
dDNA design and delivery: dDNA can be part of a vector, provided as a linear fragment, single or double-stranded, a single fragment of multiple fused repair templates or provided as multiple DNA fragments. Proper characterization of such elements (e. g. by determination of optimal size of homology sequences) for the specific host organism to be edited is required. Additionally, dDNA might be stabilized by chemical modifications (Lee et al. 2017).
Controllable expression of CRISPR elements: tight expression control of guides and nucleases, via tuning of expression either by systematic variation of constitutive promoters or by using inducible systems. Optimized promoter and terminator sequences are required to fine-tune the expression of the different CRISPR elements (Feng et al. 2018).
Innovative plasmid assembly methods: smart DNA construction schemes to develop single or multi-plasmid systems containing elements of the CRISPR-Cas system (gRNA, Cas, dDNA, selective marker), including techniques to incorporate repetitive DNA elements such as the repeat sequences of CRISPR arrays (Cress et al. 2015; Deaner, Holzman and Alper 2018; Liao et al. 2018). Recently, a cloning-free approach was developed in S. cerevisiae for the obtainment of up to 6 simultaneous deletions using SpCas9 with a efficiency of 23.3%. The strategy combined multiple successful strategies presented in this manuscript. Three transcripts (each of them containing two gRNAs flanked by tRNAgly sequences were expressed from a single plasmid encoding for SpCas9. The assembly of the plasmid via Golden Gate reaction was performed in the yeast (Zhang et al. 2019).
Organism-specific CRISPR tools: smart choice of the CRISPR-Cas expression approach depending on final application of the production strain or the target organism of choice. For instance, CRISPR-Cas tools can be combined with the introduction of dDNA containing selective markers, which makes screening and selection of positive clones more efficient. This approach can be used in proof of principle studies, whereas in other cases, marker-free strains are important. In a similar way, guides and Cas nucleases can be co-expressed from plasmid-borne expression cassettes or expressed sequentially in strains pre-expressing the Cas nuclease from a second plasmid or from a genome integrated copy.
Editing conditions: optimization of organism-specific CRISPR-Cas delivery systems and recovery protocols. Cell synchronization protocols in combination with CRISPR-Cas systems have been used in human cells to enhance HDR versus NHEJ repair (Lin et al. 2014). In some yeasts, the highest rate of HDR over NHEJ is shown during S-phase (Tsakraklides et al. 2015). Therefore, this strategy has been already proposed for its use in industrial microorganisms (Juergens et al. 2018).
Novel or improved endonucleases: these endonucleases should have alternative or less-stringent PAM recognition selection. In addition, nucleases are preferred that are smaller, more specific and more active, as reviewed by Kleinstiver et al. (2019). Cas9 variants with distinct PAM-recognizing features have been obtained by laboratory evolution (Hu et al. 2018), as well as by structure-guided protein engineering of Cas12a (Kleinstiver et al. 2019). The discovery and characterization of novel Cas endonucleases (Liu et al. 2019)- can also extend the PAM compatibility (Burstein et al. 2017; Harrington et al. 2018; Strecker et al. 2019). At the same time, multiple endonucleases can be used in orthogonal designs in order to edit the genome and regulate gene expression simultaneously (Kweon et al. 2017; Lian et al. 2017). Alternatively, orthogonal designs can incorporate modified gRNAs, as described for Cas12a by Breinig et al. (2019).
In multiplexed genome editing experiments, a negative correlation is experienced between the number of targets and the amount of obtained colonies after transformation in most microorganisms. The introduction of DSBs dramatically reduces the cell survival rate, and this causes limited numbers of simultaneous modifications as a delicate balance between DNA cleavage and repair needs to be established. Alternatively, multiplexed single-base editing does not depend on DSB generation nor dDNA supply and can be used to introduce nucleic acid base changes at a targeted window of DNA (Eid, Alshareef and Mahfouz 2018; Wu et al. 2018a). In this approach, deactivated or nickase Cas nuclease variants coupled to base editors (cytidine or adenine deaminases) are directed to a target site by the gRNA (Marx 2018). Nucleotide changes are introduced in a targeted DNA window rather than in a precise DNA position, which makes this technique less accurate for the introduction of single point mutations. Multiplexed single-base editing has been recently implemented in prokaryotic microorganisms (Banno et al. 2018; Wang et al. 2018b). Useful applications of this technique remain limited to phenotype modifications linked to single nucleotide polymorphisms (SNPs) and to the possibility of introducing stop-codons for gene inactivation in coordination with the presence of a PAM sequence in a determined window (Arazoe, Kondo and Nishida 2018). Although very promising, further development and control of this base editing approach are required in order to broaden the type of modifications and to avoid unwanted mutations (Nishida et al. 2016).
Advances in multiplexed genome editing of microorganisms can significantly accelerate future strain construction programs of cell factories with unprecedented efficiencies. Therefore, the CRISPR revolution continues: new tools and workflows are being developed to broaden the range of functionalities of currently used CRISPR-Cas systems as well as knowledge about the mechanism of these systems. As identified in this review, dedicated optimization of each of the elements involved in CRISPR-Cas genome editing is crucial for efficient multiplex genome editing and for stretching the number of simultaneous editing events.
ACKNOWLEDGEMENTS
P.D-L, J.A.R and J.v.d.O received funding from the research programme Building Blocks of Life by the Netherlands Organization for Scientific Research (NWO, Project 737.016.005), and J.A.R and R.V. also received funding from the European Union's Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie Grant agreement no 764591 (ITN-SynCrop).
Conflict of interest. None declared.