-
PDF
- Split View
-
Views
-
Cite
Cite
Amber Harper, Luis Baudouin Gonzalez, Anna Schönauer, Ralf Janssen, Michael Seiter, Michaela Holzem, Saad Arif, Alistair P McGregor, Lauren Sumner-Rooney, Widespread retention of ohnologs in key developmental gene families following whole-genome duplication in arachnopulmonates, G3 Genes|Genomes|Genetics, Volume 11, Issue 12, December 2021, jkab299, https://doi.org/10.1093/g3journal/jkab299
Close - Share Icon Share
Abstract
Whole-genome duplications (WGDs) have occurred multiple times during animal evolution, including in lineages leading to vertebrates, teleosts, horseshoe crabs, and arachnopulmonates. These dramatic events initially produce a wealth of new genetic material, generally followed by extensive gene loss. It appears, however, that developmental genes such as homeobox genes, signaling pathway components and microRNAs are frequently retained as duplicates (so-called ohnologs) following WGD. These not only provide the best evidence for WGD, but an opportunity to study its evolutionary consequences. Although these genes are well studied in the context of vertebrate WGD, similar comparisons across the extant arachnopulmonate orders are patchy. We sequenced embryonic transcriptomes from two spider species and two amblypygid species and surveyed three important gene families, Hox, Wnt, and frizzled, across these and 12 existing transcriptomic and genomic resources for chelicerates. We report extensive retention of putative ohnologs, further supporting the ancestral arachnopulmonate WGD. We also found evidence of consistent evolutionary trajectories in Hox and Wnt gene repertoires across three of the six arachnopulmonate orders, with interorder variation in the retention of specific paralogs. We identified variation between major clades in spiders and are better able to reconstruct the chronology of gene duplications and losses in spiders, amblypygids, and scorpions. These insights shed light on the evolution of the developmental toolkit in arachnopulmonates, highlight the importance of the comparative approach within lineages, and provide substantial new transcriptomic data for future study.
Introduction
The duplication of genetic material is an important contributor to the evolution of morphological and physiological innovations (Ohno 1970; Zhang 2003). The most dramatic example of this is whole-genome duplication (WGD), when gene copy numbers are doubled and retained paralogs (ohnologs) can then share ancestral functions (subfunctionalization) and/or evolve new roles (neofunctionalization; Ohno 1970; Force et al. 1999; Lynch and Conery 2000). The occurrence of two rounds (2R) of WGD in the early evolution of vertebrates has long been associated with their taxonomic and morphological diversity (e.g., Ohno 1970; Holland et al. 1994; Dehal and Boore 2005; Holland 2013a), and a subsequent 3R in teleosts is frequently linked to their success as the most diverse vertebrate group (e.g., Meyer and Schartl 1999; Glasauer and Neuhauss 2014). However, this remains controversial and difficult to test (Donoghue and Purnell 2005) and in several animal lineages, there is no clear association between WGD and diversification (Mark Welch et al. 2008; Flot et al. 2013; Havlak et al. 2014; Kenny et al. 2016; Nong et al. 2020). Along with vertebrates, chelicerates also appear to be hotspots of WGD, with up to three rounds reported in horseshoe crabs (Havlak et al. 2014; Kenny et al. 2016; Nong et al. 2020; Shingate et al. 2020), and potentially two further rounds within the spider clade Synspermiata (Král et al. 2019). Chelicerates demonstrate a highly variable body plan, occupy a wide range of habitats and ecological niches, and have evolved a variety of biologically important innovations such as venoms and silks (Schwager et al. 2015). They therefore offer an excellent opportunity for comparison with vertebrates concerning the implications of WGD for morphological and taxonomic diversity, and genome evolution in its wake.
The house spider Parasteatoda tepidariorum has emerged as a model species to study the impacts of WGD on arachnid evolution and development. Genomic and functional developmental studies have found retained ohnologs of many important genes, with evidence for neo- and subfunctionalization compared to single-copy orthologs in arachnids lacking WGD (Janssen et al. 2015; Leite et al. 2016; Turetzek et al. 2016, 2017; Schwager et al. 2017; Leite et al. 2018; Baudouin-Gonzalez et al. 2021). Work on the scorpions Centruroides sculpturatus and Mesobuthus martensii has consistently complemented findings in P. tepidariorum, with genomic studies recovering many ohnologs retained in common with spiders (Sharma et al. 2014b; Di et al. 2015; Sharma et al. 2015; Schwager et al. 2017; Leite et al. 2018). In the past few years, several additional spider genomes have become available, providing an opportunity to get a more detailed view of genome evolution following WGD. Although synteny analysis remains the gold standard for the identification of ohnologs, the required chromosome-level genomic assemblies remain relatively scarce. Work on the P. tepidariorum, C. sculpturatus, and M. martensii genomes has been complemented by targeted studies of individual gene families and transcriptomic surveys (Schwager et al. 2007; Sharma et al. 2012; Janssen et al. 2015; Leite et al. 2018; Gainett and Sharma 2020; Baudouin-Gonzalez et al. 2021). Combined with phylogenetic analyses, the identification of duplications can provide support for WGD events and their timing in arachnid evolution. Although transcriptomes can yield variant sequences of individual genes, from different alleles or individuals in mixed samples, these are generally straight-forward to filter out from truly duplicated loci owing to substantial sequence divergence in the latter. They also offer the double-edged sword of capturing gene expression, rather than presence in the genome; pseudogenized or silenced duplicates are not detected, but neither are functional genes if they are not expressed at the sampled timepoint or tissue. Such studies have produced strong additional evidence for an ancestral WGD, with patterns of duplication coinciding with our expectations for arachnopulmonate ohnologs (Clarke et al. 2014; 2015; Sharma et al. 2015; Turetzek et al. 2017; Bonatto Paese et al. 2018; Leite et al. 2018; Gainett et al. 2020; Gainett and Sharma 2020; Baudouin-Gonzalez et al. 2021).
Comparison of WGD events among arachnopulmonates, horseshoe crabs, and vertebrates indicates that despite extensive gene loss following duplication events, certain gene families are commonly retained following duplication (Holland et al. 1994; Schwager et al. 2007; Kuraku and Meyer 2009; Di et al. 2015; Sharma et al. 2015; Kenny et al. 2016; Leite et al. 2016, 2018; Schwager et al. 2017). These typically include genes from the conserved developmental “toolkit” of transcription factors (TFs), cell signaling ligands and receptors, and microRNAs (Erwin 2009). Among these, several have stood out as focal points in the study of gene and genome duplications. The Hox group of homeobox genes regulate the identity of the body plan along the antero-posterior axis of all bilaterian animals (McGinnis and Krumlauf 1992; Abzhanov et al. 1999; Carroll et al. 2005; Pearson et al. 2005; Hueber and Lohmann 2008; Holland 2013b). Four clusters of these key developmental genes were partially retained after 1R and 2R in vertebrates (Holland et al. 1994; Meyer and Schartl 1999; Kuraku and Meyer 2009; Pascual-Anaya et al. 2013), and the arachnopulmonate WGD is evident in the almost universal retention of Hox gene duplicates in sequenced genomes, with two ohnologs of all 10 arthropod Hox genes in the scorpion M. martensii (Di et al. 2015; Leite et al. 2018), all except Hox3 being represented by two copies in C. sculpturatus (Leite et al. 2018), and all except fushi tarazu (ftz) in P. tepidariorum (Schwager et al. 2017). Systematic studies of Hox gene expression patterns in the latter demonstrated that all nine pairs of Hox paralogs exhibit signs of sub- or neofunctionalization (Schwager et al. 2017). This high level of retention and expression divergence lends strong support to the importance of Hox gene duplication in the evolution of the arachnopulmonate body plan, and further consolidates the position of this gene family as a key indicator of WGD. Additionally, arachnids lacking WGD, such as ticks, mites, and harvestmen, exhibit single copies of the Hox genes, with no evidence for duplication via other mechanisms (Grbić et al. 2011; Pace et al. 2016; Leite et al. 2018; Gainett et al. 2021).
In addition to TFs, the ligands and receptors of some signaling pathways of the developmental toolkit (e.g., Hedgehog, Wnt, TGF-ß, NHR) also demonstrate higher copy numbers in vertebrates and other groups subject to WGD, including arachnopulmonates (Holland et al. 1994; Meyer and Schartl 1999; Shimeld 1999; Pires-daSilva and Sommer 2003; Cho et al. 2010; Janssen et al. 2010; Hogvall et al. 2014; Janssen et al. 2015). The Wnt signaling pathway plays many important roles during animal development, including segmentation and patterning of the nervous system, eyes, and gut (Erwin 2009; Murat et al. 2010). In the canonical pathway, Wnt ligands bind to transmembrane receptors, such as Frizzled, to trigger translocation of ß-catenin to the nucleus and mediate regulation of gene expression (Cadigan and Nusse 1997; Hamilton et al. 2001; Logan and Nusse 2004; van Amerongen and Nusse 2009). There are 13 subfamilies of Wnt genes found in bilaterians, as well as multiple receptor families and downstream components. In contrast to the extensive retention of Hox ohnologs following WGD, Wnt duplicates in P. tepidariorum appear to be restricted to Wnt7 and Wnt11, with the remaining eight subfamilies represented by single genes (Janssen et al. 2010). However, these are the only reported Wnt gene duplications in arthropods despite several recent surveys (Bolognesi et al. 2008; Murat et al. 2010; Hayden and Arthur 2013; Meng et al. 2013; Chipman et al. 2014; Hogvall et al. 2014; Janssen and Posnien 2014; Kao et al. 2016; Holzem et al. 2019), and beyond P. tepidariorum no other arachnopulmonates have been systematically searched.
Several Wnt families have also been retained after the 1R and 2R events in vertebrates, for example there are two copies each of Wnt2, Wnt3, Wnt5, Wnt7, Wnt8, Wnt9, and Wnt10 in humans (Miller 2001; Janssen et al. 2010). However, no subfamilies are represented by three or four copies in humans and so there is some consistency with arachnopulmonates in that the Wnts may be more conservative markers of WGD, to be used in combination with Hox and other homeobox genes.
Similarly, duplications within the four frizzled gene subfamilies appear to be restricted to arachnopulmonates among arthropods, wherein only fz4 is duplicated in both P. tepidariorum and M. martensii (Janssen et al. 2015).
The extensive and consistent retention of key developmental genes, like Hox genes apparent in P. tepidariorum and C. sculpturatus, and Wnt genes in P. tepidariorum, strongly support the occurrence of an ancestral WGD in arachnopulmonates. However, data are only available for a handful of species so far, resulting in very patchy taxonomic sampling. For example, only P. tepidariorum, Pholcus phalangioides and recently, Aphonopelma hentzi, have been comprehensively surveyed for homeobox genes among spiders (Leite et al. 2018; Ontano et al. 2021), omitting the large and derived retrolateral tibial apophysis (RTA) clade, which includes jumping spiders, crab spiders, and other free hunters, and the systematic identification of Wnt genes has been restricted to only P. tepidariorum. Spiders and scorpions are by far the most speciose of the arachnopulmonates, and there may be additional diversity in their repertoires of these important developmental gene families of which we are not yet aware.
In addition, and perhaps more urgently, only two of the six arachnopulmonate lineages have dominated the field thus far; sufficient genomic information for comparison is lacking beyond spiders and scorpions. Also represented in Arachnopulmonata are the amblypygids (whip spiders), relatively understudied and enigmatic animals comprising around 190 extant species. They exhibit highly derived pedipalp morphology, which are adapted to form raptorial appendages, and of the first pair of walking legs, which are antenniform and can comprise more than 100 segments (Weygoldt 2009). Despite the scarcity of transcriptomic or genomic data for amblypygids [see Gainett et al. (2020) for recent advances; Gainett and Sharma (2020)], their widely accepted position within Arachnopulmonata implies that they were also subject to an ancestral WGD. A recent survey of the Phrynus marginemaculatus transcriptome supported this in the recovery of multiple duplicate Hox and leg gap genes (Gainett and Sharma 2020). Particularly given the derived nature of their appendages, this group could shed substantial light on genomic and morphological evolution following WGD. The position of pseudoscorpions was also recently resolved within Arachnopulmonata, complemented by widespread homeobox gene duplication (Ontano et al. 2021).
To better understand the genomic consequences of WGD in a greater diversity of arachnopulmonate lineages, we sequenced de novo embryonic transcriptomes from two spiders belonging to the derived RTA clade and two amblypygids. We surveyed Hox, Wnt, and frizzled genes in these species and existing genomic and transcriptomic resources for comparison with other arachnids, both with and without an ancestral WGD, improving sampling at both the order and suborder levels.
Materials and methods
Embryo collection and fixation
Embryos of mixed ages were collected from captive females of the amblypygids Charinus acosta (Charinidae; parthenogenetic, collected at 1 day, 1 month, and 2 months after laying; equivalent to approximately 1%, 30%, and 60% of development) and Euphrynichus bacillifer (Neoamblypygi: Phrynichidae; mated, collected at approximately 30% of development), the wolf spider Pardosa amentata (collected in Oxford, UK, equivalent to stages 11/12 in P. tepidariorum) and mixed-stage embryos of the jumping spider Marpissa muscosa (kindly provided by Philip Steinhoff and Gabriele Uhl, University of Greifswald) and stored in RNAlater. Phalangium opilio were collected in Uppsala, Sweden, and developmental series of embryos ranging from egg deposition to the end of embryogenesis were collected for sequencing.
Transcriptomics
We extracted total RNA from embryos, pooled by species, of C. acosta, E. bacillifer, P. amentata, and M. muscosa using QIAzol lysis reagent and following the manufacturer’s instructions (Qiagen). Libraries were prepared using a TruSeq RNA kit (including polyA selection) and sequenced on the NovaSeq platform (100 bp PE, Edinburgh Genomics). Following quality assessment using FastQC v0.11.9 (Andrews 2010), erroneous k-mers were removed (rCorrector, default settings; Song and Florea 2015), and unfixable read pairs (from low-expression homolog pairs or containing too many errors) were discarded using a custom Python script (available at https://github.com/harvardinformatics/TranscriptomeAssemblyTools/blob/master/FilterUncorrectabledPEfastq.py courtesy of Adam Freeman). Adapter sequences were identified and removed and low-quality ends (phred score cutoff = 5) trimmed using TrimGalore! v0.6.5 (available at https://github.com/FelixKrueger/TrimGalore). De novo transcriptome assembly was performed using only properly paired reads with Trinity v2.10.0 (Haas et al. 2013) using default settings. Transcriptome completeness was assessed on the longest isoform per gene using BUSCO v4.0.2 (Seppey et al. 2019), the arachnid database (arachnida_odb10 created on 2019-11-20; 10 species, 2934 BUSCOs), and the arthropod database (arthropoda_odb10 created on 2019-11-20; 90 species, 1013 BUSCOs). Reads and assembled transcriptomes are available on SRA and TSA, respectively, under BioProject PRJNA707377.
Identification of gene candidates
To identify Hox, Wnt, and frizzled gene candidates across chelicerates, we performed BLAST searches (P-value 0.05) against existing genomic and transcriptomic resources and the four new embryonic transcriptomes generated in this study. Hox, Wnt, and Frizzled peptide sequences previously identified in P. tepidariorum and Ixodes scapularis were reciprocally blasted against the respective NCBI proteomes to confirm their identity and the top hit was selected (Supplementary File S1; Janssen et al.2010, 2015; Schwager et al. 2017; Leite et al. 2018). Hox protein sequences previously identified in C. sculpturatus (Schwager et al. 2017) and P. opilio (Leite et al. 2018) were reciprocally blasted against the respective NCBI proteomes to confirm their identity and the top hit was selected (Supplementary File S1). These sequences, along with the Hox and Fz peptide sequences identified in the M. martensii genome, were used as query sequences in our analysis (Di et al. 2015; Janssen et al. 2015).
BLASTP searches were performed against the NCBI proteomes of Stegodyphus dumicola, C. sculpturatus, Tetranychus urticae, and Limulus polyphemus. TBLASTN searches were performed against C. acosta, E. bacillifer, P. amentata, and M. muscosa (this study); the transcriptomes of P. phalangioides (Turetzek, Torres-Oliva, Kaufholz, Prpic, Posnien, in preparation), Phoxichilidium femoratum (Ballesteros et al. 2021) and P. opilio (PRJNA236471); and the genomes of Pardosa pseudoannulata (PRJNA512163), Acanthoscurria geniculata (PRJNA222716), and M. martensii (CDS sequence file; Cao et al. 2013). We then predicted the peptide sequences using the Translate ExPASy online tool (https://web.expasy.org/translate/; default settings). Protein sequence identity was confirmed by reciprocal BLAST against the NCBI database and the construction of maximum-likelihood trees. Where more than one sequence was identified as a potential candidate for a single gene, nucleotide, and protein alignments were inspected to eliminate the possibility that they were isoforms, individual variants, or fragments of the same gene. Only the longest isoforms and gene fragments were selected for phylogenetic analysis (Supplementary File S1). All Hox sequences identified contain a complete or partial homeodomain (Supplementary Files S2 and S3). The TBLASTN search in P. phalangioides identified a Ubx gene not reported previously by Leite et al. (2018). Our BLAST searches did not recover Cs-ftz-a (Schwager et al. 2017), Tu-Antp-2 (Grbić et al. 2011) or Is-Wnt8 (Janssen et al. 2010) but we included the previously identified sequences in our subsequent phylogenetic analysis. The Trinity transcript accession numbers of all identified sequences, NCBI protein accession and other sequence identifiers used in the subsequent phylogenetic analysis are found in Supplementary File S1.
Due to high levels of fragmentation in the P. opilio transcriptome, multiple nonoverlapping fragments were found to align with query Wnt sequences. To verify the identity and relationship of these fragments, primers were designed against the 5ʹ-most and 3ʹ-most ends of the aligned series (see Supplementary File S1). Total RNA was extracted using Trizol (Invitrogen) according to the manufacturer’s instructions and cDNA produced using the SuperScriptII first-strand synthesis system (Invitrogen) for RT-PCR using the designed primer pairs. PCR products were sequenced by Macrogen Europe to confirm predicted combinations of fragmented transcripts (see Supplementary File S1).
Phylogenetic analysis
Hox, Wnt, and Frizzled protein predictions were retrieved from NCBI for the insects Drosophila melanogaster, Bombyx mori, Tribolium castaneum; the crustacean Daphnia pulex; and the onychophoran Euperipatoides kanangrensis (Supplementary File S1). The Hox, Wnt, and Frizzled peptide sequences for the myriapod Strigamia maritima were retrieved from Chipman et al. (2014; Supplementary File S1). Alignments were performed in Clustal Omega using default parameters (Larkin et al. 2002; Sievers et al. 2011), with the exception of the full Hox protein sequences, which were aligned using COBALT (Papadopoulos and Agarwala 2007). Maximum-likelihood trees were generated from whole-sequence alignments to assign sequences to families and study the relationship between candidate duplicates. Phylogenetic analyses were performed in IQ-Tree (v2.0.3, Nguyen et al. 2015) using ModelFinder to identify optimal substitution models (JTT + F + R9 for full Hox sequences, LG + G4 for Hox homeodomain sequences, GTR +F + I+ G4 for Hox homeobox sequences, LG + R8 for Wnt, JTT+R8 for Fz; Kalyaanamoorthy et al. 2017) and 100,000 bootstrap replicates. Trees were visualized in FigTree v.1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/) and tidied in Adobe Illustrator. Hox sequences were additionally analyzed using RaXML v8 (Stamatakis 2014), using the same substitution models chosen by Iqtree and the automatic bootstopping algorithm (Pattengale et al. 2009). Alignments are provided in Supplementary Files S2–S4 and S9–S11.
Results
Transcriptome assemblies
To further study the outcomes of WGD in the ancestor of arachnopulmonates, we carried out RNA-Seq on embryos of two further spider species, P. amentata and M. muscosa, and two species of amblypygids, C. acosta and E. bacillifer.
RNA-Seq for the four species produced between 222,479,664 and 272,844,971 raw reads, reduced to between 95% and 96.2% after processing. Trinity assembled between 184,142 and 316,021 transcripts in up to 542,344 isoforms (Table 1). Contig N50 ranged from 592 bp in M. muscosa to 978 bp in E. bacillifer, and from 1461 bp (M. muscosa) to 2671 bp (E. bacillifer) in the most highly expressed genes (representing 90% of total normalized expression; Table 1).
Assembly metrics for transcriptomes of Charinus acosta, Euphrynichus bacillifer, Marpissa muscosa, and Pardosa amentata
| Species . | Raw reads . | Processed reads . | #Trinity genes . | #Trinity isoforms . | Contig N50 (bases)a . | Ex90N50 (bases)b . | #Ex90N50 genesc . | Arachnid BUSCO scores (C[D], F, M)d . | Arthropod BUSCO scores (C[D], F, M)e . |
|---|---|---|---|---|---|---|---|---|---|
| C. Acosta | 272,844,971 | 260,853,757 | 237,678 | 334,267 | 896 | 2,406 | 31,012 | 94.9% [12.9%], 1.0%, 4.1% | 93.2% [9.5%], 1.5%, 5.3% |
| E. bacillifer | 249,938,618 | 239,034,000 | 184,142 | 285,861 | 978 | 2,671 | 22,647 | 93.8% [7.1%], 1.5%, 4.7% | 92.9% [3.5%], 0.7%, 6.4% |
| P. amentata | 266,764,548 | 256,911,378 | 316,021 | 542,344 | 652 | 1,758 | 38,423 | 95.4% [5.3%], 0.9%, 3.7% | 92.9% [4.5%], 1.2%, 5.9% |
| M. muscosa | 222,479,664 | 211,848,357 | 276,943 | 473,878 | 592 | 1,461 | 46,196 | 94.7% [6.1%], 1.3%, 4.0% | 90.8% [3.9%], 1.9%, 7.3% |
| Species . | Raw reads . | Processed reads . | #Trinity genes . | #Trinity isoforms . | Contig N50 (bases)a . | Ex90N50 (bases)b . | #Ex90N50 genesc . | Arachnid BUSCO scores (C[D], F, M)d . | Arthropod BUSCO scores (C[D], F, M)e . |
|---|---|---|---|---|---|---|---|---|---|
| C. Acosta | 272,844,971 | 260,853,757 | 237,678 | 334,267 | 896 | 2,406 | 31,012 | 94.9% [12.9%], 1.0%, 4.1% | 93.2% [9.5%], 1.5%, 5.3% |
| E. bacillifer | 249,938,618 | 239,034,000 | 184,142 | 285,861 | 978 | 2,671 | 22,647 | 93.8% [7.1%], 1.5%, 4.7% | 92.9% [3.5%], 0.7%, 6.4% |
| P. amentata | 266,764,548 | 256,911,378 | 316,021 | 542,344 | 652 | 1,758 | 38,423 | 95.4% [5.3%], 0.9%, 3.7% | 92.9% [4.5%], 1.2%, 5.9% |
| M. muscosa | 222,479,664 | 211,848,357 | 276,943 | 473,878 | 592 | 1,461 | 46,196 | 94.7% [6.1%], 1.3%, 4.0% | 90.8% [3.9%], 1.9%, 7.3% |
Based on longest isoform per gene.
Same as contig N50 but based on top most highly expressed genes that represent 90% of the total normalized expression data.
The number of genes for which Ex90N50 is calculated.
Ten species, n = 2934 BUSCOs; C = Complete [D = Duplicated], F = Fragmented, M = Missing.
Ninety species, n = 1013 BUSCOs; C = Complete [D = Duplicated], F = Fragmented, M = Missing.
Assembly metrics for transcriptomes of Charinus acosta, Euphrynichus bacillifer, Marpissa muscosa, and Pardosa amentata
| Species . | Raw reads . | Processed reads . | #Trinity genes . | #Trinity isoforms . | Contig N50 (bases)a . | Ex90N50 (bases)b . | #Ex90N50 genesc . | Arachnid BUSCO scores (C[D], F, M)d . | Arthropod BUSCO scores (C[D], F, M)e . |
|---|---|---|---|---|---|---|---|---|---|
| C. Acosta | 272,844,971 | 260,853,757 | 237,678 | 334,267 | 896 | 2,406 | 31,012 | 94.9% [12.9%], 1.0%, 4.1% | 93.2% [9.5%], 1.5%, 5.3% |
| E. bacillifer | 249,938,618 | 239,034,000 | 184,142 | 285,861 | 978 | 2,671 | 22,647 | 93.8% [7.1%], 1.5%, 4.7% | 92.9% [3.5%], 0.7%, 6.4% |
| P. amentata | 266,764,548 | 256,911,378 | 316,021 | 542,344 | 652 | 1,758 | 38,423 | 95.4% [5.3%], 0.9%, 3.7% | 92.9% [4.5%], 1.2%, 5.9% |
| M. muscosa | 222,479,664 | 211,848,357 | 276,943 | 473,878 | 592 | 1,461 | 46,196 | 94.7% [6.1%], 1.3%, 4.0% | 90.8% [3.9%], 1.9%, 7.3% |
| Species . | Raw reads . | Processed reads . | #Trinity genes . | #Trinity isoforms . | Contig N50 (bases)a . | Ex90N50 (bases)b . | #Ex90N50 genesc . | Arachnid BUSCO scores (C[D], F, M)d . | Arthropod BUSCO scores (C[D], F, M)e . |
|---|---|---|---|---|---|---|---|---|---|
| C. Acosta | 272,844,971 | 260,853,757 | 237,678 | 334,267 | 896 | 2,406 | 31,012 | 94.9% [12.9%], 1.0%, 4.1% | 93.2% [9.5%], 1.5%, 5.3% |
| E. bacillifer | 249,938,618 | 239,034,000 | 184,142 | 285,861 | 978 | 2,671 | 22,647 | 93.8% [7.1%], 1.5%, 4.7% | 92.9% [3.5%], 0.7%, 6.4% |
| P. amentata | 266,764,548 | 256,911,378 | 316,021 | 542,344 | 652 | 1,758 | 38,423 | 95.4% [5.3%], 0.9%, 3.7% | 92.9% [4.5%], 1.2%, 5.9% |
| M. muscosa | 222,479,664 | 211,848,357 | 276,943 | 473,878 | 592 | 1,461 | 46,196 | 94.7% [6.1%], 1.3%, 4.0% | 90.8% [3.9%], 1.9%, 7.3% |
Based on longest isoform per gene.
Same as contig N50 but based on top most highly expressed genes that represent 90% of the total normalized expression data.
The number of genes for which Ex90N50 is calculated.
Ten species, n = 2934 BUSCOs; C = Complete [D = Duplicated], F = Fragmented, M = Missing.
Ninety species, n = 1013 BUSCOs; C = Complete [D = Duplicated], F = Fragmented, M = Missing.
Transcriptomes were found to be between 83.7% (C. acosta) and 89.4% (E. bacillifer) complete according to BUSCO scores compared to the arthropod database, with between 3.5% and 9.5% duplicated BUSCOs. Compared to the arachnid databases, transcriptomes were 82–90.1% complete for single-copy BUSCOs and contained between 5.3% and 12.9% duplicated BUSCOs (Table 1).
To explore the extent of duplication in these arachnopulmonates, we then surveyed the copy number of Hox, Wnt, and frizzled genes represented in their transcriptomes in comparison to other arachnids. It is important to note that the absence of genes recovered from transcriptomes does not eliminate the possibility that they are present in the genome, as the transcriptomes will only capture genes expressed at the surveyed stages of development. Mixed-stage embryonic samples (all except E. bacillifer) may therefore yield more transcripts. See Table 2 for a summary of these details for each species.
Summary of genetic resources for surveyed arachnopulmonate species
| Species . | Resource . | Stage . | No. of individuals . |
|---|---|---|---|
| Acanthoscurria geniculata | RNA | Mixed | >1 |
| Centruroides sculpturatus | DNA | Adult | 1 |
| Charinus acosta | RNA | Mixed | 1 |
| Euphrynichus bacillifer | RNA | Single | >1 |
| Marpissa muscosa | RNA | Mixed | >1 |
| Mesobuthus martensii | DNA | Adult | 1 |
| Parasteatoda tepidariorum | DNA | Adult | >1 |
| Pardosa amentata | RNA | Mixed | >1 |
| Pardosa pseudoannulata | DNA | Mixed | >1 |
| Pholcus phalangioides | RNA | Mixed | >1 |
| Stegodyphus dumicola | DNA | Adult | 1 |
| Species . | Resource . | Stage . | No. of individuals . |
|---|---|---|---|
| Acanthoscurria geniculata | RNA | Mixed | >1 |
| Centruroides sculpturatus | DNA | Adult | 1 |
| Charinus acosta | RNA | Mixed | 1 |
| Euphrynichus bacillifer | RNA | Single | >1 |
| Marpissa muscosa | RNA | Mixed | >1 |
| Mesobuthus martensii | DNA | Adult | 1 |
| Parasteatoda tepidariorum | DNA | Adult | >1 |
| Pardosa amentata | RNA | Mixed | >1 |
| Pardosa pseudoannulata | DNA | Mixed | >1 |
| Pholcus phalangioides | RNA | Mixed | >1 |
| Stegodyphus dumicola | DNA | Adult | 1 |
Summary of genetic resources for surveyed arachnopulmonate species
| Species . | Resource . | Stage . | No. of individuals . |
|---|---|---|---|
| Acanthoscurria geniculata | RNA | Mixed | >1 |
| Centruroides sculpturatus | DNA | Adult | 1 |
| Charinus acosta | RNA | Mixed | 1 |
| Euphrynichus bacillifer | RNA | Single | >1 |
| Marpissa muscosa | RNA | Mixed | >1 |
| Mesobuthus martensii | DNA | Adult | 1 |
| Parasteatoda tepidariorum | DNA | Adult | >1 |
| Pardosa amentata | RNA | Mixed | >1 |
| Pardosa pseudoannulata | DNA | Mixed | >1 |
| Pholcus phalangioides | RNA | Mixed | >1 |
| Stegodyphus dumicola | DNA | Adult | 1 |
| Species . | Resource . | Stage . | No. of individuals . |
|---|---|---|---|
| Acanthoscurria geniculata | RNA | Mixed | >1 |
| Centruroides sculpturatus | DNA | Adult | 1 |
| Charinus acosta | RNA | Mixed | 1 |
| Euphrynichus bacillifer | RNA | Single | >1 |
| Marpissa muscosa | RNA | Mixed | >1 |
| Mesobuthus martensii | DNA | Adult | 1 |
| Parasteatoda tepidariorum | DNA | Adult | >1 |
| Pardosa amentata | RNA | Mixed | >1 |
| Pardosa pseudoannulata | DNA | Mixed | >1 |
| Pholcus phalangioides | RNA | Mixed | >1 |
| Stegodyphus dumicola | DNA | Adult | 1 |
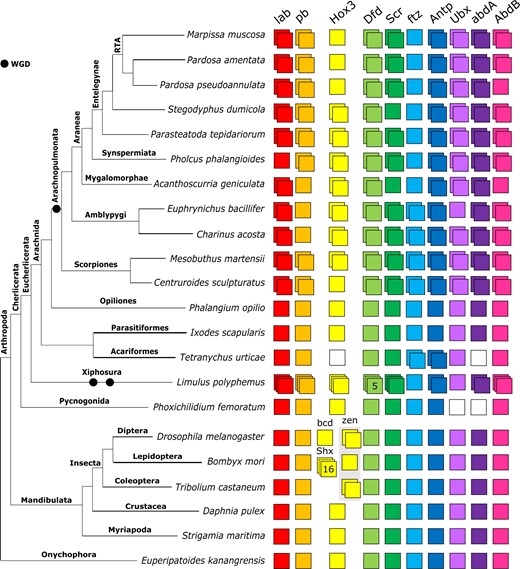
Hox repertoires and their origins
Spider Hox gene repertoires are largely consistent with P. tepidariorum, which has two copies of all except ftz (Figure 1). There are several exceptions: single copies of Hox3 in M. muscosa, P. amentata, and P. pseudoannulata, of Sex combs reduced (Scr) in A. geniculata and S. dumicola, of proboscipedia (pb) in A. geniculata, and of labial (lab) in P. phalangioides (Figure 1). Although we found two AbdB candidates in P. pseudoannulata, one did not contain a homeodomain and was excluded from phylogenetic analyses.
Repertoires of Hox genes in arachnids and other selected arthropods. Hox genes are represented by colored boxes with duplicated Hox genes represented by overlapping boxes and gene loss represented by a white box. Figure includes Hox repertoires previously surveyed in the arachnids P. tepidariorum, Centruroides sculpturatus, Mesobuthus martensii, Phalangium opilio, I. scapularis (all genomes) and Pholcus phalangioides (embryonic transcriptome), the myriapod Strigamia maritima and the insects Drosophila melanogaster, Tribolium castaneum, and Bombyx mori. The insect Hox3 homolog zen has undergone independent tandem duplications in T. castaneum to yield zen and zen2; in cyclorrhaphan flies to yield zen and bicoid; and in the genus Drosophila to yield zen2. Bombyx mori is not representative of all species of ditrysian Lepidoptera, which typically possess four distinct Hox3 genes termed Special homeobox genes (ShxA, ShxB, ShxC, and ShxD) and the canonical zen gene.
Both amblypygids exhibit extensive duplication of Hox genes, with two copies recovered for all except for pb in C. acosta and pb and Ubx in E. bacillifer (Figure 1).
The scorpions M. martensii and C. sculpturatus exhibited similar Hox repertoires, with two copies recovered for all 10 genes, except for a single copy of Hox3 in C. sculpturatus.
Among the nonarachnopulmonate arachnids, I. scapularis and P. opilio exhibited no duplication of any Hox genes, in line with previous genomic surveys (Leite et al. 2018; Gainett et al. 2021). In the mite T. urticae, we did not identify Hox3 and abdA candidates, consistent with Grbić et al. (2011) and Ontano et al. (2021). The resolution of several sequences from T. urticae was variable; for example, previously identified Tu-ftz-1, Tu-ftz-2, Tu-AbdB, and Tu-Ubx sequences (Grbić et al. 2011) did not resolve correctly in the full Hox tree (Figure 2) but did in the homeodomain and homeobox trees (Supplementary Files S5–S8).
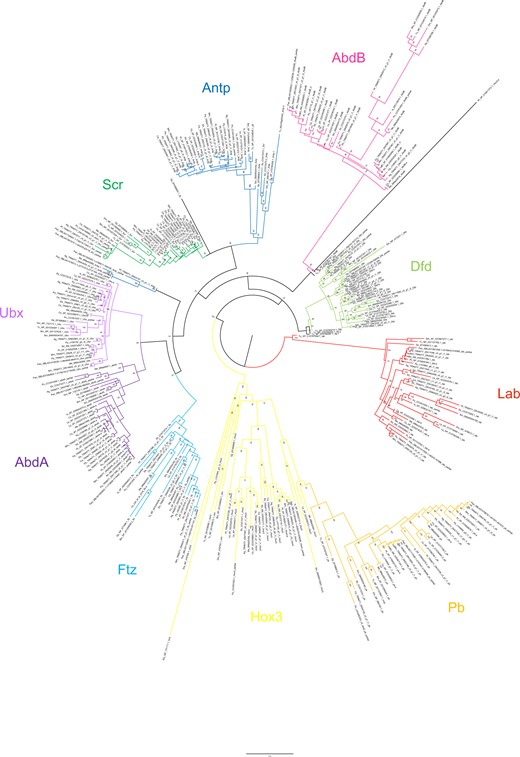
Maximum-likelihood phylogeny of Hox amino acid sequences. The Hox subfamilies are shown as different colors (after Figure 1). Panarthropods included: Acanthoscurria geniculata (Ag), Bombyx mori (Bm), Centruroides sculpturatus (Cs), Charinus acosta (Ca), Drosophila melanogaster (Dm), Euperipatoides kanangrensis (Ek), Euphrynichus bacillifer (Eb), Ixodes scapularis (Is), Limulus polyphemus (Lp), Marpissa muscosa (Mm), Mesobuthus martensii (Me), Parasteatoda tepidariorum (Pt), Pardosa amentata (Pa), Pardosa pseudoannulata (Pan), Phalangium opilio (Po), Pholcus phalangioides (Pp), Phoxichildium femoratum (Pf), Stegodyphus dumicola (Sd), Strigamia maritima (Sm), Tetranychus urticae (Tu), and Tribolium castaneum (Tc). Node labels indicate ultrafast bootstrap support values. See Supplementary File S1 for accession numbers, Supplementary File S2 for full amino acid sequence alignments.
We also surveyed two nonarachnid chelicerates, a pycnogonid P. femoratum and the horseshoe crab L. polyphemus, the latter of which has undergone multiple rounds of WGD independently of the arachnopulmonates (Kenny et al. 2016; Nong et al. 2020) we recovered single copies of all Hox genes except Ubx and abdA, which were not found, in P. femoratum, consistent with Ballesteros et al. (Ontano et al. 2021; Figure 1). Limulus polyphemus returned multiple copies of all Hox genes except ftz and Ubx, including three copies of lab, pb, Hox3, Scr, and AbdB, five potential copies of Dfd, and two copies of Antp and abdA (Figure 1).
Full protein sequences produced the best-supported phylogeny: six nodes returned support <50%. Two of these concerned the placement of outgroup (nonchelicerate) sequences and one concerned the deeper relationship of the Hox3 and Pb clades, which is beyond the scope of this study. The remaining three reflect uncertainty in the within-clade placement of chelicerate sequences (Figure 2). An additional 10 nodes returned support of 50–60%, which we consider too weak to justify interpretation. Analyses using protein sequences from homeodomains only contained insufficient phylogenetic information to resolve within-clade relationships, but confirmed gene identity (Supplementary Files S5 and S6). Nucleotide sequences of homeodomains provided more within-clade resolution, but with lower support than full protein sequences (Supplementary Files S7 and S8). Using full protein sequences, we recover Antp as a paraphyletic group, albeit with low support (61%). The homeodomain phylogenies (both protein and nucleotide) confirm the identity of these sequences, indicating substantial sequence divergence outside of this conserved region in several species. The following discussion is based on the phylogeny of the full protein sequences (Figure 2).
Overall, the resolution of duplicate Hox sequences indicates that the majority are likely to be ohnologs. In many cases, duplicates form well-supported clades of orthologs, containing sequences from multiple orders, suggesting a shared origin for duplication. The majority of Hox genes are present in duplicate across the arachnopulmonates, strengthening this pattern. Few duplicates form paralogous pairs within species, or paired clades of paralogs within lineages, as would be expected from lineage-specific duplications (although there are exceptions, such as scorpion Lab and Antp, Figure 2).
For both AbdA and AbdB, sequences broadly formed two overall clades; although these were not well supported for AbdB (49–51%), relationships within them are still informative. In both cases, spider sequences formed two distinct and well-supported (>97%) clades that reflect overall phylogenetic relationships in their topology. The amblypygid sequences resolved as two ortholog pairs, one in each of the two overall clades. These are therefore strong candidates for ohnologs. Scorpion AbdB and AbdA were not consistent with this pattern; in both cases, one pair of orthologs resolved together and one pair separately (Figure 2). Although this reflects a low likelihood of a lineage-specific duplication of abdA and AbdB in scorpions, it does not further clarify possible ohnolog relationships. Spider and scorpion Pb duplicates are candidate ohnologs, resolving in two well-supported (>99%) clades of orthologs whose topologies reflect phylogenetic relationships. Ftz duplicates in scorpions and amblypygids also resolved with orthologs of other arachnopulmonates, but one set of scorpion paralogs was placed with substantial uncertainty (support <60%). Most spider and amblypygid Antp sequences follow the pattern expected of ohnologs, forming two clades of orthologs, but three of the scorpion sequences formed a well-supported clade (91%) while the fourth resolved within a small group of sequences that fall outside the main Antp clade.
The origin of Ubx, Dfd, Lab, Hox3, and Scr duplicates in arachnopulmonates is not clear from our phylogenetic analysis alone, with neither orthologs nor paralogs resolving together consistently, and topology that is a poor reflection of phylogeny. However, spider sequences broadly formed two clades with other arachnopulmonates, and synteny analysis in both P. tepidariorum (Schwager et al. 2017) and Trichonephila antipodiana (Fan et al. 2021) demonstrate clearly that Hox cluster duplications therein are the result of WGD. The placement of the amblypygid and scorpion duplicates was more variable. In some cases, these also appear to resolve as expected of ohnologs (e.g., amblypygid Hox3), but in others, their placement could indicate lineage-specific duplication, although usually with low support (e.g., amblypygid Ubx).
Wnt repertoires and their origins
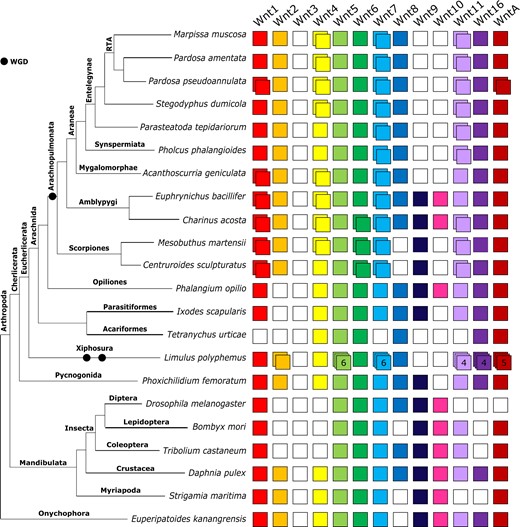
Consistent with P. tepidariorum (Janssen et al. 2010), we found representatives of 10 Wnt subfamilies in all surveyed spiders. The absence of Wnt9 and Wnt10 indicates their likely absence in the spider ancestor, while the absence of Wnt3 is consistent with all other protostomes (Janssen et al. 2010; Murat et al. 2010; Hogvall et al. 2014). Two copies of Wnt7 were retrieved from M. muscosa, P. amentata, A. geniculata, P. phalangioides, P. tepidariorum, P. pseudoannulata, and S. dumicola. All spiders except A. geniculata also yielded two copies of Wnt11. A second copy of Wnt4, which is absent in P. tepidariorum, was recovered from P. amentata, M. muscosa, A. geniculata, S. dumicola, and P. pseudoannulata. We also found duplicates of Wnt1/wg in both A. geniculata and P. pseudoannulata, and a duplicate of WntA in P. pseudoannulata.
Representation of the Wnt subfamilies in the amblypygids is higher than any other chelicerate studied to date, including those with high-quality genome assemblies (Janssen et al. 2010; Hogvall et al. 2014; Holzem et al. 2019). We recovered transcripts from 12 out of 13 subfamilies (missing Wnt3) and duplicates of Wnt1/wg, Wnt4, and Wnt7 in both species (Figure 3). Additional duplicates of Wnt6 and Wnt11 were recovered from C. acosta (Figure 3).
Repertoires of Wnt subfamilies in arthropods and an onychophoran. The Wnt subfamilies (1–11, 16, and A) are represented by colored boxes with duplicated genes represented by overlapping boxes and putatively lost subfamilies indicated by white boxes. Figure includes Wnt repertoires recovered in this study and previously surveyed in the arachnids Parasteatoda tepidariorum and Ixodes scapularis; the insects Drosophila melanogaster, Tribolium castaneum and Bombyx mori; the crustacean Daphnia pulex; the myriapod Strigamia maritima; and the onychophoran Euperipatoides kanangrensis.
Representatives of 10 Wnt subfamilies were found in the two scorpion genomes, with both missing Wnt3, Wnt8, and Wnt9. Two copies of Wnt1/wg, Wnt6, Wnt7, and Wnt11 were retrieved in both species, with an additional copy of Wnt4 in M. martensii.
We did not recover duplicates of Wnt2, Wnt5, Wnt8-10, or Wnt16 in any arachnopulmonate lineage and found no evidence of Wnt gene duplication in the nonarachnopulmonate arachnids (Figure 3). In the harvestman P. opilio, we recovered single copies of all except Wnt2 and Wnt3. Ixodes scapularis was also missing Wnt10 but was otherwise similar, whereas we did not recover these or Wnt1/wg, Wnt7, Wnt9, or Wnt11 in the mite T. urticae. In the nonarachnid chelicerates, P. femoratum and L. polyphemus, we found similar representation of the subfamilies, with all except Wnt3 and Wnt10 in P. femoratum and all except these and Wnt9 in L. polyphemus. No duplicates were recovered in P. femoratum, but large numbers of duplicates were found in L. polyphemus. These included six potential copies of Wnt5 and Wnt7, four of Wnt11 and Wnt16, and five of WntA (Figure 3).
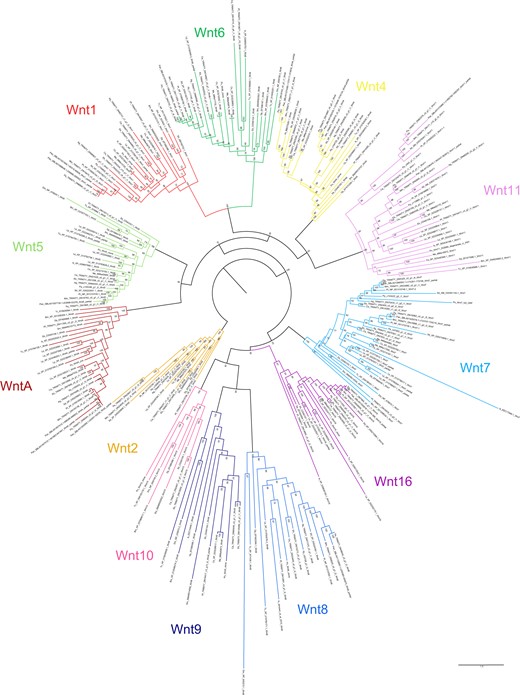
In our phylogeny using full protein sequences, only two nodes returned support <50%: one uniting Wnt8, Wnt9, Wnt10, and Wnt16 (39%), and one uniting Wnt1/wg, Wnt4, Wnt6, and Wnt11 (36%). These are positioned deep within the tree and concern the interrelationships of the Wnt subfamilies, which are beyond the scope of this study.
Most of the duplicate Wnt genes appear to be likely ohnologs; confirmation requires synteny analysis, but the relationships between paralogs resolved by phylogenetic analysis generally support duplications originating in the arachnopulmonate ancestor.
Duplicates of Wnt7 were previously identified in P. tepidariorum (Janssen et al. 2010) and are recovered from all surveyed arachnopulmonates. The spider Wnt7 duplicates formed two clades (bootstrap ≥61%) suggesting the retention of ohnologs (Figure 4). The amblypygid Wnt7 ortholog pairs resolve as sisters to the spider clades, but support for these placements is lower (50–60%), and they display slightly higher sequence similarity between paralogs (73–74%) than the spiders (60–72%). The scorpion Wnt7 ortholog pairs formed their own separate clade with strong support (92%; Figure 4), possibly indicating a lineage-specific duplication. Nonetheless, the sequence divergence between the scorpion paralog pairs (68–70%) is similar to that between putative ohnologs in spiders (60–72%).
Maximum-likelihood phylogeny of Wnt amino acid sequences. The 12 Wnt subfamilies are shown as different colors (after Figure 3). Panarthropods included: Acanthoscurria geniculata (Ag), Bombyx mori (Bm), Centruroides sculpturatus (Cs), Charinus acosta (Ca), Drosophila melanogaster (Dm), Euperipatoides kanangrensis (Ek), Euphrynichus bacillifer (Eb), Ixodes scapularis (Is), Limulus polyphemus (Lp), Marpissa muscosa (Mm), Mesobuthus martensii (Me), Parasteatoda tepidariorum (Pt), Pardosa amentata (Pa), Pardosa pseudoannulata (Pan), Phalangium opilio (Po), Pholcus phalangioides (Pp), Phoxichildium femoratum (Pf), Stegodyphus dumicola (Sd), Strigamia maritima (Sm), Tetranychus urticae (Tu), and Tribolium castaneum (Tc). Node labels indicate ultrafast bootstrap support values. See Supplementary File S1 for accession numbers, Supplementary File S9 for amino acid sequence alignments, and Supplementary File S10 for nucleotide sequence alignments of Wnt1/wg duplicates in C. acosta, C. sculpturatus, and Euphrynichus bacillifer.
Duplicates of Wnt11 previously identified in P. tepidariorum (Janssen et al. 2010) were recovered from all surveyed spiders except A. geniculata, C. acosta, and both scorpions. The spider Wnt11 duplicates formed two separate and well-supported clades (≥85%, Figure 4), and each amblypygid Wnt11 orthology group was sister to one of the spider clades (98–99%; Figure 4). Similarity between paralogs in the four new transcriptomes was very low (41–54%). We propose that the spider and amblypygid Wnt11 duplicates are probably retained from the ancestral WGD. The resolution of the scorpion Wnt11 duplicates is less clear; ortholog pairs resolve together with 100% support, but only one resolves as sister to the two clear clades occupied by the spider and amblypygid duplicates (98%; Figure 4). They exhibit similar sequence similarity (49–50%) to the putative ohnologs.
Wnt4 paralogs from M. muscosa, P. amentata, A. geniculata, P. pseudoannulata, and S. dumicola form two separate and well-supported clades with duplicates from the amblypygids (bootstrap ≥91%). They show substantial sequence divergence within species (53–64% similarity), indicating that they are again likely to represent retained ohnologs following the arachnopulmonate WGD, despite being lost in the lineage to P. tepidariorum.
The two sequences of WntA recovered from the P. pseudoannulata genome are located on the same scaffold and are dissimilar in sequence. Their peptide sequences resolve as sister to one another (Figure 4), indicating a lineage-specific tandem duplication. However, as they are partial sequences with a short overlapping region, this requires confirmation.
We identified two copies of Wnt1/wg in the transcriptomes of both amblypygids and A. geniculata, and in the genomes of P. pseudoannulata, C. sculpturatus, and M. martensii. Sequence similarity between paralogs was low (55–73%) compared to similarity between orthologs at the order level (e.g., 91% between M. muscosa and P. amentata), and comparable to orthologs at the class level (e.g., 61% between P. tepidariorum and I. scapularis). Although synteny analysis is required for conclusive confirmation, our phylogeny indicates that the amblypygid, scorpion, and A. geniculata duplicates are likely to be ohnologs retained from the arachnopulmonate WGD, as they form separate clades (≥70%, Figure 4). The putative duplicates in P. pseudoannulata, in contrast, resolve as sister to one another. These two sequences have (short) nonidentical overlapping regions, but, again, they are partial; therefore, it is equivocal whether they represent a genuine, lineage-specific duplication.
Frizzled repertoires and their origins
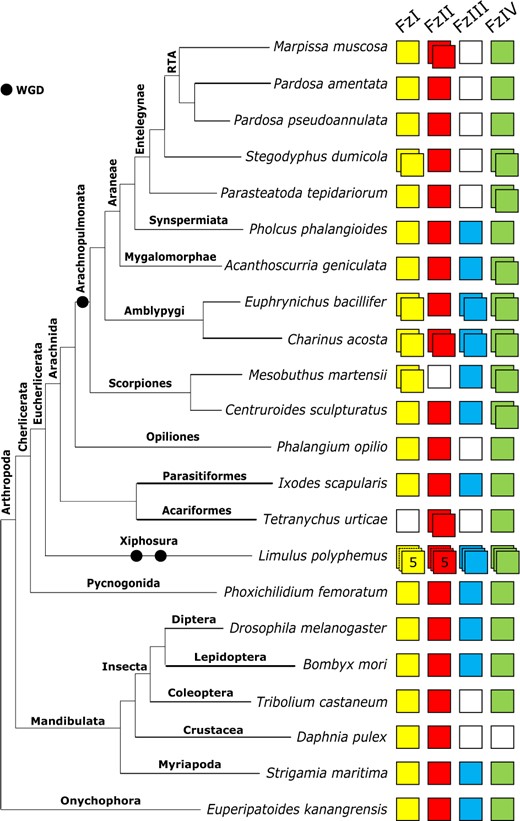
All surveyed spiders possess at least one ortholog of FzI and FzII. A second ortholog of FzI was recovered from the genome of S. dumicola and a second ortholog of FzII from the transcriptome of M. muscosa (Figure 5). FzIII was absent from the transcriptomes of M. muscosa and P. amentata, and the genomes of S. dumicola and P. pseudoannulata, consistent with P. tepidariorum (Janssen et al. 2015). However, single FzIII orthologs were recovered from A. geniculata and previously identified in P. phalangioides (Janssen et al. 2015). Consistent with P. tepidariorum (Janssen et al. 2015), we identified two FzIV sequences in S. dumicola and A. geniculata, but only one in M. muscosa, P. amentata, and P. pseudoannulata and only one was previously recovered from P. phalangioides (Janssen et al. 2015).
Repertoire of frizzled genes in arachnids and other selected arthropods. The four frizzled subfamilies (FzI, FzII, FzIII, and FzIV) are represented by colored boxes, with duplicated genes represented by overlapping boxes and gene loss represented by a white box. Figure includes frizzled repertoires previously surveyed in the arachnids Parasteatoda tepidariorum, Mesobuthus martensii, Ixodes scapularis, and Pholcus phalangioides; the myriapod Strigamia maritima; the insects Drosophila melanogaster and Tribolium castaneum; and the onychophoran Euperipatoides kanangrensis.
The two amblypygid species have a large repertoire of frizzled genes compared to other arachnids, with duplicates of FzI, FIII, and FzIV orthologs in both species, as well as a duplication of FzII in C. acosta (Figure 5).
The scorpions M. martensii (Janssen et al. 2015) and C. sculpturatus possess single FzI and FzIII orthologs and a duplication of FzIV. We also recovered a single FzII ortholog in C. sculpturatus, and an FzI duplicate in M. martensii.
Among the nonarachnopulmonate arachnids, I. scapularis possesses a single copy of all four subfamilies, but FzIII was absent in P. opilio and T. urticae. A FzII duplicate was recovered from T. urticae. Phoxichilidium femoratum and L. polyphemus possess all four subfamilies, and while no duplicates were recovered in P. femoratum, five copies of FzI and FzII and three copies of FzIII and FzIV were recovered from L. polyphemus (Figure 5).
Frizzled paralogs appear to stem from both WGD events and lineage-specific duplications. FzI duplicates were recovered from both amblypygid transcriptomes, M. martensii, and S. dumicola. The Sd-Fz1 and Me-Fz1 paralog pairs exhibit high sequence similarity (>73%) and resolved as sisters with 100% bootstrap support, indicating that these are likely the results of lineage-specific duplications. Conversely, the amblypygid Fz1-1 and Fz1-2 ortholog pairs form separate clades with sequences from spiders and scorpions, respectively (Figure 6). We therefore interpret these duplicates as ohnologs.
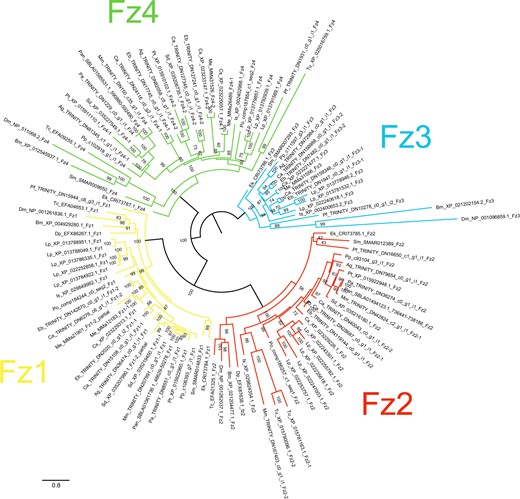
Maximum-likelihood phylogeny of Frizzled proteins. The Frizzled subfamilies are shown as different colors (after Figure 5). Panarthropods included: Acanthoscurria geniculata (Ag), Bombyx mori (Bm), Centruroides sculpturatus (Cs), Charinus acosta (Ca), Drosophila melanogaster (Dm), Euperipatoides kanangrensis (Ek), Euphrynichus bacillifer (Eb), Ixodes scapularis (Is), Limulus polyphemus (Lp), Marpissa muscosa (Mm), Mesobuthus martensii (Me), Parasteatoda tepidariorum (Pt), Pardosa amentata (Pa), Pardosa pseudoannulata (Pan), Phalangium opilio (Po), Pholcus phalangioides (Pp), Phoxichildium femoratum (Pf), Stegodyphus dumicola (Sd), Strigamia maritima (Sm), Tetranychus urticae (Tu), and Tribolium castaneum (Tc). Node labels indicate ultrafast bootstrap support values. See Supplementary File S1 for accession numbers and Supplementary File S11 for alignments.
Duplicates of FzII were recovered from M. muscosa and C. acosta. The Mm-Fz2 paralogs form a well-supported clade (79%; Figure 6), indicating that this is the result of a lineage-specific duplication followed by sequence divergence in Mm-Fz2-2 (53% sequence similarity). The origin of the FzII duplication in C. acosta is not clear. Ca-Fz2-1 resolves as a sister group to the spider sequences (99%; Figure 6) and Ca-Fz2-2 forms a clade with Eb-Fz2, which in turn resolves as sister to the Ca-Fz2-1 and spider sequences (93%; Figure 6). This topology could support an ohnolog relationship between Ca-Fz2-1 and Ca-Fz2-2 but cannot be confirmed. Sequence similarity between the C. acosta paralogs is relatively high (82%), perhaps higher than expected from ohnologs.
The origin of the amblypygid FzIII duplicates is also unclear. One ortholog pair forms a clade with L. polyphemus sequences (66%; Figure 6) and the other forms a clade with the spiders and scorpions (≥70%; Figure 6). This suggests an origin in WGD, but support for their placement is not strong (66%, Figure 6). Paralogous pairs demonstrate middling sequence similarity (65–66%).
Both amblypygids and scorpions, and the spiders P. tepidariorum, S. dumicola, and A. geniculata, possess FzIV duplicates. The spider sequences formed two separate and well-supported clades (100%; Figure 6). The amblypygid ortholog pairs resolved as sister to the two spider clades (98%; Figure 6). The scorpion ortholog pairs, however, formed a clade together (76% support; Figure 6) which is sister to all the spider and amblypygid sequences (82%; Figure 6). All paralogous pairs exhibited substantial sequence divergence (similarity 44–58%). We propose that the spider and amblypygid FzIV duplicates are retained from the ancestral WGD; however, once again, the origin of the scorpion FzIV duplicates is less clear, and may reflect a lineage-specific duplication.
Discussion
The contribution of WGD to expanding the developmental toolkit is evident across the three gene families surveyed here, but it appears that retention patterns of putative ohnologs vary substantially between them. We also see distinct phylogenetic patterns beginning to emerge in gene repertoires, with improved sampling within lineages enabling us to distinguish between possible signal and likely noise. Of course, transcriptomic data come with necessary caveats regarding gene expression and capture, and absences from transcriptomes should be regarded with caution. However, where patterns are consistent across multiple species, or better, between transcriptomic and genomic resources, we can be more confident about their authenticity. For example, comparisons of the transcriptome of P. amentata and the genome of P. pseudoannulata show identical Hox and frizzled repertoires, indicating good gene capture in the former. The apparent duplicates of Wnt1/wg and WntA in P. pseudoannulata that were not recovered in P. amentata are of uncertain status (partial sequences, phylogenetic position) and were absent from all other spider genomes. Overall, levels of gene duplication detected using BUSCO were in line with other arachnopulmonates [e.g., 3.3% in Argiope bruennichi, Sheffer et al. (2021); 5.6% in T. antipodiana, Fan et al. (2021); 11% in Araneus ventricosus, Kono et al. (2019)] but lower than in horseshoe crabs [e.g., 16.7% in Carcinoscorpius rotundicauda, Shingate et al. (2020)].
Hox duplicates are broadly retained
The widespread retention of duplicate Hox genes is consistent among the arachnopulmonate orders studied to date, and specific repertoires appear to be fairly conserved at the order level (Schwager et al. 2007; Cao et al. 2013; Di et al. 2015; Schwager et al. 2017; Leite et al. 2018; Ontano et al. 2021). Given that this is the level at which overall body plans are conserved, this is perhaps not surprising. Thanks to the relatively conserved expression patterns of Hox genes along the antero-posterior axis of chelicerates, we can begin to speculate about the possible macroevolutionary implications of duplication and loss.
The absence of a second copy of pb in both C. acosta and E. bacillifer, which are distantly related within Amblypygi, suggests a loss in the common ancestor of amblypygids. Gainett and Sharma (2020) also recovered a single copy of pb in P. marginemaculatus, but two copies in Charinus israelensis. However, one of these sequences (32 aa) had an incomplete homeodomain that was identical to the other copy; therefore, we are hesitant about its status as a duplicate. Embryos of C. acosta were collected at multiple stages of development, supporting the hypothesis that this is a genuine loss, rather than absence of expression at a particular developmental stage. In spiders, both pb ohnologs are expressed in the pedipalp and leg-bearing segments, separated temporally (Schwager et al. 2017). Given the highly derived nature of the raptorial pedipalps and the antenniform first pair of walking legs in amblypygids, it is perhaps surprising that this duplicate was not retained. However, this might indicate that other Hox genes expressed in the anterior prosomal segments (e.g., lab, Hox3, or Dfd) may contribute to these morphological innovations. Recent work by Gainett and Sharma (2020) examining the specification of the antenniform legs found little difference in Distal-less, dachshund, or homothorax expression between that and posterior leg pairs, indicating that these are not likely to be responsible. A good candidate for future study might be lab: a single ortholog is expressed in both the pedipalps and the first walking leg in the harvestman P. opilio (Sharma et al. 2012), and expression patterns and experimental manipulation provide evidence for functional divergence between the two lab ohnologs, which are expressed in the pedipalps and first walking legs, in P. tepidariorum (Pechmann et al. 2015; Schomburg et al. 2020).
While the ftz duplicate has been retained in amblypygids and scorpions, it seems likely that having a single copy of ftz is common across all spiders (Figures 1 and 2), consistent with the loss of a duplicate in their common ancestor. A recently published high-quality genome from T. antipodiana also reported a single copy of ftz (Fan et al. 2021), as did another mygalomorph transcriptome, A. hentzi (Ontano et al. 2021). In both P. opilio (Sharma et al. 2012) and P. tepidariorum (Schwager et al. 2017), a single copy of ftz is expressed in leg pairs 2–4. Any subfunctionalization or neofunctionalization that could be evident in scorpion and amblypygid ftz had therefore presumably not taken place at the point of their divergence from spiders. Expression patterns of the two paralogs in these groups would be of interest for comparison with both harvestmen and spiders.
The absence of Hox3 duplicates in M. muscosa, P. pseudoannulata, and P. amentata may indicate a lineage-specific loss in the RTA clade, which unites salticids, lycosids, and their allies. Indeed, only one copy of Hox3 was previously recovered in Cupiennius salei, which also belongs to the RTA clade (Schwager et al. 2007). Although the absence of a second copy in the embryonic transcriptomes could be attributed to failure to capture expression, both C. salei and P. pseudoannulata yielded single copies from DNA, strengthening the case for a genuine loss. In P. tepidariorum, Hox3-A and Hox3-B exhibit dramatic protein sequence divergence (20.6% similarity). Both copies are expressed in the embryo, with their expression overlapping spatially and temporally but not identical, indicating some functional divergence. Hox3-A expression was reported to be very weak (Schwager et al. 2017), and the Hox-3A homeodomain demonstrates much higher divergence from other arachnid Hox3 sequences than does Hox3-B (Supplementary File S3); together, these could reflect diminished functionality. In the other spiders for which we recovered Hox3 duplicates, paralogs also exhibited very low sequence similarities (24.7–30.6%), and these duplicates had very low similarity to each other (17.2–30.1%, compared to 33–58.2% between those resolving with other spider Hox3 sequences). In all cases, one resolved within a well-supported clade of spider Hox3 and the other was placed haphazardly. We speculate that the Hox3 duplicate is highly divergent or degenerate in spiders, leading to eventual pseudogenization in the RTA clade.
Other apparent losses of Hox gene duplicates are restricted to transcriptomes of individual species, with the exception of Scr in S. dumicola, and could reflect failure to capture additional sequences or indicate lineage-specific loss. It would be premature to conclude that they are genuinely absent from the genome. For instance, the apparent absence of a Ubx duplicate in E. bacillifer. In P. tepidariorum, the two Ubx ohnologs are expressed most strongly in close succession, at stages 8.1 and 8.2 (Schwager et al. 2017), but this may not reflect their relative expression patterns in amblypygids, and therefore one may not have been captured in this single-stage transcriptome.
Limited retention of Wnt duplicates
In contrast to the widespread retention of Hox duplicates, these new data indicate that the retention of duplicate Wnt genes is less common and restricted to certain subfamilies. Apparent ohnologs of Wnt4, Wnt7, and Wnt11 are retained in the majority of arachnopulmonates, for example, but Wnt2, Wnt5, Wnt8-10, Wnt16, and WntA are only represented by single copies. These patterns could reflect early losses of duplicates, and/or differential effects of higher copy numbers across Wnt subfamilies. The fact that retention patterns appear to differ between the arachnopulmonate and horseshoe crab independent WGD events lends support to the former hypothesis.
Our understanding of specific Wnt functions among arthropods is more limited than that of Hox genes, but Wnt expression patterns in P. tepidariorum are available for tentative comparison. For example, previous attempts to characterize the expression patterns of Wnt11 paralogs in P. tepidariorum only detected expression of Wnt11-2 (Janssen et al. 2010). Given the retention of Wnt11-1 in both spiders and scorpions, and the considerable divergence between paralogous sequences, Wnt11 could be a good candidate for sub- or neofunctionalization, but the role of Wnt11-1 remains unknown. Conversely, the presence of two apparent Wnt4 ohnologs in spiders and amblypygids (Figures 3 and 4) contrasts with the retention of only one Wnt4 in P. tepidariorum. The role of the additional copy, particularly in spiders, will be of future interest and expression patterns of Wnt4-2 in these groups will help to clarify this. Although it is possible that the second copy is redundant or in the process of pseudogenization, the fact that both ohnologs are retained in two large clades, with detectable levels of expression during development, suggests that this is not the case. Thus, its absence in P. tepidariorum unexpectedly appears to be the exception.
The discovery of duplicate Wnt1/wg in scorpions, amblypygids, and mygalomorphs is particularly exciting: duplicates of this Wnt gene have not yet been detected in any other metazoans, even following multiple rounds of WGD in vertebrates, teleosts (see https://web.stanford.edu/group/nusselab/cgi-bin/wnt/vertebrate), or horseshoe crabs. This requires critical interpretation. We can eliminate the possibility of individual variation in C. acosta, as embryos are produced by parthenogenesis and are therefore clones (de Armas 2005), and in the two scorpions, as the sequences were recovered from a single individual’s genome (Supplementary File S1). Levels of sequence divergence exceed those seen between putative tandem duplications, in both nucleotide and amino acid sequences. Wnt1/wg performs a wide variety of roles in arthropods, including in segment polarization and in appendage and nervous system development (Murat et al. 2010) and has an accordingly complex expression pattern in P. tepidariorum, appearing in the L1 and L2 segments, limb buds, and dorsal O2 and O3 segments (Janssen et al. 2010). In theory, therefore, there is ample potential for subfunctionalization. Gene expression and functional studies of Wnt1/wg duplicates in arachnopulmonates will no doubt prove extremely interesting in the future.
The presence of Wnt10 in both amblypygids and P. opilio is also intriguing because it is absent from all other chelicerates surveyed so far. This could indicate multiple losses of Wnt10 in all other arachnid lineages, the co-option of another gene in amblypygids and harvestmen, or the recovery of a lost Wnt10; however, the possible routes for co-option are unclear and pseudogene recovery has only been suggested in a handful of cases (Trabesinger-Ruef et al. 1996; e.g., Balakirev and Ayala 2003). It is notable that we did not recover Wnt10 orthologs in L. polyphemus or P. femoratum, nonarachnid chelicerates. Studies of other available horseshoe crab genomes (Shingate et al. 2020; Nong et al. 2020) will shed further light on whether Wnt10 is truly absent.
frizzled repertoires vary substantially
Previous studies of spiders, scorpions, and ticks indicated that frizzled repertoires in these groups are restricted to three or four genes, often with incomplete representation of the four subfamilies (Janssen et al. 2015). The spiders M. muscosa, S. dumicola, P. pseudoannulata, and P. amentata are consistent with this pattern, albeit with a unique duplication of FzII in the jumping spider. The presence of FzIII duplicates in P. phalangioides and A. geniculata indicates that, while entelegynes may universally lack fz3, it was likely present in the ancestor of all spiders. In contrast, all four frizzled subfamilies were recovered in both amblypygid species, with three present in duplicate in E. bacillifer and four in C. acosta. Based on our data, it appears that the frizzled repertoire of amblypygids is around twice the size of all other arachnids and may have followed a very different evolutionary trajectory to spiders and scorpions following WGD. The expanded repertoire of frizzled genes in amblypygids is intriguing since they have the largest Wnt repertoires, via both duplication and representation of the subfamilies (Wu and Nusse 2002). However, although frizzled genes encode key receptors for Wnt ligands, they have other Wnt-independent functions, so the expansion of the frizzled repertoire could be equally related to the evolution of alternative signaling roles (Janssen et al. 2015; Yu et al. 2020).
Arachnopulmonate genome evolution in the wake of WGD
Our new analyses provide a thorough survey of Hox, Wnt, and frizzled genes in arachnids, and substantially improve the density and breadth of taxonomic sampling for these key developmental genes in Arachnopulmonata. We find evidence of consistent evolutionary trajectories in Hox and Wnt gene repertoires across three of the six arachnopulmonate orders, with interorder variation in the retention of specific paralogs. We have also identified intraorder variation at the level of major clades in spiders, which could help us better understand their morphological evolution. In new data for a third arachnopulmonate lineage, the amblypygids, we find additional evidence supporting an ancestral WGD and are better able to reconstruct the chronology of gene duplications and losses in spiders and scorpions. These transcriptomic resources are among the first available for amblypygids and will aid future investigations of this fascinating group.
By improving taxonomic coverage within the spider lineage, we are better able to polarize some loss/duplication events and identify potential new trends within the spiders, particularly illustrating separations between synspermiatan and entelegyne spiders, and between the derived RTA clade and other spiders. Despite being unable to ultimately conclude that some missing transcripts reflect genuine genomic losses, it appears that the evolution of these developmental genes in spiders is more complicated than we thought. It may be that these gene repertoires are genuinely more variable within spiders than they are in amblypygids or scorpions; spiders are by far the most taxonomically diverse arachnopulmonate order, and the apparent diversity of repertoires may simply reflect this. Conversely, the higher apparent intraorder diversity of gene repertoires may be an artifact of increased sampling in spiders (up to four or five species for specific gene families) compared to the one or two available resources for scorpions and amblypygids; we may detect more diversity within these groups with increased sampling. Nonetheless, we see two notable trends within spiders, outlined below.
First, we see several characters that appear to unite the RTA clade, which contains almost half of all extant spider species (World Spider Catalog 2019), having diversified rapidly following its divergence from the orb weavers (Garrison et al. 2016; Fernández et al. 2018; Shao and Li 2018). Marpissa muscosa, P. amentata, and P. pseudoannulata all exhibit the apparent loss of Hox3 and fz4 paralogs and the retention of a Wnt4 duplicate. The identification of genetic trends potentially uniting this group is exciting, even if the macroevolutionary implications are unclear: as described above, the possible functions of a Wnt4 paralog are elusive. Members of the RTA clade are very derived compared to other araneomorph spiders, both morphologically (e.g., male pedipalp morphology and sophisticated eyes) and ecologically (most are wandering hunters), and their rapid diversification would align with clade-specific genetic divergence (Garrison et al. 2016; Fernández et al. 2018; Shao and Li 2018).
Second, although data are only available for single representatives of the plesiomorphic clades Synspermiata (P. phalangioides) and Mygalomorphae (A. geniculata), these hint at lineage-specific losses of Hox paralogs and recover the only examples of FzIII found in spiders so far. The presence of FzIII is consistent with other arachnopulmonate groups and suggests that it was present in the spider ancestor and only lost in the more derived entelegyne lineages. If selected Hox duplicates are indeed absent from the genomes of these two species, this could represent an interesting divergence between the three major groups of spiders. Though these genes are unlikely to be directly responsible, the divergence might provide a starting point for understanding the important morphological differences between mygalomorphs, synspermiatans, and entelegynes. However, genomic information for additional taxa in both groups is required to verify these potential losses.
The amblypygids emerge as a key group of interest for studying the impacts of WGD owing to their high levels of ohnolog retention. Our transcriptomes, from representatives of two major clades, provide further evidence supporting a WGD in the ancestor of arachnopulmonates and demonstrate widespread retention of ohnologs in three major families of developmental genes (consistent with the retention of many duplicated regulators of eye development in other species, Gainett et al. 2020). In all three gene families we studied, repertoires were largest in the amblypygid species. This was particularly the case in C. acosta, which belongs to the less speciose and more plesiomorphic infraorder Charinidae within living Amblypygi. Although this study represents just two amblypygid species and three gene families, this appears to contradict widespread predictions of diversification with the duplication of important developmental genes such as Hox (e.g., Van De Peer et al. 2009). Of particular interest are the amblypygid Wnt gene repertoires. We have identified from their transcriptomes, and from the published genome of C. sculpturatus, the first reported duplicates of Wnt1/wg in any animal, as well as the first reported Wnt10 in any arachnid. Future functional studies of these genes and their expression during development will be critical to understanding the evolutionary impacts of these unusual components of amblypygid gene repertoires. Amblypygids also represent a potential model group for studying the evolution of arthropod body plans, owing to the unusual and derived morphology of the pedipalps and especially the first walking legs. Thanks to a substantial existing body of work on anterior-posterior patterning, segmentation, and appendage development in spiders and other arachnids, we may have a chance to crack the genetic underpinnings of these dramatic evolutionary innovations (Pechmann et al. 2009; Sharma et al.2012, 2014a; Turetzek et al. 2016, 2017; Schwager et al. 2017; Schomburg et al. 2020; Baudouin-Gonzalez et al. 2021).
Finally, our analysis of existing genomic data for C. sculpturatus and M. martensii has recovered several Wnt and frizzled gene duplications, similar to spiders and amblypygids. However, in contrast to those groups, our phylogenies have sometimes supported within-lineage duplication in scorpions, as opposed to the retention of ohnologs following WGD, even when these are observed in spiders and amblypygids. This was the case for AbdB, Wnt1/wg, Wnt6, Wnt7, and FzIV (Figures 2, 4, and 6). However, levels of sequence similarity in these cases were comparable for C. sculpturatus paralogs and amblypygid and spider ohnologs, when we might expect within-lineage duplicates to show higher similarity. The resolution of the paralogous sequences in our phylogenetic analyses could be confounded by the early-branching position of scorpions within Arachnopulmonata, which means paralogs would be expected to appear toward the bottom of ortholog clades and are more vulnerable to movement. Nonetheless, this pattern emerged multiple times in our analyses and may be of future interest.
Both arachnopulmonates and horseshoe crabs have been subject to WGD. Comparison between these independent events is a useful tool in studying WGD, spanning smaller phylogenetic distances than arachnopulmonate-vertebrate comparisons. From the three gene families surveyed here, both patterns and inconsistencies emerge. As in arachnopulmonates, Hox gene duplicates appear to be overwhelmingly retained, even in triplet or quadruplet in some cases, in L. polyphemus. Ftz is an interesting exception that aligns with the absence of duplicates in spiders, but not scorpions or amblypygids. However, the apparent loss of all Ubx and two abdA duplicates stands at odds with the arachnopulmonates, wherein these are largely retained. As seen in arachnopulmonates, only select Wnt genes are retained in duplicate in L. polyphemus. However, it is not always the same Wnts: for example, whereas Wnt4 duplicates are common in arachnopulmonates, they are completely absent in L. polyphemus, and vice versa for Wnt5. There are, however, some Wnts that are commonly or even universally present in single copies following both independent WGD events, potentially indicating low potential for sub- or neofunctionalization, such as Wnt8, Wnt6, and Wnt1/wg.
Overall, our new data provide further evidence of an ancestral arachnopulmonate WGD, identify evolutionary patterns within gene families following WGD, reveal new diversity in spider gene repertoires, better contextualize existing data from spiders and scorpions, and broaden the phylogenetic scope of available data for future researchers. However, other arachnid groups, both with and without ancestral WGD, require further study. Recent work on two pseudoscorpions recovered duplicates of most Hox genes (Ontano et al. 2021). This not only further supports arachnopulmonate WGD but substantially improves our understanding of pseudoscorpion placement within arachnids, which has been historically problematic (e.g., Ballesteros et al. 2019; Ballesteros and Sharma 2019; Arribas et al. 2020). The sequencing of a pseudoscorpion genome provides the tantalizing chance to add a fourth lineage to future studies of WGD and its impacts (Ontano et al. 2021). The remaining arachnopulmonate orders, thelyphonids (vinegaroons or whip scorpions) and schizomids, form a clade with amblypygids (Pedipalpi) and should also have been subject to the arachnopulmonate WGD. Future work on these groups will shed light on the unusual patterns of gene retention we find in both major clades of amblypygids. Nonarachnopulmonate arachnids are invaluable for contextualizing the changes that occur following WGD, both in terms of gene repertoires and of gene function. Genomic resources and gene expression pattern studies are vital for this, and the harvestman P. opilio has emerged as the clear model species (Sharma et al. 2012; Gainett et al. 2021). The ability to compare rates of sequence divergence, within-lineage gene duplication, and, eventually, functional properties of developmental genes in these groups will provide critical comparative data for arachnopulmonates.
Data availability
Alignments and gene accession numbers are included as supplementary information files. Reads and assembled transcriptomes are available on SRA and TSA, respectively, under BioProject PRJNA707377. Supplemental material is available at figshare: https://doi.org/10.25387/g3.14456289.
Acknowledgments
The authors are very grateful to Philip Steinhoff and Gabriele Uhl (University of Greifswald) for providing embryos of M. muscosa, to Matthias Pechmann (University of Köln), Natascha Turetzek (Ludwig-Maximilians-University of Munich) and Prashant Sharma (University of Wisconsin Madison) for providing assembled transcriptomes of A. geniculata, P. phalangioides, and P. opilio, respectively, to Nathan Kenny for helpful comments on the manuscript, and to Simon Ellis (University of Oxford) for IT support.
A.H. analyzed assembled transcriptomes, identified gene candidates, performed phylogenetic analyses, interpreted data, prepared figures, and wrote sections of the manuscript. L.B.G. contributed to data analysis and interpretation. A.S. processed embryos and performed RNA extractions. R.J. provided data for P. phalangioides and P. opilio. M.S. provided amblypygid embryos. M.H. provided sequence data and contributed to phylogenetic analysis. S.A. assembled transcriptomes, analyzed assembly quality and wrote sections of the manuscript. A.P.M. designed and supervised the project, contributed to data interpretation and analysis, and wrote sections of the manuscript. L.S.R. designed and supervised the project, performed phylogenetic analyses, interpreted data, prepared figures, and wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the John Fell Fund, University of Oxford (award 0005632 to L.S.R.), Natural Environment Research Council (NERC) (NE/T006854/1 to A.P.M. and L.S.R.), the Leverhulme Trust (RPG-2016-234 and RPG-2020-237 to A.P.M. and L.S.R., respectively), and the Biotechnology and Biological Sciences Research Council (BBSRC) (grant number BB/M011224/1). These funding bodies were not involved in the design of this study, data collection or analysis, or in writing the manuscript.
Conflicts of interest
The authors declare that there is no conflict of interest.
Literature cited
Kono N, Nakamura H, Ohtoshi R, Moran DAP, Shinohara A, Yoshida Y, Fujiwara M, Mori M, Tomita M, Arakawa K. 2019. Orb-weaving spider Araneus ventricosus genome elucidates the spidroin gene catalogue. Sci Rep. 9:1–13. doi:10.1038/s41598-019-44775-2.
Sheffer MM, Hoppe A, Krehenwinkel H, Uhl G, Kuss AW, Jensen L, Jensen C, Gillespie RG, Hoff KJ, Prost S. 2021. Chromosome-level reference genome of the European wasp spider Argiope bruennichi: A resource for studies on range expansion and evolutionary adaptation. Gigascience. 10:112. doi:10.1093/gigascience/giaa148.