-
PDF
- Split View
-
Views
-
Cite
Cite
Jiyong Su, Jin Gao, Yunlong Si, Linlin Cui, Chenyang Song, Yue Wang, Runjie Wu, Guihua Tai, Yifa Zhou, Galectin-10: a new structural type of prototype galectin dimer and effects on saccharide ligand binding, Glycobiology, Volume 28, Issue 3, March 2018, Pages 159–168, https://doi.org/10.1093/glycob/cwx107
Close - Share Icon Share
Abstract
Galectin-10 (Gal-10) which forms Charcot-Leyden crystals in vivo, is crucial to regulating lymph cell function. Here, we solved the crystal structures of Gal-10 and eight variants at resolutions of 1.55–2.00 Å. Structural analysis and size exclusion chromatography demonstrated that Gal-10 dimerizes with a novel global shape that is different from that of other prototype galectins (e.g., Gal-1, -2 and -7). In the Gal-10 dimer, Glu33 from one subunit modifies the carbohydrate-binding site of another, essentially inhibiting disaccharide binding. Nevertheless, glycerol (and possibly other small hydroxylated molecules) can interact with residues at the ligand binding site, with His53 being the most crucial for binding. Alanine substitution of the conserved Trp residue (Trp72) that is crucial to saccharide binding in other galectins, actually leads to enhanced erythrocyte agglutination, suggesting that Trp72 negatively regulates Gal-10 ligand binding. Overall, our crystallographic and biochemical results provide insight into Gal-10 ligand binding specificity.
Introduction
Charcot-Leyden crystals (CLCs) that have hexagonal and bipyramidal forms, are found as distinct species in the spleen, especially in eosinophils, as well as in basophils and macrophages and in the sputum of individuals with bronchial asthma (Charcot and Robin 1853; Leyden 1872; Gleich et al. 1976; Ackerman et al. 1982). CLCs are considered hallmarks of eosinophil involvement in several diseases, such as asthma, acute myeloid leukemia, mastocytoma (Lao et al. 1998); periapical lesion (Silver and Simon 2000); allergic rhinitis (Bryborn et al. 2010); celiac disease (De Re et al. 2009); eosinophilic cystitis (Staribratova et al. 2010); colorectal cancer (Ågesen et al. 2011); atopic dermatitis (Noh et al. 2015) and parasitic infection in liver (Ackerman et al. 1982; Thakral et al. 2015; Beeson and Bass 1977; Vermeersch et al. 2007; Radujkovic et al. 2011; Manny and Ellis 2012; Taylor et al. 2013).
The CLC gene was first mapped to human chromosome 19 (Mastrianni et al. 1992) and later more accurately to chromosome 19q13.2 (http://www.genecards.org). CLC isolated from human eosinophils contains 1.2% carbohydrate content (Gleich et al. 1976), implying that it has lectin attributes. In fact, the amino acid sequence of CLC with 142 amino acid residues is homologous to the carbohydrate-binding domain (CRD) of galectins (Leffler et al. 1989; Ackerman et al. 1993; Barondes et al. 1994). Moreover, the crystal structure of CLC (Leonidas et al. 1995) shows that it has a highly similar fold to that found in galectins (Ackerman et al. 1993). In addition, the intron-exon architecture of the CLC gene is analogous to that of galectins, with the CRD encoded by a single exon (Dyer et al. 1997). In light of these observations, CLC was re-named galectin-10 (Gal-10) (Leffler et al. 2002).
The mRNA of Gal-10 is highly transcribed in bone marrow (Than et al. 2014), indicating that Gal-10 may play a key role in lymph cell maturation. Gal-10 is found in the cytoplasm, nucleus, cell membrane, periplasm and various granules of eosinophils (Dvorak et al. 1991). Mast cells do not express Gal-10, but can acquire the lectin by endocytosis. Gal-10 spontaneously forms crystals in phagosomes of macrophages (Dvorak et al. 1990), and has been reported to be a novel biomarker essential for anergy and suppressive function of human CD4+CD25+ regulatory T cells (Kubach et al. 2007). The mRNA level of Gal-10 has also been shown to be a marker for CRTH2 activation in human whole blood in vitro (Lin et al. 2010).
Galectins are a family of proteins with binding specificity for β-galactosides (Barondes et al. 1994; Lahm et al. 2004; Liu and Rabinovich 2005). To date, twelve galectin genes (Gal-1, -2, -3, -4, -7, -8, -9, -10, -12, -13, -14 and -16) have been identified in the human genome (Liu and Rabinovich 2005; Yang et al. 2008). Galectins are generally classified as prototype, chimeric type, and tandem-repeat-type (Kasai and Hirabayashi 1996; Cooper 2002; Yang et al. 2008; Tasumi and Vasta 2007). Gal-10 falls into the prototype category. Initial biochemical studies with simple sugars in the solid phase (Leonidas et al. 1995; Dyer and Rosenberg 1996) suggested that Gal-10 has weak, yet specific, binding for N-acetyl-D-glucosamine and lactose (Leonidas et al. 1995; Dyer and Rosenberg 1996). However, contradictory results have indicated that this weak carbohydrate-binding activity may be attributed to Gal-10 binding to the cross-linked agarose (or Sepharose) matrix (Dvorak et al. 1991; Savage et al. 1997). In addition, a previously reported crystal structure of Gal-10 shows that this lectin has no affinity for β-galactosides, but rather can bind mannose in a unique fashion (Swaminathan et al. 1999). To date, the natural carbohydrate ligand(s) for CLC remains unknown.
Although all galectins have a structurally conserved sugar binding pocket (Barondes et al. 1994), there are some variations of conserved sugar binding residues, differences that may contribute to galectin-specific substrate specificity. Within the carbohydrate-binding site, there is a highly conserved Trp residue (Trp72 in Gal-10) that interacts with the pyranoside ring of galactose residues via CH-π interactions. In addition, there is a conserved His residue (His53 in Gal-10) at the base of the binding site that forms hydrogen bonds with O4 of galactose, and an Asn residue (Asn65 in Gal-10) that is proximal to this His that forms hydrogen bonds with O3 and O4 of galactose. These three residues are crucial to stabilizing binding to galactose. However, because knowledge of these interactions is based solely on crystallographic studies, site-directed mutagenesis studies are required to validate these structural models.
In the present study, we have mutated eight residues (C29A, H53A, C57A, N65A, W72A, K73A, Q74A and Q75A) in and around the carbohydrate-binding site of Gal-10 and solved their crystal structures. In all instances, structural resolution was at the 1.55–2.00 Å level. Gel filtration chromatography demonstrated that Gal-10 indeed forms dimers. Furthermore, we used the hemagglutination assay and affinity chromatography to investigate the importance of these residues for carbohydrate binding. In addition, glycerol was found to co-crystallize in the carbohydrate-binding sites of these Gal-10 variants.
Results
Preparation of Gal-10 variants
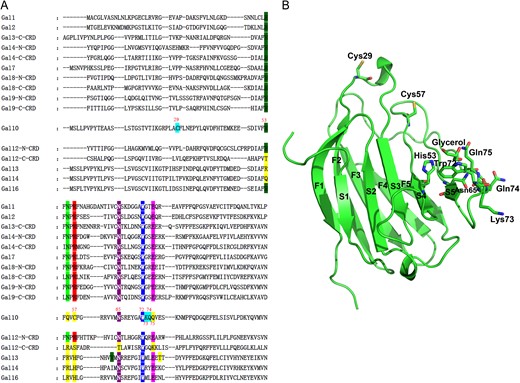
Comparison of the amino acid sequences of Gal-10 with other galectins (Figure 1A) shows that residues in the ligand binding site of Gal-10 are somewhat different from those in other galectins. Therefore, the importance of these residues in determining ligand binding in Gal-10 are worth investigation. Three residues (His53, Asn65 and Trp72) are highly conserved in the CRDs of Gal-1, -2, -3, -4, -7, -8, -9, -12 N-terminal CRD and -16 (Figure 1A and B). Three other Gal-10 residues (Lys73, Gln74, and Gln75) are not conserved as in other galectins (Figure 1A and B). Two cysteine residues (Cys29 and Cys57) have been suggested to be important for the physiological function of Gal-10 (Ackerman et al. 2002) (Figure 1B). Therefore, we mutated all of these eight residues to alanine for insight into their roles in Gal-10 ligand binding.
Primary and 3-D structure of Gal-10. (A) Alignment of the primary structures of Gal-10 with other human galectin CRDs is shown. The alignment was generated using the program ClustalX 2.1. For Gal-10, mutated residues are highlighted in different colors. (B) Positions of the eight mutated residues in Gal-10 are indicated in the crystal structure.
Following site-directed mutagenesis, we expressed all Gal-10 variants in E. coli and concentrated them to ~3–5 mg/mL. For crystallization, we used 288 (i.e., 96 × 3) different conditions, with wild type Gal-10 crystallizing readily under most of them. The hexagonal bipyramid crystal forms were found under all crystallization conditions, implying that the space group of these crystals is the same. The optimal solution condition (0.1 M Bis-Tris pH 6.5 and 2.0 M (NH4)2SO4) produced crystals suitable for data collection. In order to better compare structures with wild type Gal-10, all variants were crystallized under similar conditions (0.1 M Bis-Tris pH 5–7 and 1.8–2.0 M (NH4)2SO4).
Description of wide-type galectin-10 and variants structures
The space group of our Gal-10 structure is same as previously reported for Gal-10 (Leonidas et al. 1995; Swaminathan et al. 1999; Ackerman et al. 2002). Structural statistics for wild type Gal-10 and all variants are provided in Table I. Differences in Cα RMSD values compared to those reported for other Gal-10 structures (PDB codes: 1G86, 1HDK, 1LCL and 1QKQ) are relatively small (0.045 Å to 0.296 Å). This indicates that our Gal-10 structures are similar to each other and that site-directed mutagenesis did not influence Gal-10 structures. However, RMSD values for N-terminal residues Ser2-Pro5 and C-terminal residues Ser138-Arg142 are larger, indicating that these sequences are more flexible.
Data collection and refinement statistics
| PDB code . | Gal-10 with glycerol . | Gal-10 . | C29A . | H53A . | C57A . | N65A . | W72A . | K73A . | Q74A . | Q75A . | H53A soaking 5 min . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 5XRG . | 5XRH . | 5XRI . | 5XRJ . | 5XRK . | 5XRL . | 5XRM . | 5XRN . | 5XRO . | 5XRP . | 5YT4 . | |
| Resolution (Å) | 19.54–1.55 (1.58–1.55) | 19.50–1.55 (1.58–1.55) | 19.61–1.68 (1.71–1.68) | 19.49–1.90 (1.94–1.90) | 19.59–1.70 (1.73–1.70) | 19.65–2.00 (2.05–2.00) | 19.59–2.00 (2.05–2.00) | 19.62–1.60 (1.63–1.60) | 19.57–1.60 (1.63–1.60) | 19.60–2.00 (2.05–2.00) | 19.53–2.00 (2.05–2.00) |
| Space group | P6522 | P6522 | P6522 | P6522 | P6522 | P6522 | P6522 | P6522 | P6522 | P6522 | P6522 |
| Unit cell parameters (a, b, c) (Å) | 48.70 48.70 260.14 | 48.56 48.56 260.33 | 48.93 48.93 258.67 | 48.60 48.64 257.56 | 48.68 48.69 262.19 | 49.06 49.06 259.03 | 48.83 48.83 260.33 | 48.96 48.96 259.23 | 48.74 48.74 260.85 | 48.78 48.78 262.64 | 48.67 48.77 257.66 |
| No. of measured reflections | 483,424 (17,790) | 489,727 (17,756) | 397,789 (20,529) | 272,973 (19,833) | 378,610 (21,111) | 229,755 (10,087) | 241,367 (15,605) | 473,070 (24,068) | 459,532 (23,430) | 119,230 (7992) | 131,183 (10,031) |
| No. of unique reflections | 27,890 (1303) | 27,731 (1288) | 22,254 (1082) | 15,392 (1018) | 21,154 (1078) | 13,413 (865) | 13,511 (907) | 25,733 (1240) | 25,544 (1254) | 13,570 (951) | 13,190 (949) |
| Completeness (%) | 99.7 (97.2) | 99.7 (96.9) | 99.9 (100.0) | 99.9 (100.0) | 98.9 (97.9) | 99.0 (88.5) | 99.3 (92.7) | 99.9 (100.0) | 99.7 (98.8) | 99.6 (96.6) | 99.7 (99.5) |
| Multiplicity | 17.7 (13.7) | 17.7 (13.8) | 17.9 (19.0) | 17.7 (19.5) | 17.9 (19.6) | 17.1 (11.7) | 17.9 (17.2) | 18.4 (19.4) | 18.0 (18.7) | 8.8 (8.4) | 9.9 (10.6) |
| Rmerge (%) | 7.1 (27.2) | 8.0 (45.3) | 9.8 (82.1) | 9.2 (27.5) | 9.2 (20.7) | 15.0 (40.6) | 6.4 (10.8) | 14.1 (64.0) | 6.4 (23.9) | 11.1 (27.6) | 7.8 (30.7) |
| <I/δ (I)> | 24.6 (6.9) | 24.3 (6.7) | 21.6 (5.4) | 22.6 (10.3) | 21.4 (9.9) | 14.2 (5.2) | 34.2 (21.4) | 14.5 (5.0) | 28.3 (10.2) | 13.7 (7.5) | 13.8 (6.8) |
| Rmodel (%) | 16.74 | 17.15 | 18.33 | 17.92 | 16.90 | 17.05 | 16.96 | 18.56 | 16.27 | 17.07 | 18.54 |
| Rfree (%) | 19.66 | 19.57 | 20.37 | 20.19 | 19.20 | 20.89 | 20.78 | 21.82 | 18.90 | 21.42 | 21.61 |
| Rmsd bond lengths (Å) | 0.007 | 0.006 | 0.008 | 0.007 | 0.006 | 0.009 | 0.009 | 0.006 | 0.005 | 0.006 | 0.008 |
| Rmsd bond angles (°) | 0.980 | 0.905 | 0.982 | 0.920 | 0.886 | 1.011 | 0.931 | 0.930 | 0.916 | 0.912 | 0.940 |
| Ramachandran plotf residues in favored regions (%) | 97.2 | 97.9 | 96.4 | 97.8 | 97.9 | 98.6 | 96.4 | 97.1 | 97.9 | 97.1 | 97.8 |
| Ramachandran outliers (%) | 0.7 | 0.7 | 0.7 | 0.7 | 0.7 | 0.7 | 0.7 | 0.7 | 0.7 | 0.7 | 0.7 |
| Ligand | Glycerol | No ligand | Glycerol | No ligand | Glycerol | Glycerol | Glycerol | Glycerol | Glycerol | Glycerol | Glycerol |
| PDB code . | Gal-10 with glycerol . | Gal-10 . | C29A . | H53A . | C57A . | N65A . | W72A . | K73A . | Q74A . | Q75A . | H53A soaking 5 min . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 5XRG . | 5XRH . | 5XRI . | 5XRJ . | 5XRK . | 5XRL . | 5XRM . | 5XRN . | 5XRO . | 5XRP . | 5YT4 . | |
| Resolution (Å) | 19.54–1.55 (1.58–1.55) | 19.50–1.55 (1.58–1.55) | 19.61–1.68 (1.71–1.68) | 19.49–1.90 (1.94–1.90) | 19.59–1.70 (1.73–1.70) | 19.65–2.00 (2.05–2.00) | 19.59–2.00 (2.05–2.00) | 19.62–1.60 (1.63–1.60) | 19.57–1.60 (1.63–1.60) | 19.60–2.00 (2.05–2.00) | 19.53–2.00 (2.05–2.00) |
| Space group | P6522 | P6522 | P6522 | P6522 | P6522 | P6522 | P6522 | P6522 | P6522 | P6522 | P6522 |
| Unit cell parameters (a, b, c) (Å) | 48.70 48.70 260.14 | 48.56 48.56 260.33 | 48.93 48.93 258.67 | 48.60 48.64 257.56 | 48.68 48.69 262.19 | 49.06 49.06 259.03 | 48.83 48.83 260.33 | 48.96 48.96 259.23 | 48.74 48.74 260.85 | 48.78 48.78 262.64 | 48.67 48.77 257.66 |
| No. of measured reflections | 483,424 (17,790) | 489,727 (17,756) | 397,789 (20,529) | 272,973 (19,833) | 378,610 (21,111) | 229,755 (10,087) | 241,367 (15,605) | 473,070 (24,068) | 459,532 (23,430) | 119,230 (7992) | 131,183 (10,031) |
| No. of unique reflections | 27,890 (1303) | 27,731 (1288) | 22,254 (1082) | 15,392 (1018) | 21,154 (1078) | 13,413 (865) | 13,511 (907) | 25,733 (1240) | 25,544 (1254) | 13,570 (951) | 13,190 (949) |
| Completeness (%) | 99.7 (97.2) | 99.7 (96.9) | 99.9 (100.0) | 99.9 (100.0) | 98.9 (97.9) | 99.0 (88.5) | 99.3 (92.7) | 99.9 (100.0) | 99.7 (98.8) | 99.6 (96.6) | 99.7 (99.5) |
| Multiplicity | 17.7 (13.7) | 17.7 (13.8) | 17.9 (19.0) | 17.7 (19.5) | 17.9 (19.6) | 17.1 (11.7) | 17.9 (17.2) | 18.4 (19.4) | 18.0 (18.7) | 8.8 (8.4) | 9.9 (10.6) |
| Rmerge (%) | 7.1 (27.2) | 8.0 (45.3) | 9.8 (82.1) | 9.2 (27.5) | 9.2 (20.7) | 15.0 (40.6) | 6.4 (10.8) | 14.1 (64.0) | 6.4 (23.9) | 11.1 (27.6) | 7.8 (30.7) |
| <I/δ (I)> | 24.6 (6.9) | 24.3 (6.7) | 21.6 (5.4) | 22.6 (10.3) | 21.4 (9.9) | 14.2 (5.2) | 34.2 (21.4) | 14.5 (5.0) | 28.3 (10.2) | 13.7 (7.5) | 13.8 (6.8) |
| Rmodel (%) | 16.74 | 17.15 | 18.33 | 17.92 | 16.90 | 17.05 | 16.96 | 18.56 | 16.27 | 17.07 | 18.54 |
| Rfree (%) | 19.66 | 19.57 | 20.37 | 20.19 | 19.20 | 20.89 | 20.78 | 21.82 | 18.90 | 21.42 | 21.61 |
| Rmsd bond lengths (Å) | 0.007 | 0.006 | 0.008 | 0.007 | 0.006 | 0.009 | 0.009 | 0.006 | 0.005 | 0.006 | 0.008 |
| Rmsd bond angles (°) | 0.980 | 0.905 | 0.982 | 0.920 | 0.886 | 1.011 | 0.931 | 0.930 | 0.916 | 0.912 | 0.940 |
| Ramachandran plotf residues in favored regions (%) | 97.2 | 97.9 | 96.4 | 97.8 | 97.9 | 98.6 | 96.4 | 97.1 | 97.9 | 97.1 | 97.8 |
| Ramachandran outliers (%) | 0.7 | 0.7 | 0.7 | 0.7 | 0.7 | 0.7 | 0.7 | 0.7 | 0.7 | 0.7 | 0.7 |
| Ligand | Glycerol | No ligand | Glycerol | No ligand | Glycerol | Glycerol | Glycerol | Glycerol | Glycerol | Glycerol | Glycerol |
Data collection and refinement statistics
| PDB code . | Gal-10 with glycerol . | Gal-10 . | C29A . | H53A . | C57A . | N65A . | W72A . | K73A . | Q74A . | Q75A . | H53A soaking 5 min . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 5XRG . | 5XRH . | 5XRI . | 5XRJ . | 5XRK . | 5XRL . | 5XRM . | 5XRN . | 5XRO . | 5XRP . | 5YT4 . | |
| Resolution (Å) | 19.54–1.55 (1.58–1.55) | 19.50–1.55 (1.58–1.55) | 19.61–1.68 (1.71–1.68) | 19.49–1.90 (1.94–1.90) | 19.59–1.70 (1.73–1.70) | 19.65–2.00 (2.05–2.00) | 19.59–2.00 (2.05–2.00) | 19.62–1.60 (1.63–1.60) | 19.57–1.60 (1.63–1.60) | 19.60–2.00 (2.05–2.00) | 19.53–2.00 (2.05–2.00) |
| Space group | P6522 | P6522 | P6522 | P6522 | P6522 | P6522 | P6522 | P6522 | P6522 | P6522 | P6522 |
| Unit cell parameters (a, b, c) (Å) | 48.70 48.70 260.14 | 48.56 48.56 260.33 | 48.93 48.93 258.67 | 48.60 48.64 257.56 | 48.68 48.69 262.19 | 49.06 49.06 259.03 | 48.83 48.83 260.33 | 48.96 48.96 259.23 | 48.74 48.74 260.85 | 48.78 48.78 262.64 | 48.67 48.77 257.66 |
| No. of measured reflections | 483,424 (17,790) | 489,727 (17,756) | 397,789 (20,529) | 272,973 (19,833) | 378,610 (21,111) | 229,755 (10,087) | 241,367 (15,605) | 473,070 (24,068) | 459,532 (23,430) | 119,230 (7992) | 131,183 (10,031) |
| No. of unique reflections | 27,890 (1303) | 27,731 (1288) | 22,254 (1082) | 15,392 (1018) | 21,154 (1078) | 13,413 (865) | 13,511 (907) | 25,733 (1240) | 25,544 (1254) | 13,570 (951) | 13,190 (949) |
| Completeness (%) | 99.7 (97.2) | 99.7 (96.9) | 99.9 (100.0) | 99.9 (100.0) | 98.9 (97.9) | 99.0 (88.5) | 99.3 (92.7) | 99.9 (100.0) | 99.7 (98.8) | 99.6 (96.6) | 99.7 (99.5) |
| Multiplicity | 17.7 (13.7) | 17.7 (13.8) | 17.9 (19.0) | 17.7 (19.5) | 17.9 (19.6) | 17.1 (11.7) | 17.9 (17.2) | 18.4 (19.4) | 18.0 (18.7) | 8.8 (8.4) | 9.9 (10.6) |
| Rmerge (%) | 7.1 (27.2) | 8.0 (45.3) | 9.8 (82.1) | 9.2 (27.5) | 9.2 (20.7) | 15.0 (40.6) | 6.4 (10.8) | 14.1 (64.0) | 6.4 (23.9) | 11.1 (27.6) | 7.8 (30.7) |
| <I/δ (I)> | 24.6 (6.9) | 24.3 (6.7) | 21.6 (5.4) | 22.6 (10.3) | 21.4 (9.9) | 14.2 (5.2) | 34.2 (21.4) | 14.5 (5.0) | 28.3 (10.2) | 13.7 (7.5) | 13.8 (6.8) |
| Rmodel (%) | 16.74 | 17.15 | 18.33 | 17.92 | 16.90 | 17.05 | 16.96 | 18.56 | 16.27 | 17.07 | 18.54 |
| Rfree (%) | 19.66 | 19.57 | 20.37 | 20.19 | 19.20 | 20.89 | 20.78 | 21.82 | 18.90 | 21.42 | 21.61 |
| Rmsd bond lengths (Å) | 0.007 | 0.006 | 0.008 | 0.007 | 0.006 | 0.009 | 0.009 | 0.006 | 0.005 | 0.006 | 0.008 |
| Rmsd bond angles (°) | 0.980 | 0.905 | 0.982 | 0.920 | 0.886 | 1.011 | 0.931 | 0.930 | 0.916 | 0.912 | 0.940 |
| Ramachandran plotf residues in favored regions (%) | 97.2 | 97.9 | 96.4 | 97.8 | 97.9 | 98.6 | 96.4 | 97.1 | 97.9 | 97.1 | 97.8 |
| Ramachandran outliers (%) | 0.7 | 0.7 | 0.7 | 0.7 | 0.7 | 0.7 | 0.7 | 0.7 | 0.7 | 0.7 | 0.7 |
| Ligand | Glycerol | No ligand | Glycerol | No ligand | Glycerol | Glycerol | Glycerol | Glycerol | Glycerol | Glycerol | Glycerol |
| PDB code . | Gal-10 with glycerol . | Gal-10 . | C29A . | H53A . | C57A . | N65A . | W72A . | K73A . | Q74A . | Q75A . | H53A soaking 5 min . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 5XRG . | 5XRH . | 5XRI . | 5XRJ . | 5XRK . | 5XRL . | 5XRM . | 5XRN . | 5XRO . | 5XRP . | 5YT4 . | |
| Resolution (Å) | 19.54–1.55 (1.58–1.55) | 19.50–1.55 (1.58–1.55) | 19.61–1.68 (1.71–1.68) | 19.49–1.90 (1.94–1.90) | 19.59–1.70 (1.73–1.70) | 19.65–2.00 (2.05–2.00) | 19.59–2.00 (2.05–2.00) | 19.62–1.60 (1.63–1.60) | 19.57–1.60 (1.63–1.60) | 19.60–2.00 (2.05–2.00) | 19.53–2.00 (2.05–2.00) |
| Space group | P6522 | P6522 | P6522 | P6522 | P6522 | P6522 | P6522 | P6522 | P6522 | P6522 | P6522 |
| Unit cell parameters (a, b, c) (Å) | 48.70 48.70 260.14 | 48.56 48.56 260.33 | 48.93 48.93 258.67 | 48.60 48.64 257.56 | 48.68 48.69 262.19 | 49.06 49.06 259.03 | 48.83 48.83 260.33 | 48.96 48.96 259.23 | 48.74 48.74 260.85 | 48.78 48.78 262.64 | 48.67 48.77 257.66 |
| No. of measured reflections | 483,424 (17,790) | 489,727 (17,756) | 397,789 (20,529) | 272,973 (19,833) | 378,610 (21,111) | 229,755 (10,087) | 241,367 (15,605) | 473,070 (24,068) | 459,532 (23,430) | 119,230 (7992) | 131,183 (10,031) |
| No. of unique reflections | 27,890 (1303) | 27,731 (1288) | 22,254 (1082) | 15,392 (1018) | 21,154 (1078) | 13,413 (865) | 13,511 (907) | 25,733 (1240) | 25,544 (1254) | 13,570 (951) | 13,190 (949) |
| Completeness (%) | 99.7 (97.2) | 99.7 (96.9) | 99.9 (100.0) | 99.9 (100.0) | 98.9 (97.9) | 99.0 (88.5) | 99.3 (92.7) | 99.9 (100.0) | 99.7 (98.8) | 99.6 (96.6) | 99.7 (99.5) |
| Multiplicity | 17.7 (13.7) | 17.7 (13.8) | 17.9 (19.0) | 17.7 (19.5) | 17.9 (19.6) | 17.1 (11.7) | 17.9 (17.2) | 18.4 (19.4) | 18.0 (18.7) | 8.8 (8.4) | 9.9 (10.6) |
| Rmerge (%) | 7.1 (27.2) | 8.0 (45.3) | 9.8 (82.1) | 9.2 (27.5) | 9.2 (20.7) | 15.0 (40.6) | 6.4 (10.8) | 14.1 (64.0) | 6.4 (23.9) | 11.1 (27.6) | 7.8 (30.7) |
| <I/δ (I)> | 24.6 (6.9) | 24.3 (6.7) | 21.6 (5.4) | 22.6 (10.3) | 21.4 (9.9) | 14.2 (5.2) | 34.2 (21.4) | 14.5 (5.0) | 28.3 (10.2) | 13.7 (7.5) | 13.8 (6.8) |
| Rmodel (%) | 16.74 | 17.15 | 18.33 | 17.92 | 16.90 | 17.05 | 16.96 | 18.56 | 16.27 | 17.07 | 18.54 |
| Rfree (%) | 19.66 | 19.57 | 20.37 | 20.19 | 19.20 | 20.89 | 20.78 | 21.82 | 18.90 | 21.42 | 21.61 |
| Rmsd bond lengths (Å) | 0.007 | 0.006 | 0.008 | 0.007 | 0.006 | 0.009 | 0.009 | 0.006 | 0.005 | 0.006 | 0.008 |
| Rmsd bond angles (°) | 0.980 | 0.905 | 0.982 | 0.920 | 0.886 | 1.011 | 0.931 | 0.930 | 0.916 | 0.912 | 0.940 |
| Ramachandran plotf residues in favored regions (%) | 97.2 | 97.9 | 96.4 | 97.8 | 97.9 | 98.6 | 96.4 | 97.1 | 97.9 | 97.1 | 97.8 |
| Ramachandran outliers (%) | 0.7 | 0.7 | 0.7 | 0.7 | 0.7 | 0.7 | 0.7 | 0.7 | 0.7 | 0.7 | 0.7 |
| Ligand | Glycerol | No ligand | Glycerol | No ligand | Glycerol | Glycerol | Glycerol | Glycerol | Glycerol | Glycerol | Glycerol |
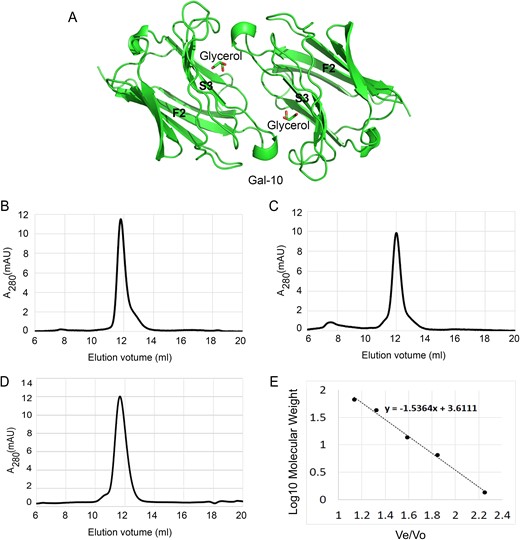
Analysis of our Gal-10 structure suggested that the lectin could form dimers (Figure 2A). Therefore, we used PISA (protein-protein interaction interface server) to predict the dimer interface formed by two S-faces, and found that the solvent-accessible surface of this interface is ~805 Å2, with fourteen hydrogen bonds and two salt bridges being formed. This supports the suggestion that Gal-10 dimerizes through this interface. We then characterized Gal-10 aggregation by size exclusion chromatography (Figure 2B–E). Using the elution volumes of four standard proteins (bovine serum albumin, ovalbumin, ribonuclease A, aprotinin) and vitamin B12, we calculated the apparent molecular weights of our galectin samples (Figure 2B). For Gal-10, we observed a single elution peak at 11.77 mL (calculated to be 28 kDa) (Figure 2C). This peak occurs essentially at the same position as that for dimeric Gal-1 (12.00 mL, calculated to 26.5 kDa) (Figure 2D). Furthermore, in the presence of 500 mM lactose, Gal-10 elutes from the gel as a dimer (11.89 mL, calculated to 28.7 kDa) (Figure 2E), indicating that Gal-10 dimer state is insensitive to the presence of lactose.
Gal-10 forms dimers. (A) Gal-10 crystallized as a dimer. An α-helix connecting S3 and F2 strand from one monomer directly interacts with another monomer. This α-helix is close to the carbohydrate-binding site. The global structure of the Gal-10 dimer is different from that of other prototype galectins. (B) The gel filtration profile of Gal-10. The elution peak of Gal-10 falls at 11.77 mL. (C) The gel filtration profile of Gal-1. The elution peak of Gal-1 falls at 12.00 mL. (D) The gel filtration profile of Gal-10 in the presence of 500 mM lactose. The elution peak of Gal-10 falls at 11.89 mL. (E) Four known proteins and a compound were used to generated a standards equation (y = −1.5364x + 3.6111) for calculating Gal-1 and Gal-10 molecular weights. Ve indicates the protein elution volume. Vo indicates the void volume of column determined by thyroglobulin (669.0 kDa).
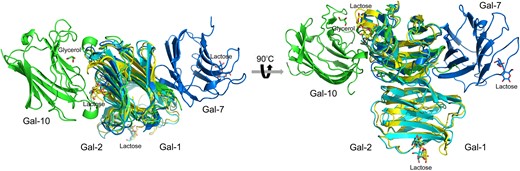
Structural comparison of Gal-10 with Gal-1, -2 and -7 shows that the jelly-roll structure of the Gal-10 monomer is mostly the same as in other galectins (Figure 3). However, the Gal-10 dimer structure is significantly different from that of other galectins. The S-face of Gal-10 plays a key role in dimer formation. However, it appears that the S-face of one monomer subunit could modify the ligand binding site of the other subunit. The two ligand binding sites in Gal-10 are close to each other, closer than with any other galectin dimer (Figure 2A). In contrast, Gal-1 and Gal-2 form dimers via hydrogen bonds and electrostatic interactions contributed by β-strands F1 and S1 (Lobsanov et al. 1993; Lopez-Lucendo et al. 2004), with their carbohydrate-binding sites being located at the far end of each monomer subunit (S5 and S6 β-strands, Figure 3A and B). Gal-7 forms the dimers via F-face interactions of two CRDs that are stabilized by hydrogen bonds and van der Waals interactions (Leonidas et al. 1998), with their carbohydrate-binding sites being located at the opposing sides of the Gal-7 dimer (Figure 3A and B). In Gal-10, two pseudo carbohydrate-binding sites, which cannot bind disaccharides, are located with S5 and S6 β-strands (Figure 3A and B). Overall, the Gal-10 dimer structure is significantly different from those found in other prototype galectins. This in turn implies that Gal-10 evolved its 3-D structure following duplication of its ancestor galectin gene, and that the multivalence and ligand binding specificity of Gal-10 might be different from other galectins.
Overlay of the structures of Gal-1, -2, -7 and -10. (PDB codes: Gal-1, 1LCL; Gal-2, 5DG2; Gal-7, 4GAL; Gal-10, 5XRG) (A) Top view of the overlay. (B) Side view of the overlay. Besides Gal-1 and -2, all prototype galectins show different global homodimer structures comparing to each other.
Carbohydrate-binding sites of Gal-10 and its variants
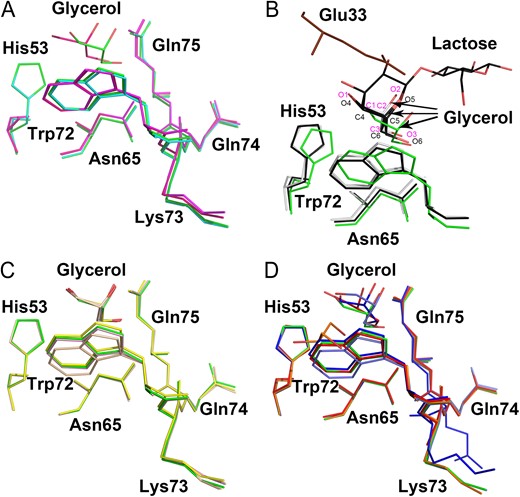
Prior to X-ray data collection, crystals of Gal-10 and its variants were soaked in the reservoir solution supplemented with 20% (v/v) glycerol as a cryoprotectant. All variants were found to be complexed with one molecule of glycerol (Figure 4). Interestingly, whereas most variants required soaking for only 1 min, the H53A variant required 5 min. Moreover, we found that a crystal of wild type Gal-10 (crystallized in the presence of 50 mM lactose without glycerol) had water molecules (and not lactose) in the carbohydrate-binding site where glycerol would have been bound. Apparently, Gal-10 cannot bind lactose. Carbohydrate-binding sites of two wild type Gal-10 structures with and without glycerol are completely superimposable (Figure 4A), with the exception of the H53A variant where Trp72 is oriented differently (Figure 4A). This finding suggests that His53 is crucial for Gal-10 ligand binding.
Comparison of carbohydrate-binding sites of Gal-10 variants. (A) Overlay of carbohydrate-binding sites of two wild type Gal-10 structures and two H53A variant structures. One wild type Gal-10 (colored green) co-crystallized with glycerol and another wild type Gal-10 (colored cyan) did not. H53A (colored purple) could not co-crystallize with glycerol upon 1 min soaking with glycerol. H53A (colored hot pink) could co-crystallize with glycerol upon 5 min soaking with glycerol. (B) Overlay of carbohydrate-binding sites of Gal-10 (colored green) with Gal-3 (white) and Gal-8 (black). Glycerol in the carbohydrate-binding site of Gal-10 adopts the same conformation as glycerol in Gal-3 and Gal-8. A glutamate residue (Glu33, colored brown) from another Gal-10 monomer occupies some portion of the normal lactose binding site. This position of this glutamate prevents larger saccharides from binding to the Gal-10 carbohydrate-binding site. (C) Overlay of carbohydrate-binding sites of C29A (colored yellow), C57A (colored wheat), Q74A (colored pink) and wild type Gal-10 (colored green). Glycerol molecules in their carbohydrate-binding sites could perfectly merge with each other. This indicates that these mutations do not negatively affect carbohydrate binding. (D) Overlay of carbohydrate-binding sites of N65A (colored blue), W72A (colored orange), K73A (colored red), Q75A (colored marine) and wild type Gal-10 (colored green). All glycerol molecules in these variants showed different conformations comparing to wild type Gal-10. W72A also could co-crystallized with glycerol. However, the glycerol molecule in W72A shifts a large distance compared with other variants. This indicates that Trp72 may play a role in regulating Gal-10 ligand binding specificity.
Careful inspection of the Gal-10 dimer interface shows that Glu33 from an opposing subunit occupies a large area at the top of the Gal-10 carbohydrate-binding site (Figure 4B). This may explain why we could not co-crystallize or soak-in any sugar molecules (including lactose, mannose, glucose, sucrose, N-acetyl-D-glucosamine and N-acetyl-D-lactosamine) into the Gal-10 carbohydrate-binding site. Therefore, for further insight into this quandry, we overlaid the structures of Gal-3 (PDB: 2NMO; Collins et al. 2007), Gal-8 (PDB: 5GZC; Si, Wang et al. 2016) and Gal-10. Gal-3 can bind both lactose and glycerol, with glycerol showing only partial occupancy with the C1, C2, C3, O1, O2 and O3 atoms of glycerol fully overlapping with the C4, C5, C6, O4, O5 and O6 atoms of lactose (Figure 4B). The carbohydrate-binding site of Gal-8 N-CRD also could bind glycerol (Si, Wang et al. 2016) just like Gal-3. The overlay of these three galectins showed that glycerol in Gal-10 is similarly oriented, but for the carboxyl group of Glu33 from an opposing subunit that prevents saccharides from interacting at the carbohydrate-binding site.
In Gal-10 variants C29A, C57A and Q74A, glycerol molecules perfectly overlapped with those in wild type Gal-10 (Figure 4C). Cys29 and Cys57 are distant from the carbohydrate-binding site, and although mutation of Cys57 to alanine does influence the orientation of Trp72, it did not prevent glycerol from binding in the same way as in wild type Gal-10. However, we proved that Trp72 is crucial to determining Gal-10 ligand binding specificity (see the following sections), suggesting that the redox state of the Cys57 thiol side chain may indirectly modify Gal-10 ligand binding specificity by influencing the orientation of Trp72.
On the other hand, glycerol in Gal-10 variants N65A, W72A, K73A and Q75A was found to adopt different conformations compared to wild type Gal-10 (Figure 4D). Unexpectedly, the W72A variant could also trap a glycerol molecule with the same orientation as in wild type Gal-10. This was surprising because this tryptophan is highly conserved and crucial to stabilizing the pyranoside ring of galactose in other galectins. The position and conformation of glycerol in W72A were different from glycerol in the carbohydrate-binding site of other Gal-10 variants (Figure 4D). In addition, mutation of Asn65 to alanine altered the position of the ketone oxygen atom of Lys73, suggesting that H-bonding with the Asn65 amide group is essential for maintaining the orientation of Lys73 (Figure 4D).
Gal-10 induces erythrocyte agglutination
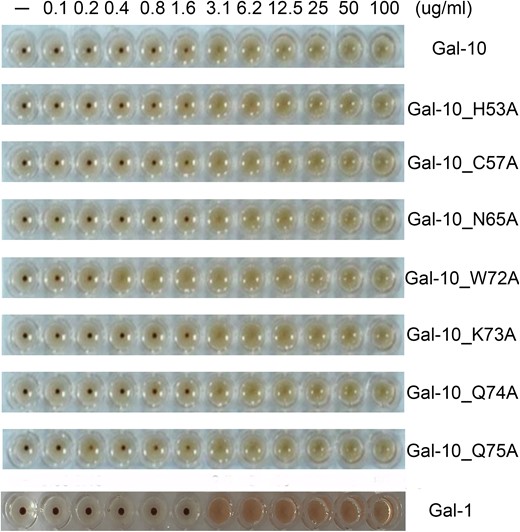
The hemagglutination assay is a classic method used to evaluate inhibitory effects of carbohydrates on galectins. Here, we used chicken erythrocytes to determine the minimum inhibitory concentration (MIC) of each carbohydrate inhibit Gal-10-induced hemagglutination. Gal-10 alone induced agglutination with a minimum agglutination concentration (MAC) of 3.1 ug/mL, the same as control Gal-1 (Figure 5) consistent with our previous results (Si, Feng et al. 2016). Moreover, the MAC was the same for all Gal-10 variants, with the exception of W72A which was considerably lower, i.e., 0.4 ug/mL (Figure 5). This was surprising, because it lacked the conserved Trp residue normally required for β-galactoside binding. We also found that none of the disaccharides (i.e., lactose, mannose, glucose, fructose, sucrose, N-acetyl-D-glucosamine and N-acetyl-D-lactosamine) that we used to inhibit Gal-10-mediated agglutination, worked (Supplementary Figure 1A–D). This contradicts a previous report that some saccharides could inhibit Gal-10 activity (Leonidas et al. 1995). As a control, 2.0 mM lactose could inhibit agglutination induced by Gal-1 (Supplementary Figure 1B).
Hemagglutination assay with all Gal-10 species and Gal-1. With the exception of W72A, Gal-10 and its other variants induce agglutination of chicken erythrocytes with a MAC (Minimum Agglutination Concentration) value of 3.1 ug/mL. The MAC value for the W72A variant is much lower, i.e., 0.4 ug/mL. The MAC value for Gal-1 is 3.1 ug/mL.
Assessment of direct interactions between Gal-10 and saccharides
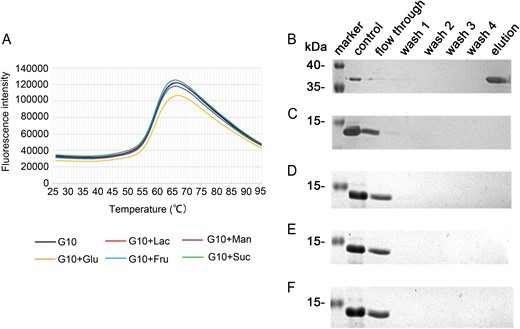
We used the thermal shift assay to assess whether saccharides could influence the denaturation temperature (TM) of Gal-10. Although we expected the TM value to increase upon sugar binding, the disaccharides lactose, mannose, glucose, fructose, sucrose, N-acetyl-D-glucosamine and N-acetyl-D-lactosamine, had no effect on TM values (Figure 6A). This finding is consistent with our agglutination studies and indicates that these saccharides do not stabilize the structure of Gal-10, and likely do not bind to the lectin. Affinity chromatography also demonstrated that Gal-10 does not bind these saccharides, and although control Gal-3 could interact with lactose-modified sepharose 6B (Figure 6B), Gal-10 did not (Figure 6C). In addition, Gal-10 did not interact with mannose- or glucose-modified sepharose-6B (Figure 6D–F). These results are consistent with previous studies (Leonidas et al. 1995; Dyer and Rosenberg 1996).
Binding of disaccharides to Gal-10. (A) Thermal shift assay. Lactose, mannose, glucose, fructose and sucrose did not increase the melting temperature of Gal-10, indicating that these saccharides cannot bind to Gal-10. (B) Lactose-modified sepharose 6B could recover Gal-3. (C) sepharose-6B could not unspecifically recover Gal-10. (D)–(F) Lactose sepharose-6B, Mannose sepharose-6B and Glucose-sepharose 6B also could not recover Gal-10. These results indicate that Gal-10 cannot directly interact with these saccharides.
Discussion
In eosinophils, Gal-10 is the second most abundant protein, comprising about 10% of all proteins present (Leonidas et al. 1995). Our gel filtration data indicate that Gal-10 is a dimer as seen in our crystal structure of the lectin. As we know, prototype galectins Gal-1, -2 and -7 also form dimers, albeit in different ways. Our crystal structure of Gal-10 shows that it forms a novel prototype galectin dimer structure with the α-helix connecting S3 and F2 β-strands directly contacting S1, S2 and S3 β-strands of an opposing monomer subunit. In addition, our Gal-10 structures show that this lectin can bind glycerol which is flexible and can adopt various conformations (Diehl et al. 2010). Previously, glycerol co-crystallized in the carbohydrate-binding sites of Gal-3 and Gal-8 N-CRD (Collins et al. 2007; Si, Wang et al. 2016). In those structures, glycerol mimicked part of the galactose ring, taking the place of its C4, C5, C6, O2, O5 and O6 atoms. Here, we found that even though the positioning of Glu33 in Gal-10 inhibits binding of disaccharides, it does not impede binding of glycerol. This implies that Gal-10 may be able to bind different ligands compared to other lectins. Here, we showed that glycerol binding to Gal-10 is specific, because we could cryo-cool a crystal of Gal-10 in liquid nitrogen in the absence of the cryoprotectant glycerol and observed that four bound water molecules took the place of glycerol. All other variants could also co-crystallize with glycerol. Overall, our results suggest that the carbohydrate-binding site of Gal-10 is a natural site for binding or trapping molecules with hydroxyl groups. Mutation of any single amino acid could not fully abolish affinity of Gal-10 for some ligands.
Unexpectedly, the conformation of glycerol in the W72A variant was quite different compared to that in the other Gal-10 variants. And whereas we expected W72A to be inactive in the agglutination assay, its activity was actually greater than that with wild type Gal-10. In several glycoside hydrolase families, a tryptophan is at the catalytic center to assist the enzyme by interacting with the pyran ring of monosaccharides via van der Waals forces (Pozzo et al. 2010). This interaction is similar to that observed between the conserved tryptophan and pyran ring of galactose in galectins. Mutation of this tryptophan in glycoside hydrolases does not abolish activity, but rather modifies substrate specificity (Pozzo et al. 2010). Similar to Gal-10, mutation of Trp72 to alanine may significantly change/increase its ligand binding specificity, which could explain why the W72A variant is more effective at promoting hemagglutination.
All of our variants could induce agglutination with similar activities as wild type Gal-10. This is consistent with our crystallographic data that demonstrate that all variants can bind glycerol. Although H53A requires a longer soaking time to bind glycerol, this variant could still effectively induce erythrocyte agglutination. Because all Gal-10 variants cannot bind simple disaccharides, it is likely that Gal-10 induces hemagglutination via an unknown mechanism. Actual galectin ligands on cells are generally more complex than simple disaccharides, and this could explain why small disaccharides could not inhibit the biological activity of Gal-10. The thermal shift assay showed that sugars cannot stabilize the Gal-10 structure, and our affinity chromatography studies indicate that Gal-10 cannot bind to any disaccharide-modified sepharose 6B examined here. In conclusion, our results have provided new structural and biochemical information on Gal-10 and its carbohydrate-binding specificity.
Materials and methods
Cloning, expression, and purification of wild type Gal-10
The Gal-10 gene was synthesized by SynBio Technologies (Monmouth Junction, USA), and amplified by using primers that contain NdeI and XhoI restriction sites. PCR products were digested and cloned into a pET28a vector (Novagen, Gibbstown, USA). For overexpression of the recombinant proteins, the construct was transformed into E. coli BL21 (DE3) cells and grown in LB medium supplemented with kanamycin (100 μg/mL). When the optical density of the cultures reached 0.6–0.8, IPTG was added to a final concentration of 0.5 mM to induce protein expression. After 16 h of induction at 25°C, the cells were harvested by centrifugation and lysed by sonification in a lysis buffer consisted of 10 mM Tris/HCl, pH 7.5, 300 mM NaCl, 2 mM β-mercaptoethanol, 20 mM imidazole. The clarified cell extract was used for protein purification with Ni-NTA Agarose (Qiagen, Hilden, Germany). After purification, the His-tagged protein was dialyzed in 10 mM Tris-HCl, pH 7.5; 150 mM NaCl; 2 mM β-mercaptoethanol. During dialysis, thrombin was added to remove the His tag with 5 units (National Institutes of Health unit) per milligram protein. As determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), protein purity was >90%. Finally, the protein was concentrated to 5 mg/mL and stored at −80°C.
Site-directed mutagenesis and protein purification
Site-directed mutagenesis of tPphA was performed according to the manual in the QuickChange XL site directed mutagenesis kit (Stratagene, La Jolla, Canada). All constructs were checked by DNA sequencing. For over production of Gal-10 variants, constructs were transformed into E. coli BL21 (DE3) cells and grown in LB medium. Purifications of Gal-10 variants were performed in the same way as for the wild type protein. As determined by SDS-PAGE, all protein purities were >80%. Proteins were concentrated to approximately 3–5 mg/mL and stored at −80°C.
Hemagglutination assay
The hemagglutination assay was performed according to published protocols (Gao et al. 2013; Si, Wang et al. 2016) with some minor modifications. Briefly, chicken erythrocytes were prepared and maintained at 4°C. The hemagglutination assay was performed in microtiter V plates, with each well containing Gal-1 or Gal-10 in 75 μL Tris buffer (10 mM Tris-HCl,150 mM NaCl, pH 7.5) and 25 μL 3% (v/v) chicken erythrocyte suspensions. Cells were added last, followed by mild shaking, and agglutination was allowed to occur for 60 min on ice to ensure a consistent temperature. Lactose, mannose, glucose, fructose, sucrose, N-acetyl-glucosamine and N-acetyl-glucosamine were used to inhibit biological activity of wild type protein and Gal-10 variants. Lactose was used to inhibit Gal-1 biological activity.
Crystallization, data collection and structure determination
Hampton Research packs (PEGRx1, PEGRx2, Index, Crystal Screen and Crystal Screen 2) were used for the initial crystallization screen (sitting-drop vapor diffusion method). After one day of incubation at 20°C, crystals appeared under many conditions. To obtain a crystal suitable for X-ray diffraction, we optimized the crystal by using the initial condition (Crystal screen 2 No. 45) containing 0.1 M Bis-Tris pH 5.5, 2 M (NH4)2SO4. Larger crystals were obtained after 1–7 days from drops that contained 1 μL protein and 1 μL solution from the well containing 0.1 M Bis-Tris pH 5.5–7, 2–2.2 M(NH4)2SO4 (hanging-drop method) at 15°C. Prior to X-ray data collection, crystals were soaked for approximately 1 min in the reservoir solution supplemented with 20% (v/v) glycerol as a cryoprotectant, and then flash cooled in liquid nitrogen. H53A crystal was also soaked for 5 min in glycerol solution. Data sets were collected at 100 K at the Shanghai Synchrotron Radiation Facility (Shanghai, China).
Data sets were indexed and integrated using XDS and scaled using Aimless from the CCP4 package (6.4.0, Science and Technology Facilities Council, Rutherford Appleton Laboratory, Didcot, Oxon, UK, 2015). Structures were determined by Phaser with a molecular replacement method using the structure of Gal-10 (PDB: 1LCL) as the search model. Structure refinement and water updating were performed using Phenix refine and manual adjustment. Final structure validations were performed using MolProbity. Figures for all structures were generated using PyMOL or Coot.
Gel filtration chromatography
Gel filtration was performed at 25°C using the Äkta purifier 10 system (GE Healthcare, Uppsala, Sweden), with a running buffer of PBS. A total of 400-μg protein sample was loaded onto a Superdex 75, 10/300 column and eluted at a flow rate of 1 mL/min. The absorbance was monitored at 280 nm.
Assessment of carbohydrate-binding activity
The experiment was performed according to the published protocol (Fornstedt and Porath 1975) with some minor modifications. We prepared Lac sepharose 6B, Man sepharose 6B and Glu sepharose 6B, and performed the experiment using 100 μg Gal-10 and 10 uL sepharose 6B in an Eppendorf tube at 20°C for 1 h. The conjugate was washed four times with Tris buffer, and subsequently eluted with Tris buffer containing 0.5 M ligand. Each supernatant was collected and prepared in loading buffer and heated to 98°C for 10 min prior to running SDS-PAGE.
Thermal shift assay
The thermal shift assay was performed using the ABI StepOne/StepOnePlus Real Time Detection System (ABI, Foster City, USA). For this, 15 μg of Gal-10, 0.3 μL of 250 × SyPRO orange, 6 μL of Tris buffer and the disaccharides (20 mM lactose, mannose, glucose, fructose or sucrose) were added to wells in 96-well PCR plates (BIOplastics, Landgraaf, The Netherlands). Plates were heated from 25°C to 95°C at a heating rate of 4°C/min. Fluorescence intensity was measured at Ex/Em = 490/530 nm.
Supplementary data
Supplementary data are available at Glycobiology online.
Funding
This work was supported by the National Natural Science Foundation of China [31500637 to J.S., 31500274 to Y.Z.], the Drug Discovery Found of Jilin Province [No. 20150311057YY], the Natural Science Foundation of Jilin Province of China [No. 20160101343JC].
Conflict of interest statement
None declared.
Acknowledgements
We thank Kevin H. Mayo for critical reading and editing of this manuscript. We thank the staff from BL17B/BL18U/BL19U1/BL19U2/BL01B beamline of National Facility for Protein Science Shanghai (NFPS) at Shanghai Synchrotron Radiation Facility, for assistance during data collection.
Abbreviations
CLC, Charcot-Leyden crystal; CRD, carbohydrate-binding domain; Gal, Galectin.