-
PDF
- Split View
-
Views
-
Cite
Cite
Stormy J. Chamberlain, Karen A. Johnstone, Amanda J. DuBose, Thomas A. Simon, Marisa S. Bartolomei, James L. Resnick, Camilynn I. Brannan, Evidence for genetic modifiers of postnatal lethality in PWS-IC deletion mice, Human Molecular Genetics, Volume 13, Issue 23, 1 December 2004, Pages 2971–2977, https://doi.org/10.1093/hmg/ddh314
Close - Share Icon Share
Abstract
Prader–Willi syndrome (PWS), most notably characterized by infantile hypotonia, short stature and morbid obesity, results from deficiencies in multiple genes that are subject to genomic imprinting. The usefulness of current mouse models of PWS has been limited by postnatal lethality in affected mice. Here, we report the survival of the PWS-imprinting center (IC) deletion mice on a variety of strain backgrounds. Expression analyses of the genes affected in the PWS region suggest that while there is low-level expression from both parental alleles in PWS-IC deletion pups, this expression does not explain their survival on certain strain backgrounds. Rather, the data provide evidence for strain-specific modifier genes that support the survival of PWS-IC deletion mice.
INTRODUCTION
Prader–Willi syndrome (PWS) is characterized by infantile hypotonia, gonadal hypoplasia, a moderate delay in physical and mental development, short stature, obsessive/compulsive behavior and neonatal feeding difficulties, later followed by hyperphagia which contributes to profound obesity (1). PWS is an imprinting disorder caused by loss of paternal gene expression from chromosome 15q11–q13. The predominant causes are paternal inheritance of a 4 Mb deletion of 15q11–q13 or maternal uniparental disomy (UPD) for chromosome 15 (2). A second neurobehavioral disorder, Angelman syndrome (AS), also maps to 15q11–q13 but is caused by the loss of maternal contribution (3). A small number of cases of both PWS and AS are due to imprinting defects. These can be caused by microdeletions involving the imprinting center (IC), which regulates imprinting across an ∼2 Mb region (4). The IC has a bipartite structure, comprising the PWS-IC, located around exon 1 of SNRPN (5), and the AS-IC, located 35 kb upstream of SNRPN (6). No case of PWS has been attributed to a single gene defect, strongly suggesting that it is a contiguous gene syndrome (7). A number of paternally expressed PWS candidate genes have been identified, including MKRN3 (8), MAGEL2 (9), NDN (10), SNRPN (11,12), IPW (13), a number of snoRNAs (HBII-436, HBII-13, HBII-437, HBII-438A, HBII-85, HBII-52 and HBII-438B) (14,15) and an antisense transcript to the UBE3A gene (16). Recent evidence suggests that SNRPN, the snoRNAs and the UBE3A antisense are products of a single transcriptional unit (17).
Both gene content and imprinting are conserved in a region of mouse chromosome 7C. The first mouse model of PWS to be reported was a model for UPD (18). Mice with maternal duplication of a region of chromosome 7 show poor feeding, failure to thrive and die between 2 and 8 days after birth. A model for PWS deletions arose fortuitously as a transgene-induced deletion of the PWS/AS orthologous region in mice (19). These mice are reported to have failure to thrive, feeding difficulties, reduced movement, irregular respiration and all die within 1 week of birth. We previously generated a model for PWS-IC deletions by targeted deletion of 35 kb (originally reported to be 42 kb) including 16 kb upstream of Snrpn and exons 1–6 (PWS-ICdel) (20). Breeding male chimeric founders (129S1/Sv) harboring a maternal deletion mutation to wild-type females (C57BL/6J) resulted in mutant offspring that usually died within 48 h and never survived beyond 7 days after birth. These mice exhibited several phenotypes similar to those found in PWS infants, including poor feeding, failure to thrive and small size. In addition, the progeny lacked expression of the local paternally expressed genes Mkrn3, Ndn and Ipw (20), as well as Magel2 (21), MBII-13, MBII-85 and MBII-52 (14). Additional mouse models have been employed to analyze the contribution of subsets or individual genes to PWS phenotypes. The most notable of these are a deletion from Snrpn to Ube3a, which results in hypotonia, growth retardation and 80% postnatal lethality (22), and deletion of Ndn, which presents with variable phenotype (see Discussion), but two reports identify substantial postnatal lethality in Ndn-deficient mice (23–25).
The phenotypes consistently reported in mouse models of PWS include feeding difficulties, failure to thrive and growth deficiency. These phenotypes are also evident in PWS infants. However, the usefulness of mouse models to study later onset obesity and behavioral phenotypes has been limited by the 100% postnatal lethality observed in the deletion, UPD and PWS-IC deletion models. Here, we report survival of PWS-ICdel mice dependent upon maternal strain. Surviving PWS-ICdel mice still present with failure to thrive and growth deficiency, but display a strain-dependent reduction in postnatal lethality. We also present evidence for the existence of a modifier locus that is dependent on the maternal strain.
RESULTS
Survival of PWS-IC+/del mice is strain-dependent
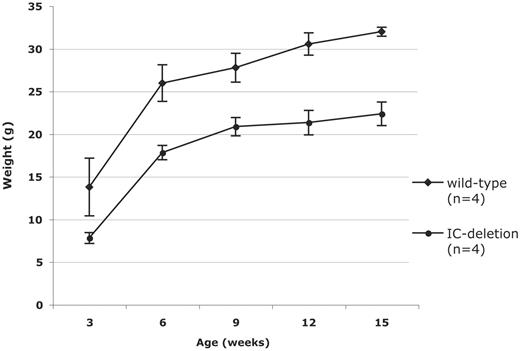
In contrast to the fully penetrant neonatal lethality phenotype observed in crosses with wild-type C57BL/6J females, crosses between wild-type female FVB/NJ mice and PWS-ICdel carrier males (on both 129S1/Sv and C57BL/6J backgrounds) produced several mutant offspring that survived to 2 weeks of age. Furthermore, offspring inheriting the PWS-ICdel allele paternally (PWS-IC+/del), could survive to adulthood if most of the wild-type competitor sibs were removed. Although surviving PWS-IC+/del mice are significantly smaller than wild-type sibs (Fig. 1), both males and females are fertile and never become obese on a standard diet (∼9% fat).
To determine whether survival is due to superior mothering by FVB/NJ females as compared with C57BL/6J females, newborn pups were fostered to mothers of the opposite strain. In addition, an FVB/NJ female and a C57BL/6J females were set up in a trio mating with a heterozygous PWS-ICdel male. In either case, only PWS-IC+/del mice born to FVB/NJ females survived, demonstrating that survival is a consequence of the genetic background of either the offspring or the mother and does not result from superior mothering skills (data not shown).
Females from several strains (C57BL/6J, FVB/NJ, C3H/HeJ, BALB/cJ, 129S1/Sv and DBA/2J) were mated to C57BL/6J PWS-ICdel/+ males to determine whether survival was restricted to offspring of the FVB/NJ strain. In each case, all but one of the wild-type littermates were removed from the litters within two days of birth and the survival of PWS-IC+/del pups was monitored. Pups born to C57BL/6J or DBA/2J females had the shortest survival time, with the longest surviving affected individual from either strain living only for 7 days. Mothers of all other strains produced at least two affected pups that survived to 3 weeks of age (data not shown). PWS-IC+/del mice on all strains displayed failure to thrive and weakness, and if death occurred it was generally within 14 days of birth. Almost all animals surviving for 14 days also survived to adulthood. (C57BL/6J×C3H/HeJ)F1 and either (C57BL/6J×FVB/NJ)F1 or (FVB/NJ×C57BL/6J)F1 females were also mated with heterozygous PWS-ICdel males to ascertain survival of PWS-IC+/del pups from F1 mothers. Similarly, all wild-type pups except one were culled within 2 days of birth. Both F1 mothers produced offspring that survived to 3 weeks of age. However, when compared with FVB/NJ mothers, the rate of survival was considerably lower (Table 1). This further suggests that a genetic factor is involved in survival and that the C57BL/6J background is non-permissive.
Incomplete silencing of PWS genes on all strain backgrounds
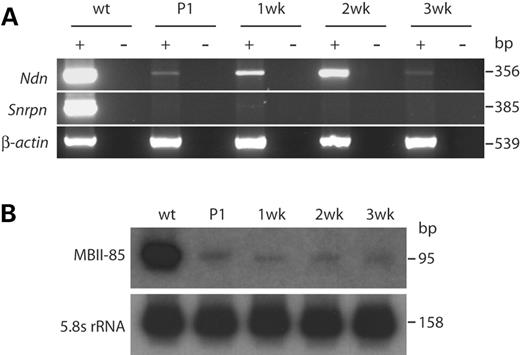
As PWS-IC+/del offspring of FVB/NJ mothers survive with a high frequency, we sought to determine whether gene expression of imprinted genes was altered in surviving mice. (FVB/NJ×C57BL/6J)F1 PWS-IC+/del mice were sacrificed as neonates and at 1, 2 and 3 weeks of age. Expression of several PWS candidate genes in the brain was examined by northern blot and RT–PCR. While Ndn and Snrpn were not detectable by northern blot (data not shown), we detected expression of Ndn by RT–PCR (Fig. 2A). Snrpn expression was also evident, but at a very low level. Expression seemed to be variable between mutant offspring, perhaps because of the non-linear nature of PCR, and showed no correlation with age. Comparable low-level expression of the MBII-85 snoRNA was also detectable by northern blot in all ages of PWS-IC+/del mice (Fig. 2B). These results were unexpected because our previous data indicated that these genes were completely silenced following paternal inheritance of the PWS-IC deletion (20). Low-level expression of Snrpn is surprising because the RT–PCR amplifies sequences removed by the PWS-IC deletion, indicating that Snrpn is being expressed from the maternal chromosome.
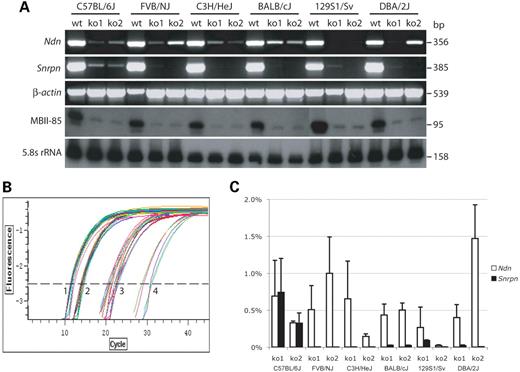
As low-level expression of paternal genes had not been previously observed in pups of C57BL/6J mothers, we tested the hypothesis that residual expression could mitigate the severe lethality phenotype associated with paternal inheritance of the PWS-IC deletion. Northern blot, RT–PCR and real-time quantitative PCR (qPCR) were used to study expression of Ndn, Snrpn and MBII-85 in total brain RNA from PWS-IC+/del pups born to mothers of survival non-permissive (C57BL/6J and DBA/2J) and permissive (FVB/NJ, BALB/cJ, C3H/HeJ and 129S1/Sv) strains. Northern blot and RT–PCR showed low-level expression of Ndn, Snrpn and MBII-85 in all strains (Fig. 3A) including non-permissive strains. In addition, qPCR analysis showed that these genes were expressed at a very low level compared with wild-type expression levels (Fig. 3B and C). The amplification plots for Ndn expression showed that it was expressed with similar kinetics in PWS-IC+/del pups from all strains (Fig. 3B), and expression was generally <1% of wild-type expression levels (Fig. 3C). While there was some variability both within and between strains, there was no correlation with differences in survival. On the other hand, Snrpn expression was higher in offspring from C57BL/6J mothers (0.5% of wild-type), while all other strains expressed with similar kinetics (<0.07% of wild-type). The data suggests that the low-level expression of PWS transcripts does not play a role in strain-dependent survival of PWS-IC deletion mice. It is interesting to note that although Snrpn expression must originate from the maternal chromosome, expression is most evident with C57BL/6J mothers, suggesting some relaxation of imprinting in this strain.
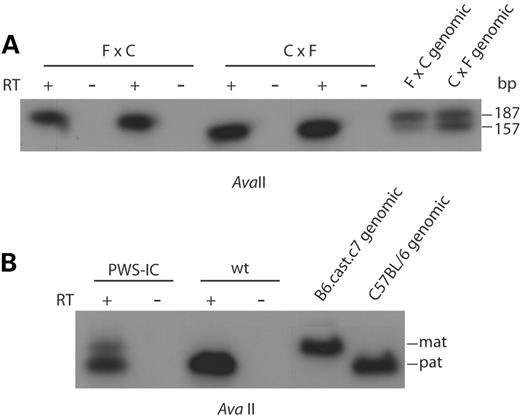
As there is some evidence of maternal expression of Snrpn, we sought to determine whether maternal expression contributed to low-level expression of Ndn. FVB/NJ females were mated with wild-type males from the B6.CAST.c7 strain, congenic for a region of Mus musculus castaneus chromosome 7 on a C57BL/6J background. The reciprocal mating was also performed. Total brain RNA was isolated from (FVB/NJ×B6.CAST.c7)F1 or (B6.CAST.c7×FVB/NJ)F1 mice at birth. The parental origin of Ndn expression was distinguished by RT–PCR and restriction digestion to detect an expressed AvaII polymorphism between domesticus and castaneus alleles. Both crosses demonstrate that expression is paternal in origin (Fig. 4A). B6.CAST.c7 females were also mated with PWS-ICdel/+ males, and parental expression of Ndn in (B6.CAST.c7×PWS-ICdel)F1 pups was analyzed using the same polymorphism. Although Ndn expression was limited to the paternal domesticus allele in wild-type pups, expression in pups that had inherited the PWS-ICdel allele paternally was biallelic, representing both domesticus and castaneus alleles (Fig. 4B). This indicates that a low level of maternal expression is only observed or detectable in the absence of an active paternal PWS-IC.
Evidence for a maternal genetic factor influencing survival
The most likely interpretation of these results is that survival does not correlate with residual transcription of PWS genes. The current evidence supports the existence of a modifier locus that influences survival in PWS-IC+/del mice. The inability to successfully foster mutant offspring between strains also points to the involvement of genetic factors. This is further supported by the decreased survival observed in crosses with F1 mothers. C57BL/6J (non-permissive) and FVB/NJ or C3H/HeJ (both permissive) F1 mothers produced significantly fewer surviving offspring than FVB/NJ mothers (Table 1).
As reported here and previously (20), crosses between C57BL/6J females and PWS-ICdel/+ males on the 129S1/Sv background generate B6129SF1 offspring that never survive beyond 7 days of age (n=30 litters, no PWS-IC+/del mice survived, 53 dead pups with confirmed PWS-IC+/del genotype). In contrast, in this study, we have shown that the reciprocal cross between a 129S1/Sv female and a PWS-ICdel/+ male on the C57BL/6J background produces 129SB6F1 offspring that can survive (n=2 litters, 2 PWS-IC+/del mice survived) when the wild-type sibs are culled. While the offspring from these crosses are genetically identical, the effects on survival are not. The results indicate that survival of PWS-IC+/del pups is dependent upon the maternal strain.
DISCUSSION
Heterozygous PWS-ICdel males bred to C57BL/6J or DBA/2J females produce mutant offspring that do not survive until weaning, even if wild-type siblings are removed to reduce competition. However, long-term survival of PWS-IC+/del mice was achieved with mothers from several other strains, including FVB/NJ, C3H/HeJ, 129S1/Sv and BALB/cJ. On all of these strains, surviving PWS-IC+/del mice are significantly smaller than wild-type littermates. Unlike affected children, they do not become obese on a standard diet, both males and females are fertile, and they appear to be grossly normal in all other respects. The absence of an obesity phenotype in PWS-IC+/del mice is perplexing, since obesity is a major component of the PWS phenotype in humans. There may be significant physiological differences between humans and mice in this respect. Alternatively, the failure to thrive phenotype may be relatively severe and may not subside sufficiently to permit PWS-IC+/del mice to become obese on a standard diet. Behavioral studies of adult PWS-IC+/del mice are currently underway.
Expression analysis of Ndn, Snrpn and MBII-85 snoRNA reveals residual expression of these imprinted genes in PWS-IC+/del mice, which was not previously observed (20). The original mutation was on a 129S1/Sv strain background. Altered transcription may be a reflection of relaxation of imprinting as the mutation has been progressively backcrossed to the C57BL/6 strain. Originally detected in PWS-IC+/del pups from FVB/NJ mothers, this expression is also present at comparable levels in pups from all other maternal strains, including C57BL/6J and DBA/2J, which do not support survival. This suggests that the increased survival of PWS-IC+/del pups is not a direct result of the low-level expression of genes in the PWS region.
Previous data indicated that, in the absence of the PWS-IC, the paternal chromosome is completely repressed (20). In this study, we detect low-level expression from the paternal chromosome in PWS-IC+/del mice. The PWS-IC is required for the establishment and maintenance of expression of the upstream genes (26), which lie more than 1 Mb away from the IC. Expression of Ndn could reflect residual expression in the absence of enhancers or other elements normally present in the PWS-IC. The parental origin of MBII-85 expression is unknown. Although expression could be maternal in origin, it could also arise from the paternal chromosome. In humans, the main promoter for MBII-85 expression is believed to be the SNRPN promoter (17,27). Although the Snrpn promoter is physically deleted in PWS-IC+/del mice, transcription could initiate at alternative upstream exons of Snrpn, which are associated with much weaker expression than the major promoter (28,29).
Residual expression originates, in part, from the normally repressed, maternal chromosome. However, this expression is only detectable when the paternal PWS-IC is deleted. It is possible that the maternal allele normally produces a few transcripts, but these are not detectable in the presence of abundant paternal transcripts. An alternative explanation might be that the maternal chromosome is only active in the absence of the paternal PWS-IC. This would require a trans effect where the PWS-ICdel allele causes deregulation of the maternal allele allowing low-level transcription from the maternal chromosome. Trans effects have only rarely been documented in mice (30). There are rare reports of normally repressed genes being expressed in PWS individuals with maternal UPD or a paternal 15q11–q13 deletion (31,32) suggesting in rare cases that there may also be some relaxation of imprinting in humans allowing expression from the maternal chromosome.
Why do PWS-IC+/del pups survive on some strains and not others, when low-level expression occurs in all strains? While we have not excluded the possibility that a temporal or spatial variation in expression could account for survival, our data support the existence of modifier loci that influence the survival of affected pups. Heterozygous males on the 129S1/Sv genetic background bred to C57BL/6J females produce PWS-IC+/del pups that never survive. Conversely, carrier males on the C57BL/6J genetic background bred with 129S1/Sv females produce PWS-IC+/del pups that are capable of surviving to adulthood, even though the genetic background is identical. The evidence suggests that the maternal strain dictates survival. This could be due to differences in the uterine environment or slight differences in gestation time, which affects the ability of pups to survive. This possibility could be tested by embryo transfer. An alternative explanation is that the modifier gene or genes responsible for survival of PWS-IC+/del mice are required in the oocyte. A third explanation could be that the modifier gene or genes are also imprinted. Pronuclear transplantation, in which pronuclei from a (C57BL/6J×PWS-ICdel)F1 oocyte are moved into enucleated fertilized FVB/NJ or (FVB/NJ×PWS-ICdel)F1 oocytes, could be used to distinguish between these two possibilities (33,34). If the transplanted pronuclei give rise to pups that are able to survive, then the modifier gene is most likely oocyte specific; however, if the resultant pups still do not survive, then the modifying gene is most likely imprinted.
Strain-specific differences have previously been implicated in phenotypic variation seen in Ndn-deficient mice (23–25). One report documented increased survival with FVB/N mothers as compared with C57BL/6 mothers (24), and a second suggested increased penetrance as the contribution of the C57BL/6 genetic background increased (25), suggesting a role for modifier genes influencing the Ndn-deficient phenotype. Our data supports the existence of modifier loci affecting the penetrance of postnatal lethality in PWS-IC+/del mice. Furthermore, we have demonstrated that the modifier gene or genes are maternal in origin. Both studies show that penetrance of postnatal lethality is increased on the C57BL/6 background. Is the modifying effect on the PWS-IC+/del mice the same as that seen in Ndn-deficient mice? The contribution of individual genes to the PWS phenotype in mice remains unclear. Postnatal lethality in Ndn-deficient mice occurs within the first 30 h (24,25) and is reported to be due to a deficiency in central respiratory drive (35). UPD, deletion and PWS-IC deletion mouse models of PWS show survival up to 8 days. The rate of early postnatal lethality is only documented for PWS-IC deletion mice, with 72% dying within 48 h (20). Initial lethality may be caused by respiratory failure due to Ndn deficiency, and mice making it through this phase subsequently die from other causes. Identification of modifier loci will help in understanding the individual gene contribution to the PWS phenotypes in mice.
Mouse models of PWS, including the PWS-IC deletion mouse, demonstrate a severe failure to thrive phenotype that usually results in fully penetrant postnatal lethality. This lethality precludes the use of these mouse models to study the late-onset features of PWS. The survival of PWS-IC+/del mice reported here will allow us to assess whether these mice have obesity and behavioral phenotypes, and to what extent the mouse is a model for the later clinical features associated with PWS. This study demonstrates that strain background affects the phenotype of PWS-IC+/del mice and long-term survival of these mice can be achieved on certain strain backgrounds by culling wild-type sibs. Furthermore, we have shown that low-level expression occurs from both alleles in PWS-IC+/del mice and is not correlated with increased survival. The findings demonstrate that genetic modifiers specific to the maternal strain influence the ability of PWS-IC+/del mice to survive.
MATERIALS AND METHODS
Mouse strains
The strains of mice used in this study were C57BL/6J, FVB/NJ, C3H/HeJ, DBA/2J, Balb/cJ (The Jackson Laboratory) and 129S1/Sv. A C57BL/6J strain congenic for a region of M. musculus castaneus chromosome 7 (B6.CAST.c7) was also used (36). PWS-IC deletion mice were generated on a 129S1/Sv background (20,29) and were backcrossed to a C57BL/6J background (>10 backcross generations). Unless otherwise stated, the PWS-ICdel strain used is C57BL/6J. In all crosses, the female strain is listed first and the male strain second, as is conventional. Mice were fed Pico-Vac mouse diet (Purina Mills, LLC).
Culling and fostering
Within 2 days of birth, it is generally possible to distinguish PWS-IC+/del mice from their wild-type littermates. Wild-type pups were removed from the litter within 2 days, except one that was left to stimulate milk production. Mice were fostered by removing the PWS-IC+/del pups from one mother (usually from the C57BL/6J strain) and giving them to the other mother (usually from the FVB/NJ strain) whose pups had been removed. Additionally, urine from the foster mother was wiped on the foster pups to aid in her acceptance of the new pups.
RT–PCR
Total RNA was isolated from whole brains obtained from neonatal, 1-, 2- and 3-week-old mice. RNA was extracted using RNAzol (Tel-Test) according to the manufacturer's instructions. RNA (10 µg) was pretreated with DNase I (Roche) and half of the RNA was reverse-transcribed (RT) with random hexamers using Superscript II (Invitrogen). The other half of the RNA was manipulated in parallel in the absence of enzyme. One-twentieth of the reactions was used to seed PCR with Taq polymerase (Roche) for 30 cycles of 94°C for 30 s, 60°C for 45 s and 72°C for 45 s, using the following primers. Ndn 9F (5′-GTATCCCAAATCCACAGTGC-3′), Ndn10R (5′-CTTCCTGTGCCAGTTGAAGT-3′), Snrpn N1.1 (5′-CTGAGGAGTGATTTGCAACGC-3′), Snrpn N2.2 (5′-CAAGATCCTTAATACTCGGGG-3′), β-actin F (5′-GTGGGCCGCTCTAGGCACCAA-3′) and β-actin R (5′-CTCTTTGATGTCACGCACGATTTC-3′).
Northern blot analysis
Total RNA was separated on 8% acrylamide–7 M urea gels and transferred to nylon membranes (Hybond N+, Amersham) using a semi-dry blotting apparatus (Trans-blot SD, BioRad) as described by Cavaillé et al. (14). RNA was fixed to the membrane by baking at 80°C in a vacuum oven. The membranes were hybridized overnight at 58°C, according to Church and Gilbert (37). Oligonucleotides for MBII-85 (5′-TTCCGATGAGAGTGGCGGTACAGA-3′) and 5.8S rRNA (5′-TCCTGCAATTCACATTAATTCTCGCAGCTAGC-3′) were end-labeled using γ32P-ATP (Perkin–Elmer) and T4 polynucleotide kinase (Invitrogen). Probes were purified from free nucleotides using the nucleotide removal kit (QIAGEN). Membranes were washed twice for 15 min at room temperature in 2×SSC, 0.1% SDS, and exposed to Kodak XAR film at −80°C for 4–36 h.
qPCR
qPCR reactions were performed with the Dynamo Hot Start SYBR Green qPCR kit (MJResearch) on an Opticon Monitor II (MJResearch) according to instructions. Cycling conditions were 95°C for 15 min followed by 40 cycles of 94°C for 10 s, 60°C for 30 s and 72°C for 30 s, using the following primer sets: Ndn (5′-GGGACTGATGATCTGTATCG-3′ and 5′-CTTCCTGTGCCAGTTGAAGT-3′), Snrpn (5′-GGAGTGATTTGCAACGCAATG-3′ and 5′-AGGCTGTCCTTCGACGTTTG-3′) and β-actin (5′-TGTGACGTTGACATCCGTAAA-3′ and 5′-CCACCGATCCACACAGAGTA-3′). RT+/− reactions were amplified initially to check efficiencies of RT reactions and screen for genomic DNA contamination. Samples were amplified in triplicate. Raw CT values were obtained using Opticon Monitor II software. Relative expression levels were calculated using the 2−ΔΔCT method (38,39) and analysis was performed in Microsoft Excel.
Allele-specific expression
A polymorphic AvaII site between M. musculus castaneus and M. musculus domesticus was used to distinguish allele-specific expression of Ndn (M. Mann and M.S. Bartolomei, unpublished data). RT–PCR was performed on total brain RNA with NdnpolyF (5′-ACAAAGTAAGGACCTGAGCGACC-3′) and NdnpolyR (5′-CAACATCTTCTATCCGTTCTTCG-3′) resulting in a product of 200 bp. AvaII generates products of 187 and 13 bp from castaneus and 157, 30 and 13 bp from domesticus. PCR products were digested overnight with AvaII, resolved on 4.8% agarose gels, Southern blotted and hybridized with 32P-labeled PCR product.
ACKNOWLEDGEMENTS
The authors wish to thank Edwin Peery and Chris Futtner for helpful discussions and critical reading of the manuscript. This work was supported by a grant from the National Institutes of Health to C.I.B. (R01 HD37872). This article is dedicated to the memory of Cami Brannan.
Present address: Department of Genetics, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA.
Deceased.
Figure 1. Surviving PWS-IC+/del mice are smaller than their wild-type littermates. PWS-IC+/del (IC-deletion) and wild-type male (FVB/N×C57BL/6J)F1 mice were weighed at 3, 6, 9, 12 and 15 weeks of age.
Figure 2. Low-level expression of paternal genes in PWS-IC+/del offspring of FVB/NJ mothers is not age-dependent. PWS-IC+/del offspring from FVB/NJ×PWS-ICdel matings were sacrificed within 24 h of birth (P1) and at 1, 2 and 3 weeks of age. A wild-type littermate was sacrificed at P1. Total brain RNA was prepared and subjected to RT–PCR (Ndn and Snrpn) or northern analysis (MBII-85). (A) Expression of Ndn is clearly detectable in PWS-IC+/del mice by RT–PCR. Detection of expression is variable between animals but does not appear to correlate with age. Snrpn expression is barely detectable in mutant animals by RT–PCR. β-actin was used as an amplification control, and no RT controls were performed for each amplification. (B) MBII-85 was analyzed by northern blot with 5.8S rRNA as a loading control. Comparable low-level expression is observed at all ages.
Figure 3. Low-level gene expression does not correlate with strain-dependent differences in survival. Heterozygous PWS-ICdel males were mated with C57BL/6J, FVB/NJ, C3H/HeJ, Balb/cJ, DBA/2J or 129/Sv females. Total RNA was prepared from the brains of two PWS-IC+/del mice (ko) and one wild-type (wt) littermate from each cross. (A) Ndn and Snrpn expression were analyzed by RT–PCR using β-actin as an amplification control. Ndn expression is detected in PWS-IC+/del mice on all strain backgrounds. While detection of expression shows some variation between animals, expression differences do not correlate with survival as non-permisive strains (C57BL/6J and DBA/2J) also show low-level expression. MBII-85 was analyzed by northern blot using 5.8S rRNA as a loading control. Comparable low-level expression is observed on all strain backgrounds. (B) A representative qPCR plot showing amplification of Ndn, Snrpn and β-actin in offspring from survival non-permissive (C57BL/6J) and permissive (FVB/NJ) maternal strains. Reactions were performed in triplicate. The amplification classes are: 1. β-actin amplification of all samples; 2, Ndn and Snrpn amplification in wild-type pups from both maternal strains; 3, Ndn amplification in PWS-IC+/del pups from both maternal strains and Snrpn amplification in PWS-IC+/del pups from C57BL/6J mothers; 4, amplification of Snrpn in PWS-IC+/del offspring from FVB/NJ mothers. Amplification profiles for Ndn were similar in PWS-IC+/del pups from all other maternal strains, and Snrpn profiles in other strains were comparable to FVB/NJ. (C) Graphical representation of relative expression levels in mutants. Mutant expression levels relative to wild-type were calculated for each strain using the 2−ΔΔCT method (38,39), normalized against β-actin. Reactions were performed in triplicate and the data is an average of three independent experiments. Expression is graphed as a percentage of wild-type expression levels.
Figure 4. Residual gene expression is biallelic in PWS-IC+/del pups. RT–PCR was performed on total brain RNA and parental alleles were distinguished by AvaII digestion, which cuts the M. musculus domesticus (FVB/NJ and C57BL/6J) allele twice and the M. musculus castaneus (B6.CAST.c7) allele once. The products detected are 187 bp (castaneus) and 157 bp (domesticus). (A) Analysis of Ndn expression in reciprocal crosses between wild-type FVB/NJ (F) and B6.CAST.c7 (C) mice shows that expression is always from the paternally inherited chromosome (paternal strain is listed second). (B) Analysis of Ndn expression in crosses between B6.CAST.c7 females and heterozygous PWS-ICdel males shows that in wild-type offspring expression is paternal in origin, but in PWS-IC+/del offspring expression occurs from both paternal and maternal alleles. The PWS-IC+/del lane contains 10 times more input RNA and a 10-fold greater exposure time than the wt lane.
Percentage survival of PWS-IC+/del mice. Heterozygous PWS-ICdel males on the C57BL/6J strain background were mated to FVB/NJ, C57BL/6J, (C57BL/6J×FVB/NJ)F1 (B6FVBF1) and (C57BL/6J×C3H/HeJ)F1 (B6C3F1) females.
| Maternal strain . | Number of PWS-IC+/del mice born . | Number of PWS-IC+/del survivors . | Percentage survival . |
|---|---|---|---|
| C57BL/6J | 75 | 0 | 0 |
| FVB/NJ | 51 | 30 | 59 |
| B6C3F1 | 56 | 5 | 9 |
| B6FVBF1 | 102 | 10 | 10 |
| Maternal strain . | Number of PWS-IC+/del mice born . | Number of PWS-IC+/del survivors . | Percentage survival . |
|---|---|---|---|
| C57BL/6J | 75 | 0 | 0 |
| FVB/NJ | 51 | 30 | 59 |
| B6C3F1 | 56 | 5 | 9 |
| B6FVBF1 | 102 | 10 | 10 |
Wild-type littermates were culled before 2 days of age. Survival represents the percentage of mutants which survived to 21 days of age.
Percentage survival of PWS-IC+/del mice. Heterozygous PWS-ICdel males on the C57BL/6J strain background were mated to FVB/NJ, C57BL/6J, (C57BL/6J×FVB/NJ)F1 (B6FVBF1) and (C57BL/6J×C3H/HeJ)F1 (B6C3F1) females.
| Maternal strain . | Number of PWS-IC+/del mice born . | Number of PWS-IC+/del survivors . | Percentage survival . |
|---|---|---|---|
| C57BL/6J | 75 | 0 | 0 |
| FVB/NJ | 51 | 30 | 59 |
| B6C3F1 | 56 | 5 | 9 |
| B6FVBF1 | 102 | 10 | 10 |
| Maternal strain . | Number of PWS-IC+/del mice born . | Number of PWS-IC+/del survivors . | Percentage survival . |
|---|---|---|---|
| C57BL/6J | 75 | 0 | 0 |
| FVB/NJ | 51 | 30 | 59 |
| B6C3F1 | 56 | 5 | 9 |
| B6FVBF1 | 102 | 10 | 10 |
Wild-type littermates were culled before 2 days of age. Survival represents the percentage of mutants which survived to 21 days of age.
References
Holm, V.A., Cassidy, S.B., Butler, M.G., Hanchett, J.M., Greenswag, L.R., Whitman, B.Y. and Greenberg, F. (
Nicholls, R.D., Saitoh, S. and Horsthemke, B. (
Jiang, Y., Lev-Lehman, E., Bressler, J., Tsai, T.F. and Beaudet, A.L. (
Brannan, C.I. and Bartolomei, M.S. (
Ohta, T., Gray, T.A., Rogan, P.K., Buiting, K., Gabriel, J.M., Saitoh, S., Muralidhar, B., Bilienska, B., Krajewska-Walasek, M., Driscoll, D.J. et al. (
Buiting, K., Lich, C., Cottrell, S., Barnicoat, A. and Horsthemke, B. (
Nicholls, R.D. and Knepper, J.L. (
Jong, M.T., Gray, T.A., Ji, Y., Glenn, C.C., Saitoh, S., Driscoll, D.J. and Nicholls, R.D. (
Boccaccio, I., Glatt-Deeley, H., Watrin, F., Roeckel, N., Lalande, M. and Muscatelli, F. (
MacDonald, H.R. and Wevrick, R. (
Glenn, C.C., Porter, K.A., Jong, M.T., Nicholls, R.D. and Driscoll, D.J. (
Reed, M.L. and Leff, S.E. (
Wevrick, R., Kerns, J.A. and Francke, U. (
Cavaillé, J., Buiting, K., Kiefman, M., Lalande, M., Brannan. C.I., Horsthemke, B., Bachellerie, J.-P., Brosius, J. and Hüttenhoffer, A. (
de los Santos, T., Schweizer, J., Rees, C.A. and Francke, U. (
Rougeulle, C., Cardoso, C., Fontés, M., Colleaux, L. and Lalande, M. (
Runte, M., Hüttenhofer, A., Gross, S., Kiefmann, M., Horsthemke, B. and Buiting, K. (
Cattanach, B.M., Barr, J.A., Evans, E.P., Burtenshaw, M., Beechey, C.V., Leff, S.E., Brannan, C.I., Copeland, N.G., Jenkins, N.A. and Jones, J. (
Gabriel, J.M., Merchant, M., Ohta, T., Ji, Y., Caldwell, R.G., Ramsey, M.J., Tucker, J.D., Longnecker, R. and Nicholls, R.D. (
Yang, T., Adamson, T.E., Resnick, J.L., Leff, S., Wevrick, R., Francke, U., Jenkins, N.A., Copeland, N.G. and Brannan, C.I. (
Lee, S., Kozlov, S., Hernandez, L. Chamberlain, S.J., Brannan, C.I., Stewart, C.L. and Wevrick, R. (
Tsai, T.F., Jiang, Y.H., Bressler, J., Armstrong, D. and Beaudet, A.L. (
Tsai, T.F., Armstrong, D. and Beaudet, A.L. (
Gérard, M., Hernandez, L., Wevrick, R. and Stewart, C.L. (
Muscatelli, F., Abrous, D.N., Massacrier, A., Boccaccio, I., Le Moal, M., Cau, P. and Cremer, H. (
Bielinska, B., Blaydes, S.M., Buiting, K., Yang, T., Krajewska-Walasek, M., Horsthemke, B. and Brannan, C.I. (
Runte, M., Kroisel, P.M., Gillessen-Kaesbach, G., Varon, R., Horn, D., Cohen, M.Y., Wagstaff, J., Horsthemke, B. and Buiting, K. (
Bressler, J., Tsai, T., Ramirez, M., Armstrong, D. and Beaudet, A. (
Landers, M., Bancescu, D.L., Le Meur, E., Rougeulle, C., Glatt-Deeley, H., Brannan, C., Muscatelli, F. and Lalande, M. (
Herman, H., Lu, M., Anggraini, M., Sikora, A., Chang, Y., Yoon, B.J. and Soloway, P.D. (
Rogan, P.K., Seip, J.R., White, L.M., Wenger, S.L., Steele, M.W., Sperling, M.A., Menon, R. and Knoll, J.H. (
Muralidhar, B., Marney, A. and Butler, M.G. (
Latham, K.E. (
Latham, K. and Sapienza, C. (
Ren, J., Lee, S., Pagliardini, S., Gérard, M., Stewart, C.L., Greer, J.J. and Wevrick, R. (
Wakeland, E.K., Morel, L., Achey, K., Yui, M. and Longmate, J. (
Church, G.M. and Gilbert, W. (
Pfaffl, M.W. (