-
PDF
- Split View
-
Views
-
Cite
Cite
M Enciso, J P Carrascosa, J Sarasa, P A Martínez-Ortiz, S Munné, J A Horcajadas, J Aizpurua, Development of a new comprehensive and reliable endometrial receptivity map (ER Map/ER Grade) based on RT-qPCR gene expression analysis, Human Reproduction, Volume 33, Issue 2, February 2018, Pages 220–228, https://doi.org/10.1093/humrep/dex370
Close - Share Icon Share
Abstract
Is it possible to determine the receptivity status of an endometrium by combined quantitative reverse transcription PCR (RT-qPCR) expression analysis of genes involved in endometrial proliferation and immunity?
The new ER Map®/ER Grade® test can predict endometrial receptivity status by RT-qPCR using a new panel of genes involved in endometrial proliferation and the maternal immune response associated to embryonic implantation.
The human endometrium reaches a receptive status adequate for embryonic implantation around Days 19–21 of the menstrual cycle. During this period, known as the window of implantation (WOI), the endometrium shows a specific gene expression profile suitable for endometrial function evaluation. The number of molecular diagnostic tools currently available to characterize this process is very limited. In this study, a new system for human endometrial receptivity evaluation was optimized and presented for the first time.
ER Map®/ER Grade® validation was achieved on 312 endometrial samples including fertile women and patients undergoing fertility treatment between July 2014 and March 2016. Expression analyses of 184 genes involved in endometrial receptivity and immune response were performed. Samples were additionally tested with an independent endometrial receptivity test.
A total of 96 fertile women and 120 assisted reproduction treatment (ART) patients participated in the study. Endometrial biopsy samples were obtained at LH + 2 and LH + 7 days in fertile subjects in a natural cycle and at the window of implantation (WOI) in patients in a hormone-replacement therapy (HRT) cycle. Total RNA was purified, quality-checked and reverse-transcribed. Gene expression was quantified by high-throughput RT-qPCR and statistically analyzed. Informative genes were selected and used to classify samples into four different groups of endometrial receptivity status.
Significantly different gene expression levels were found in 85 out of 184 selected genes when comparing LH + 2 and LH + 7 samples (paired t-test, P < 0.05). Gene ontology analyses revealed that cell division and proliferation, cell signaling and response, extracellular organization and communication, immunological activity, vascular proliferation, blood pressure regulation and embryo implantation are the most over-represented biological terms in this group of genes. Principal component analysis and discriminant functional analysis showed that 40 of the differentially expressed genes allowed accurate classification of samples according to endometrial status (proliferative, pre-receptive, receptive and post-receptive) in both fertile and infertile groups.
N/A.
To evaluate the efficacy of this new tool to improve ART outcomes, further investigations such as non-selection studies and randomized controlled trials will also be required.
A new comprehensive system for human endometrial receptivity evaluation based on gene expression analysis has been developed. The identification of the optimal time for embryo transfer is essential to maximize the effectiveness of ART. This study is a new step in the field of personalized medicine in human reproduction which may help in the management of endometrial preparation for embryo transfer, increasing the chances of pregnancy for many couples.
The authors have no potential conflict of interest to declare. No external funding was obtained for this study.
Introduction
One of the key processes for the establishment of a successful pregnancy is embryonic implantation into the endometrium. Implantation is a complex process that involves an intricate dialog between the embryo and the endometrial cells (Singh et al., 2011). This interaction is essential for the apposition, adhesion and invasion of the blastocyst in the human endometrium (Giudice and Irwin, 1999).
The human endometrium is a highly dynamic structure, which undergoes periodical changes during the menstrual cycle in order to reach a receptive status adequate for embryonic implantation. This period of receptivity is known as the window of implantation (WOI) and occurs between Days 19 and 21 of the menstrual cycle (Navot et al., 1991; Harper, 1992). In any other phase of the menstrual cycle, the endometrium is not receptive to pregnancy (Garrido-Gómez et al., 2013). Successful implantation requires therefore a viable embryo and synchrony between it and the receptive endometrium (Teh et al., 2016). The correct identification and prediction of the period of uterine receptivity is essential to maximize the effectiveness of assisted reproduction treatments.
The study of endometrial receptivity is not new as histological analysis has been traditionally used for endometrial dating (Noyes et al. 1950); however, the accuracy of this method to predict endometrial receptivity has been shown to be limited (Coutifaris et al., 2004; Murray et al., 2004). Some alternative methods to evaluate endometrial receptivity have been developed in the last decade: biochemical markers (Zhang et al., 2012), soluble ligands (Thouas et al., 2015), hormone receptors (Aghajanova et al., 2009), cytokines (Paiva et al., 2011), microRNAs (Sha et al., 2011) or HOX-class homeobox genes (Kwon and Taylor, 2004).
Other studies, focused on the understanding of the molecular mechanisms underlying the histological changes of the endometrium during the menstrual cycle, have identified specific genes responsible for the alterations observed (Talbi et al., 2006). Some other reports have addressed this molecular analysis from a wider perspective, performing a global screening of the transcriptome at different moments of the menstrual cycle (Carson, 2002; Ponnampalam et al., 2004), or under different infertility conditions (Koler et al., 2009; Altmäe et al., 2010), pathologies (Kao et al., 2003) or ovarian stimulation protocols (Horcajadas et al., 2005). Valuable information about the process of endometrial proliferation can be extracted from these studies. However, even though the list of studies published in this topic is long, the number of molecular diagnostic tools to identify the moment of uterine receptivity is short (Lessey et al., 1995,Dubowy et al., 2003; Díaz-Gimeno et al., 2011). Some studies looking at the utility of single molecule markers for endometrial receptivity have concluded that a single molecule may not be suffice to describe a complex phenomenon like receptivity and, in this sense, transcriptomic profiles may be a more reliable tool (Zhang et al., 2013).
Most global transcriptomic analyses of the endometrium have been performed using an unselected source of genes involved in many biological processes, but not specifically expressed in the endometrial tissue or related to the process of endometrial receptivity acquisition. We decided that a selection of genes, specifically described to be expressed in the endometrium during the WOI and involved in the process of embryonic implantation, would be a better strategy to accurately define the transcriptomic signature of the receptive endometrium and also to develop a reliable diagnostic tool for endometrial receptivity. Processes such endometrial proliferation and immune response have been described as essential for endometrial preparation and embryonic implantation, so a selection of genes involved in those processes could provide interesting biological and clinical information about the process of endometrial receptivity (Singh et al., 2011; Haller-Kikkatalo et al., 2014).
For global endometrial transcriptomic analyses, the preferred technique has been gene expression microarrays. Although useful for high-throughput transcriptomics, microarrays have a limited dynamic range of detection and the results need to be validated using RT-qPCR (Wang et al., 2009). Similarly for NGS RNA-Seq, while being very effective for large scale studies and the detection of novel transcripts, genes identified still need to be examined and validated using RT-qPCR, as in the case of microarrays (Mortazavi et al., 2008; Costa et al., 2013). RT-qPCR has been shown to have the widest dynamic range, the lowest quantification limits and the least biased results and hence it is considered the gold standard method for gene expression analysis. In this context, we believe the use of RT-qPCR may be a more robust and reliable technique for the analysis of the expression of genes relevant for the process of endometrial receptivity and, also, for the development of diagnostic tools based on the identification of specific signatures associated with different endometrial status.
In the present study, we define a new system for human endometrial receptivity evaluation based on the analysis of the expression of genes related to endometrial proliferation and the immunological response associated with embryonic implantation. We use a high-throughput RT-qPCR platform for the accurate and reliable measurement of gene expression in endometrial samples and specific transcriptomic profile identification.
Materials and Methods
Study design
In order to define the new ER Map®/ER Grade® method for endometrial receptivity evaluation, gene expression data from endometrial biopsies obtained at different moments of the menstrual cycle from healthy fertile donors (Group A) and subfertile women (Group B) were analyzed. Endometrial biopsies from Group A were used to define ER Map®/ER Grade® endometrial receptivity transcriptomic signature. Endometrial samples from Group B were tested and diagnosed for receptivity with the new ER Map®/ER Grade® tool and the endometrial receptivity array ERA® (Igenomix, Spain). Receptivity status concordance between ER Map®/ER Grade® and ERA classification was evaluated in this group of samples.
Patient selection and sample collection
Group A consisted of 96 healthy fertile donors (18–34 years), with regular menstrual cycles and normal BMI (25–30). Endometrial biopsies from this group were obtained on two different days of the same natural menstrual cycle: LH + 2, i.e. 2 days after the luteinizing hormon surge and LH + 7, i.e. 7 days after the LH surge. Group B consisted of 120 subfertile patients (30–42 years) undergoing hormone-replacement therapy cycles. Endometrial biopsies from this group of patients were obtained after five full days of progesterone impregnation (P4+5).
Endometrial biopsies of approximately 30 mg were obtained from the uterine fundus using a Pipelle catheter (Gynetics, Namont-Achel, Belgium) under sterile conditions. Tissue was then placed in a CryoTube® (Nunc, Roskilde, Denmark) containing 1 ml RNAlater® (Sigma-Aldrich, St Louis, MO, USA) and stored at −20°C until further processing.
Ethical approval
Ethical approval for the study was obtained from Centro Hospital Universitario Virgen del Rocío (Sevilla, Spain, CEI#2014PI/025). All fertile donors and subfertile patients signed an informed consent document.
WOI gene selection
Extensive review of the literature using PubMed (https://www.ncbi.nlm.nih.gov/pubmed/) was carried out since 2005, focusing on genes related to endometrial receptivity and the maternal immune response associated with embryonic implantation. The topics used for the search were: ‘endometrial receptivity markers’, ‘endometrial receptivity’, ‘embryonic implantation’, ‘window of implantation’, ‘immune response in embryonic implantation’, ‘markers of embryonic implantation’, ‘gene expression & endometrium’, ‘gene expression microarray & endometrium’ (Carrascosa et al., 2017). Eight candidate reference genes for expression analysis were selected: actin (ACTN), beta-2 microglobulin (B2M), cytochrome C1 (CYC1), EMG1 N1-specific pseudouridine methyltransferase (EMG1), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), TATA-box binding protein (TBP), topoisomerase (DNA) I (TOPI) and tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta (YWHAZ). The expression stability of these reference genes was calculated using the two freeware Microsoft Excel-based applications geNorm (Vandesompele et al., 2002) and NormFinder (Andersen et al., 2004) by following the software developer’s manual.
RNA extraction and cDNA preparation
Total RNA was extracted using RNeasy mini kit (Qiagen, London, UK); RNA purity and concentration was confirmed by NanoDrop 2000 Spectophotometer (Thermo Scientific, Waltham, MA, USA) and RNA integrity was assessed using Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA) following the manufacturer’s instructions. RNAs were reverse-transcribed into cDNA using Fluidigm Reverse Transcription Master Mix (Fluidigm, San Francisco, CA, USA) and either immediately used or stored at −20°C until further downstream processing.
Gene expression analysis
Pairs of primers targeting the selected and reference genes were designed using the software platform D3 Assay Design. Specific target amplification (STA) was carried out on cDNA samples using Fluidigm PreAmp Master Mix and DELTAgene assays following the manufacturer’s instructions (Fluidigm, San Francisco, CA). RT-qPCR reactions were performed following the Fast Gene Expression Analysis Using Evagreen on the Biomark HD Systemand 96.96 Dynamic Array™ IFC (Fluidigm, San Francisco, CA). Data was collected with Fluidigm® Real-Time PCR analysis software using linear baseline correction method and global auto Cq threshold method. Data were then exported to Excel as.csv files and Cq values were normalized using the mean of the reference genes selected.
Principal component analysis and discriminant functional analysis
Differential expression of genes from LH + 2 and LH + 7 groups was assessed by comparing ΔCq values (paired t-test (P < 0.05)). Fold change (−ΔΔCq) was calculated to determine upregulated and downregulated genes in the WOI. In order to assess if receptivity status could be established with a reduced number of genes, a principal component analysis (PCA) of the genes showing significant fold change between LH + 2 and LH + 7 was performed. Discriminant functional analysis (DA) was then used to evaluate the ability of the genes with the highest absolute coefficient value from each of the leading principal components to accurately discriminate samples into the following states: proliferative, receptive, pre-receptive and post-receptive. A Split-Sample validation of the DA was performed to assess the reliability and robustness of discriminant findings. Both fertile and infertile patient samples were split into two subsets. One data set (70% of the samples) was used as a training set and the other one as testing set (remaining 30% of the samples). The percentage of correct classifications was calculated to determine the reliability of the DA model. Data analyses were performed by using IBM SPSS Statistics software version 19.0.
Gene function analysis
To study the biological functions and pathways of the genes selected, DAVID v.6.7 bioinformatics resources were used. Assessment and integration of protein–protein interactions was performed by the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING v.10.0 database, http://string-db.org).
Results
WOI gene selection
A total of 184 genes related to endometrial receptivity and embryonic implantation were carefully chosen after an extensive literature review (Supplementary Table SI). Several biological processes mainly related to cellular proliferation, response to wounding, defense and immune response were found to be statistically over-represented as analyzed by DAVID (Supplementary Table SII). Exploration of the interactions of proteins codified by the selected genes revealed a total of 1334 protein–protein interactions when the expected was 425 (clustering coefficient = 0.616) (Supplementary Fig. S1). The set of proteins codified by the selected genes have more interactions among themselves than what would be expected for a random set of similar size, drawn from the genome. Such enrichment indicates that these proteins are biologically connected as a group.
Gene expression analysis
Expression stability analysis of the eight selected reference genes showed that CYC1, GAPDH, TBP and YWHAZ were the most stable genes (Supplementary Fig. S2) and hence these genes were selected and used for normalization of gene expression values.
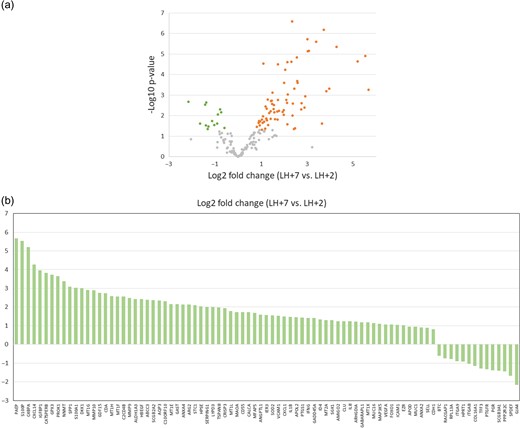
Comparison of gene expression data of the selected WOI genes on days LH + 2 and LH + 7 showed a total of 85 genes presenting significant differences in the fold change (P < 0.05; paired t-test). Most genes were upregulated (n = 71) rather than downregulated (n = 14) (Fig. 1). Gene ontology (GO) analysis revealed that these 85 genes were related to cell division and proliferation, cell signaling and response, extracellular organization and communication, immunological activity, vascular proliferation, blood pressure regulation and embryo implantation (Supplementary Table SIII). Additionally, comprehensive analysis of protein–protein interactions showed a total of 23 interactions when the expected number was 10 (clustering coefficient = 0.218, P = 0.000344).
Differentially expressed genes in LH + 2 and LH + 7. (a) Volcano plot of gene expression differences for the 184 WOI genes on days LH + 2 and LH + 7 of fertile subjects menstrual cycles. The log2-fold change is plotted on the x-axis and the negative log10 P-value is plotted on the y-axis. Green dots represent gene probes with P-value < 0.05 by paired t-test and downregulated fold change (log2FC < −0.5). Orange dots represent gene probes with P-value < 0.05 by paired t-test and upregulated fold change (log2FC > 0.5). (b) Bar graph showing log2-fold changes of the 85 differentially expressed mRNAs (paired t-test, P < 0.05) in LH + 7 vs. LH + 2. There were 71 upregulated mRNAs and 14 downregulated mRNAs in LH + 7 compared to LH + 2.
Principal component analysis and discriminant functional analysis
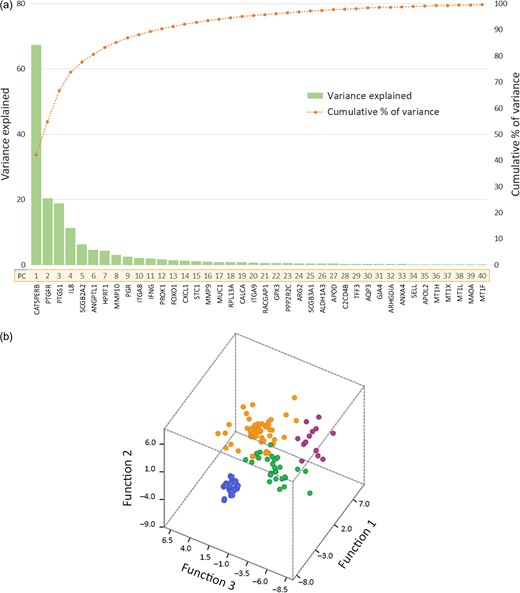
PCA of the 85 genes showing significant fold change between LH + 2 and LH + 7 revealed that 40 components explained more than 99.5% of total sample variance. The variance provided by each component and the cumulative percentage along the 40 components together with the genes with the highest absolute coefficient value for each of the leading principal components (ER Map®/ER Grade® genes) are represented in Fig. 2a. These genes were selected for further discriminant function analysis (DA) (Jolliffe, 1973). DA assessed the effectiveness of the selected genes to accurately classify the receptivity status of endometrial biopsies from both fertile donors and subfertile patients (Fig. 2b).
(a) Principal component analysis. Chart for the variance (eigenvalue) provided by each principal component (PC) from the PCA and the cumulative percentage along the 40 PCs. The green bars illustrate the variance of each PC, and the orange line illustrates the cumulative variance explained by the retaining PCs. The genes with the highest coefficient value from each component (ER Map®/ER Grade® genes) are detailed below each PC number. (b) Discriminant Functional Analysis. Canonical plot using DA with the 40 genes selected to classify 312 endometrial samples. X, Y and Z axes represent the discriminant function scores for the first three dimensions. Non-receptive samples are represented as blue circles, pre-receptive as green circles, receptive as orange circles and post-receptive as purple circles.
Within the group of donors, ER Map®/ER Grade® genes allowed accurate classification of samples into two endometrial receptivity status: proliferative (non-receptive) and receptive. Using a DA model based on the 40 genes selected, 100% of fertile donor samples were correctly classified, 100% of LH + 2 samples were categorized as non-receptive, and all LH + 7 samples were classified as receptive in both the training and testing sets (Table I). Within the patient group, ER Map®/ER Grade® genes classification matched the endometrial biopsy status prediction provided by an independent endometrial receptivity test (ERA) in 97.59% samples in the training set and 91.67% in the testing set. In the training set, two samples were classified differently by the two tests and, in the testing set, there were three.
Discriminant functional analysis classification results. Table summary of the discriminant classification and split-sample validation results of fertile donors and patients’ endometrial samples into different endometrial receptive status. Using a discriminant model based on the 40 selected ER Map®/ER Grade® genes, 100% fertile donor samples were correctly classified in both training and testing sets. Within the patient group, 97.59% of cases in the training set and 91.67% in the testing set matched the original group classification.
| . | Original group membership . | N . | Predicted group membership (%)a,b,c,d . | |||
|---|---|---|---|---|---|---|
| Non-receptive . | Pre-receptive . | Receptive . | Post-receptive . | |||
| Donors | ||||||
| Training set | LH + 2 | 67 | 100.0 | 0.0 | ||
| LH + 7 | 67 | 0.0 | 100.0 | |||
| Testing set | LH + 2 | 29 | 100.0 | 0.0 | ||
| LH + 7 | 29 | 0.0 | 100.0 | |||
| Patients | ||||||
| Training set | Pre-receptive | 29 | 100.0 | 0.0 | 0.0 | |
| Receptive | 41 | 2.4 | 95.1 | 2.4 | ||
| Post-receptive | 13 | 0.0 | 0.0 | 100.0 | ||
| Testing set | Pre-receptive | 13 | 92.3 | 7.7 | 0.0 | |
| Receptive | 18 | 5.6 | 94.4 | 0.0 | ||
| Post-receptive | 6 | 0.0 | 16.7 | 83.3 | ||
| . | Original group membership . | N . | Predicted group membership (%)a,b,c,d . | |||
|---|---|---|---|---|---|---|
| Non-receptive . | Pre-receptive . | Receptive . | Post-receptive . | |||
| Donors | ||||||
| Training set | LH + 2 | 67 | 100.0 | 0.0 | ||
| LH + 7 | 67 | 0.0 | 100.0 | |||
| Testing set | LH + 2 | 29 | 100.0 | 0.0 | ||
| LH + 7 | 29 | 0.0 | 100.0 | |||
| Patients | ||||||
| Training set | Pre-receptive | 29 | 100.0 | 0.0 | 0.0 | |
| Receptive | 41 | 2.4 | 95.1 | 2.4 | ||
| Post-receptive | 13 | 0.0 | 0.0 | 100.0 | ||
| Testing set | Pre-receptive | 13 | 92.3 | 7.7 | 0.0 | |
| Receptive | 18 | 5.6 | 94.4 | 0.0 | ||
| Post-receptive | 6 | 0.0 | 16.7 | 83.3 | ||
aDonors training set: 100% of original grouped cases correctly classified.
bDonors testing set: 100% of original grouped cases correctly classified.
cPatients training set: 97.59% of original grouped cases correctly classified.
dPatients testing set: 91.67% of original grouped cases correctly classified.
Discriminant functional analysis classification results. Table summary of the discriminant classification and split-sample validation results of fertile donors and patients’ endometrial samples into different endometrial receptive status. Using a discriminant model based on the 40 selected ER Map®/ER Grade® genes, 100% fertile donor samples were correctly classified in both training and testing sets. Within the patient group, 97.59% of cases in the training set and 91.67% in the testing set matched the original group classification.
| . | Original group membership . | N . | Predicted group membership (%)a,b,c,d . | |||
|---|---|---|---|---|---|---|
| Non-receptive . | Pre-receptive . | Receptive . | Post-receptive . | |||
| Donors | ||||||
| Training set | LH + 2 | 67 | 100.0 | 0.0 | ||
| LH + 7 | 67 | 0.0 | 100.0 | |||
| Testing set | LH + 2 | 29 | 100.0 | 0.0 | ||
| LH + 7 | 29 | 0.0 | 100.0 | |||
| Patients | ||||||
| Training set | Pre-receptive | 29 | 100.0 | 0.0 | 0.0 | |
| Receptive | 41 | 2.4 | 95.1 | 2.4 | ||
| Post-receptive | 13 | 0.0 | 0.0 | 100.0 | ||
| Testing set | Pre-receptive | 13 | 92.3 | 7.7 | 0.0 | |
| Receptive | 18 | 5.6 | 94.4 | 0.0 | ||
| Post-receptive | 6 | 0.0 | 16.7 | 83.3 | ||
| . | Original group membership . | N . | Predicted group membership (%)a,b,c,d . | |||
|---|---|---|---|---|---|---|
| Non-receptive . | Pre-receptive . | Receptive . | Post-receptive . | |||
| Donors | ||||||
| Training set | LH + 2 | 67 | 100.0 | 0.0 | ||
| LH + 7 | 67 | 0.0 | 100.0 | |||
| Testing set | LH + 2 | 29 | 100.0 | 0.0 | ||
| LH + 7 | 29 | 0.0 | 100.0 | |||
| Patients | ||||||
| Training set | Pre-receptive | 29 | 100.0 | 0.0 | 0.0 | |
| Receptive | 41 | 2.4 | 95.1 | 2.4 | ||
| Post-receptive | 13 | 0.0 | 0.0 | 100.0 | ||
| Testing set | Pre-receptive | 13 | 92.3 | 7.7 | 0.0 | |
| Receptive | 18 | 5.6 | 94.4 | 0.0 | ||
| Post-receptive | 6 | 0.0 | 16.7 | 83.3 | ||
aDonors training set: 100% of original grouped cases correctly classified.
bDonors testing set: 100% of original grouped cases correctly classified.
cPatients training set: 97.59% of original grouped cases correctly classified.
dPatients testing set: 91.67% of original grouped cases correctly classified.
Discussion
For successful embryo implantation, a healthy embryo at blastocyst state and a functional endometrium ready to receive it are required. There is growing evidence that shows the importance of embryonic-endometrial synchrony for the achievement of a successful pregnancy (Prapas et al., 1998; Wilcox et al., 1999; Shapiro et al., 2008, 2014, 2016; Franasiak et al., 2013; Healy et al., 2017). This concept, however, has yet to be taken into the IVF clinical practice. Much effort is put into the production and selection of the most competent embryo to be transferred (Chen et al., 2013; Fragouli and Wells, 2012; Cruz et al., 2011; Forman et al., 2013), but little attention is paid to the other essential part of the pregnancy; no detailed analysis of the functionality of the endometrium or the period of uterine receptivity is routinely performed in IVF centers. The identification of the optimal time for embryo transfer is essential to maximize the effectiveness of ART.
In the present study, we describe the expression analysis of a new panel of genes able to determine the receptivity status of an endometrium. In contrast to previous studies aimed at developing tools for endometrial receptivity evaluation (Horcajadas et al., 2008; Díaz-Gimeno et al., 2011), we chose to perform a selection of genes which are involved in biological processes taking place on the endometrium during the WOI and which are related to endometrial preparation for embryonic implantation.
Upon the selection performed based on the literature (Supplementary Table SI), we found an over-representation of processes very relevant to the phenomenon of endometrial receptivity acquisition such as cellular proliferation, response to wounding, defense and immune response. Within this group of genes, we found a subset of 85 especially interesting as they showed significant differences in expression between the proliferative and secretory phases. These genes GO analyses revealed cellular components, biological processes and molecular functions related to cell signaling and response, extracellular organization, cell division and proliferation, immunological activity, vascular proliferation and embryo implantation. Interestingly an over-representation of processes involving vesicles and exosomes was also found. These terms match with previously described processes known to occur at the time of implantation. Cellular matrix remodeling and an increase in vascular permeability and angiogenesis at the implantation site are one of the earliest prerequisites for embryo implantation (Zhang et al., 2013). Also intense communication through cell signaling between the embryo and the endometrial cells has been described as part of the embryo-endometrial crosstalk essential for adequate embryonic implantation involving, in some cases, extracellular vesicles/exosomes (Ng et al., 2013). Also, immune responses have been proven to play important roles in early pregnancy (Altmäe et al., 2010; Haller-Kikkatalo et al., 2014).
PCA analysis, a dimension-reduction tool that can be used to reduce a large set of variables to a small set that still contains most of the information in the large set, revealed that a subset of 40 of the 85 differentially expressed genes, called ER Map®/ER Grade® genes, could accurately differentiate between LH + 2 and LH + 7. These genes, listed in Fig. 2, allow 100% correct classification of endometrial samples from donors into these two status groups. The ER Map®/ER Grade® gene panel is also able to assess the receptivity status of samples from infertile patients obtained at the secretory phase, classifying samples into: ‘receptive’, this means the WOI matches the day on which the biopsy was taken; ‘pre-receptive’, meaning that the endometrium has not reached its WOI yet or ‘post-receptive’, i.e. this endometrium has already passed its WOI.
Focusing on the technical aspects of the development here described, we chose high-throughput RT-qPCR for the analysis of such panel in endometrial biopsies. RT-qPCR is the most robust and reliable technique currently available for gene expression analysis. Alternative methodologies output such as microarray results and RNA-seq expression data need to be validated using RT-qPCR methods (Mortazavi et al., 2008; Costa et al., 2013).
When compared to the ERA® test, the only other endometrial receptivity test based on gene expression analysis currently available, ER Map®/ER Grade® classification matched the ERA® results in 97.59% of samples in the training set and 91.67% in the testing set. These small differences may be due to either technical or experimental design differences. The experimental design of the present study may have reduced interpatient noise since LH + 2 and LH + 7 samples were obtained from the same donor and within the same cycle, in contrast to the ERA® gene selection experimental design where samples were obtained from different women on different natural cycle days (Horcajadas et al., 2008; Díaz-Gimeno et al., 2011). Also, the fact that the ER Map®/ER Grade® genes selection panel only shares seven genes with ERA® (ANXA4, AQP3, ARG2, GPX3, MAOA, MT1H and SCGB2A2) may explain the discrepancies. Other reason for inconsistencies may be, as pointed out above, the use of different gene expression analysis techniques. Although it has been demonstrated that results of microarray experiments correlate with the results of RT-qPCR, microarrays have a limited dynamic range of detection and may often be affected by background hybridization and probe saturation (MAQC, 2006; Wang et al., 2009). The limited accuracy of microarrays compared to RT-qPCR may be one of the reasons for the differences observed. In any case, the discrepancy is considerably small (5 out of 120 samples analyzed) and both tests seem to produce similar results.
The accurate identification of the period of endometrial receptivity could be key for the achievement of a successful pregnancy in many couples. The importance of embryonic-endometrial synchrony for successful implantation has been reported in several studies. Shapiro et al. (2008) showed that the lower implantation rates observed in Day 6 embryos transferred fresh compared to Day 5 embryos were not due to an embryonic factor but rather to the endometrial moment where embryos were transferred. Similar results were reported by Franasiak et al. (2013), showing that the diminished ART outcomes from embryos with delayed blastulation, traditionally attributed to reduced embryo quality, result from an embryonic-endometrial dissynchrony. These studies highlight the importance of embryo-endometrial synchrony to increase implantation rates.
Reports exploring the concept of the WOI show that the timing of implantation can also influence pregnancy loss. Wilcox et al. (1999) showed a strong increase in the risk of early pregnancy loss with late implantation. Further studies looking at the impact of endometrial-embryo asynchrony on ART outcomes have found that the combination of elevated progesterone on the day of trigger (advanced endometrium) and slow growing embryos results in low live birth rates (Healy et al., 2017). This problem seems to be influenced by maternal age. Shapiro et al. in a recent study (2016) reported an elevated incidence of factors associated with embryo-endometrium asynchrony in women over 35 years, high pre-ovulatory serum progesterone levels and increased numbers of delayed-growth embryos. This, together with the already well known decrease in gamete quality of women of advanced reproductive age (Fragouli et al., 2013), underlines the importance of women’s age for reproductive success and the need for the development of diagnostic and therapeutic tools to increase the chances of these women becoming a mother.
The implementation of endometrial receptivity tests such as the one developed in the present study into the clinical practice routine may help guide embryo transfers to be performed at the best endometrial moment, guaranteeing embryo-endometrial synchrony and thus, allowing for the achievement of better ART results. Couples with repeated implantation failure or previously failed IVF cycles or couples with recurrent miscarriage would benefit from the detailed analysis of endometrial receptivity and embryo-endometrial synchronization. This study is a new step in the field of personalized medicine in human reproduction in the management of the endometrium in preparation for embryo transfer, with the final goal of achieving better ART results, increasing embryo implantation rates and the likelihood of successful pregnancies.
Acknowlegments
The authors thank all the patients and donors for donating their samples to this study.
Supplementary data
Supplementary data are available at Human Reproduction online.
Authors’ roles
M.E. participated in study design, sample processing, data analysis and manuscript preparation. J.P.C was involved in sample processing, data analysis and manuscript preparation. J.S. participated in study design, sample processing, data analysis and manuscript preparation. P.A.M.O. was involved in data analysis and figures preparation. S.M. participated in the design of the study and reviewed the manuscript. J.A.H participated in the design of the study, data analysis and manuscript preparation. J.A. participated in the design and supervision of the study.
Funding
No external funding was obtained for this study.
Conflict of interest
None declared.
References
Author notes
The authors consider that the first two authors should be regarded as joint First Authors.
- pregnancy
- signal transduction
- gene expression
- conflict of interest
- immune response
- cell growth
- embryo
- embryo transfer
- fertility
- genes
- infertility
- menstrual cycle
- mothers
- ovum implantation
- principal component analysis
- reproductive physiological process
- endometrium
- rna
- endometrial biopsy
- clinical diagnostic instrument