-
PDF
- Split View
-
Views
-
Cite
Cite
Michael Davenport, Jordan Poles, Jacqueline M. Leung, Martin J. Wolff, Wasif M. Abidi, Thomas Ullman, Lloyd Mayer, Ilseung Cho, P'ng Loke, Metabolic Alterations to the Mucosal Microbiota in Inflammatory Bowel Disease, Inflammatory Bowel Diseases, Volume 20, Issue 4, 1 April 2014, Pages 723–731, https://doi.org/10.1097/MIB.0000000000000011
Close - Share Icon Share
Inflammation during inflammatory bowel disease may alter nutrient availability to adherent mucosal bacteria and impact their metabolic function. Microbial metabolites may regulate intestinal CD4+ T-cell homeostasis. We investigated the relationship between inflammation and microbial function by inferred metagenomics of the mucosal microbiota from colonic pinch biopsies of patients with inflammatory bowel disease.
Paired pinch biopsy samples of known inflammation states were analyzed from ulcerative colitis (UC) (23), Crohn's disease (CD) (21), and control (24) subjects by 16S ribosomal sequencing, histopathologic assessment, and flow cytometry. PICRUSt was used to generate metagenomic data and derive relative Kyoto Encyclopedia of Genes and Genomes Pathway abundance information. Leukocytes were isolated from paired biopsy samples and analyzed by multicolor flow cytometry. Active inflammation was defined by neutrophil infiltration into the epithelium.
Carriage of metabolic pathways in the mucosal microbiota was relatively stable among patients with inflammatory bowel disease, despite large variations in individual bacterial community structures. However, microbial function was significantly altered in inflamed tissue of UC patients, with a reduction in carbohydrate and nucleotide metabolism in favor of increased lipid and amino acid metabolism. These differences were not observed in samples from CD patients. In CD, microbial lipid, carbohydrate, and amino acid metabolism tightly correlated with the frequency of CD4+Foxp3+ Tregs, whereas in UC, these pathways correlated with the frequency of CD4+IL-22+ (TH22) cells.
Metabolic pathways of the mucosal microbiota in CD do not vary as much as UC with inflammation state, indicating a more systemic perturbation of host–bacteria interactions in CD compared with more localized dysfunction in UC.
Inflammatory bowel diseases (IBD) are characterized by chronic inflammation of the human gastrointestinal tract, driven by abnormal interactions between the immune system and the intestinal microbiota.1,–3 For both ulcerative colitis (UC) and Crohn's disease (CD), the 2 main subtypes of IBD, recent studies using 16S ribosomal sequencing have shown disease associations with dysbiosis or abnormal compositions of the gut microbiota.4,–8 Dysbiosis includes increases in bacterial numbers, decreased bacterial biodiversity, increased abundance of the phyla Proteobacteria and Bacteroidetes, and a decreased abundance of the phylum Firmicutes.9,–11 This dysbiosis is observed in colonic pinch biopsy samples and fecal samples.6,12 Characterization of the mucosal microbiota from pinch biopsies reflects the communities adhering to the intestinal epithelium, which is quite distinct from the luminal bacteria. Analyses of pinch biopsies enable investigation of the microbiota at different locations in the colon, which can also be directly compared with pathologic samples to correlate disease activity with adherent bacterial communities in IBD. There are large differences in the microbiota between stool and mucosal biopsies,6,13,14 and differences in colonic pH may result in differences at various locations of the gut.6,15
Although sequencing 16S ribosomal RNA reveals alterations to taxonomic groups and species composition of IBD patients, it does not inform us of the metabolic activity and function of the microbial communities. Function of the microbiota in communities can be assessed using shotgun metagenomics,13,16 but this approach is dependent on the isolation of sufficient quantities of bacterial DNA, which is not possible from colonic pinch biopsy samples. Recently, an approach was developed to investigate microbiota functional profiles by inferring the metagenome of the closest available whole-genome sequences using 16S gene sequence profiles.6,17 Although this approach has some limitations,18 it provides a way to detect changes to microbial function when the quantity of bacterial DNA is very limited (e.g., in pinch biopsies). Studies on microbial function in healthy individuals have shown that although bacterial communities vary considerably among individuals, overall microbial functions of these communities are largely conserved.13 However, in IBD, functional differences were more pronounced than taxonomic differences.6 Although a few studies have now analyzed the changes in microbial function in IBD based on metagenomic sequencing,6,19 alterations in microbial function during different inflammation states of the sampled tissues has not been examined.
In this study, we used inferred metagenomics by PICRUSt17 to investigate functional differences in the microbiota of patients with UC and CD with biopsies taken from regions of known inflammation states. By pairing biopsies used for analyses of microbial communities with pathologic results and flow cytometry profiles of CD4+ effector cytokine production, we compared the effect of inflammation state on the mucosal microbiota metabolic function for UC and CD patients and related this information to the homeostasis of CD4+ T-cell populations in the same sampled tissue.
Materials and Methods
Participants
Pinch biopsies were taken from IBD patients and healthy subjects undergoing surveillance colonoscopies at the Mount Sinai Medical Center and the Manhattan campus of the VA New York Harbor Healthcare System. The CD and UC patient cohorts were of similar age (CD, 38 ± 11 yr and UC, 41 ± 11 yr), race (CD, 84.5% whites and UC, 85.7% whites), sex (CD, 84.6% men; UC, 64% men), and disease duration (CD, 15.8 ± 11.1 yr and UC, 15.6 ± 9.2 yr) (see Table, Supplemental Digital Content 1, http://links.lww.com/IBD/A448). The healthy subjects were significantly older (61 ± 7 yr) as they were undergoing surveillance colonoscopy for colorectal cancer. Institutional review board approval was obtained before involving patients in the study.
16S Ribosomal RNA Analyses, QIIME, and PICRUSt
DNA was isolated from pinch biopsy material. The V4 region of the 16S ribosomal RNA (rRNA) gene was PCR amplified for sequencing with region-specific barcoded primers20 and sequenced on a MiSeq sequencer21 (Illumina, San Diego, CA) along with other barcoded samples. Data files were demultiplexed and converted from the FASTQ format to the .fna format using QIIME, and reads shorter than 140 base pairs were discarded.22 Sequences were assigned to operational taxonomic units (OTUs) with a threshold of 97% pairwise identity and classified taxonomically according to the Ribosomal Database Project for use in taxonomic analysis. OTUs were subsequently picked by UCLUST against a closed reference table, the latest version of the Greengenes OTU database.23,24 PICRUSt was then used on the Greengene-picked OTUs to generate metagenomic data and derive relative Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway abundance.17
KEGG data were analyzed with QIIME and LEfSe.25 Cluster was used to generate cladograms and heat maps of functional data. Beta diversity was calculated using the Bray–Curtis method and visualized using principal coordinate analysis. LEfSe analysis was performed with a linear discriminant analysis (LDA) score threshold of >3.0, a 0.05 alpha value for the factorial Kruskal–Wallis test, and a one-against-all multiclass analysis strategy. LEfSe uses the Kruskal–Wallis sum-rank test to detect features with significantly different abundances between assigned classes and performs LDA to estimate the effect size of each feature.25 Statistical analysis was performed using Prism 6.0 (GraphPad Software; La Jolla, CA). The unpaired t test was used to assess statistical significance for all the samples.
Isolation of Cells
Pinch biopsies from the colonic mucosa of healthy individuals and IBD patients were digested at 37°C for 1 hour in 100 units per milliliter of collagenase type VIII (Sigma, St. Louis, MO) and 0.150 mg/mL DNase (Sigma) in complete Roswell Park Memorial Institute medium 1640 (Invitrogen, Carlsbad, CA) containing 10% foetal calf serum, 2 mM L-glutamine, 100 units/mL penicillin, 0.1 mg/mL streptomycin, and 0.05 mM 2-mercaptoethanol. Cells were filtered through a 50-μm filter, washed with 5 mL phosphate-buffered saline, and pelleted. Cells were then resuspended in 40% Percoll (GE Healthcare, Port Washington, NY) in complete Roswell Park Memorial Institute medium, underlaid with 80% Percoll, and centrifuged at 2200 rpm (974 g) for 20 minutes at room temperature. Lamina propria mononuclear cells were collected at the interface and used for subsequent flow cytometry analyses.
Fluorescence-Activated Cell Sorting Staining
Cells were stimulated with 50 ng/mL phorbol 12-myristate 13-acetate and 500 ng/mL ionomycin for 4 hours at 37°C in the presence of brefeldin A (GolgiPlug; BD, East Rutherford, NJ). After this in vitro stimulation, cells were stained with anti-CD3, anti-CD4, and anti-CD8 and fixed in 4% paraformaldehyde in phosphate-buffered saline. Cells were permeabilized in Perm/Wash buffer (BD) and stained with anti-IL-22 and anti-Foxp3. Cells were acquired on an LSRII flow cytometer (BD) and analyzed with FlowJo (Tree Star, Inc, Ashland, OR) software. Positive gates for cytokine and nuclear antigen expression are drawn based on control experiments performing Fluorescence Minus One controls with peripheral blood mononucleated cell samples stained with the same intracellular cytokine panels.
Results
Functional Variation and Taxonomic Abundances in Mucosal Biopsies
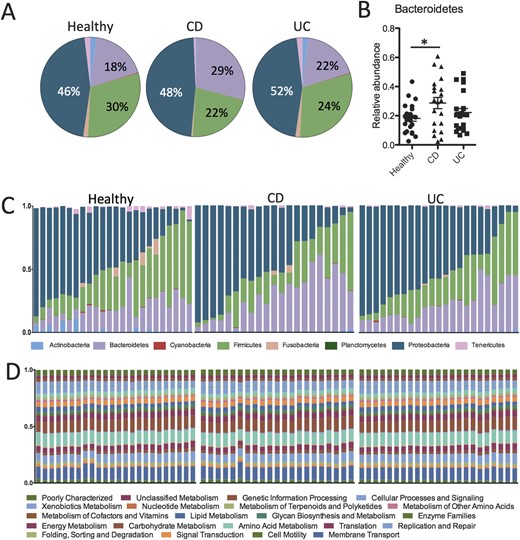
To investigate the mucosal microbiota communities in pinch biopsies from UC (n = 21) and CD (n = 23) patients relative to healthy subjects (n = 24), we performed deep sequencing analysis on the variable region 4 (V4) of bacterial 16S rRNA. A total of 868,044 quality-filtered sequences were obtained from the samples with a mean of 12,226 ± 4493 sequences per sample. At the phylum level, the relative abundance of the 3 most prevalent bacterial phyla of Bacteroidetes, Firmicutes, and Proteobacteria was slightly different between UC, CD, and healthy control subjects (Fig. 1A). Samples from CD patients had a significantly higher proportion of Bacteroidetes compared with healthy control subjects (Fig. 1B). Although UC and CD biopsies showed slightly decreased abundance of Firmicutes and slightly increased abundance of Proteobacteria, these differences were not statistically significant (Fig. 1A; Table, Supplemental Digital Content 2, http://links.lww.com/IBD/A449, and Fig., Supplemental Digital Content 3, http://links.lww.com/IBD/A450).
Taxonomic and functional diversity of the mucosal microbiota in IBD and healthy patients. A, Pie charts representing the mean relative abundances of the major (>1%) phyla present in the mucosal microbiota from healthy subjects and UC and CD patients. B, Scatter plot showing significantly higher relative abundance of the Bacteroidetes phylum in biopsies from CD patients (P < 0.05). C, Stacked bar charts showing the individual variability of the relative abundances of the major bacterial phyla among individual patients. D, Relative abundances of metabolic pathways encoded in the mucosal microbiota are evenly distributed and similar across both individuals and disease states. *P < 0.05.
The relative abundance of different clades among individual samples within the same group varied considerably for IBD patients and healthy individuals (Fig. 1C). However, when we assessed the microbial metabolic and functional KEGG pathways of these communities by inferred metagenomics using PICRUSt,6,17 the pathways were more evenly distributed and consistent between individuals (Fig. 1D). This observation is consistent with results from the Human Microbiome Project, demonstrating the stability of metabolic pathways despite variability of microbial taxa.13
Metabolic Alterations to the Mucosal Microbiota Based on Inflammation Status in UC and CD
The pinch biopsies from patients with IBD were taken from various locations in the colon and small intestine, and for each location, a separate biopsy was sent to pathologic assessment for the characterization of inflammation state. Active chronic colitis was defined by neutrophil infiltration into the epithelium, in the setting of epithelial cell damage.26 To determine the relationship between the relative abundance of KEGG metabolic pathways present in the mucosal biopsies in relation to the inflammation state of the tissues, we used both unsupervised and supervised approaches.
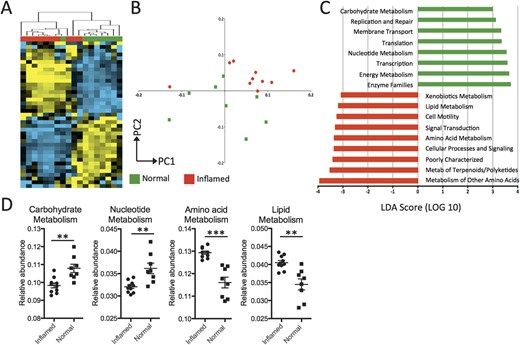
By performing unsupervised hierarchical clustering analysis on the relative abundance values of KEGG pathways represented from the mucosal microbiota of UC patients, we found that there is a clear distinction between the clustering of inflamed samples and noninflamed samples (Fig. 2A; see Fig., Supplemental Digital Content 4, http://links.lww.com/IBD/A451). The separate clustering of inflamed and noninflamed samples was also observed by principal component analysis (Fig. 2B). To identify which groups of microbial functional pathways were driving the differences based on inflammation status, we determined the KEGG pathways that were enriched or underrepresented in inflamed tissue using the LEfSe approach25 (Fig. 2C).
Functional divergence between the mucosal microbiota of inflamed and noninflamed samples from UC patients. A, Unsupervised hierarchical clustering analysis of abundance values for KEGG pathways shows distinctive clustering of inflamed (red) and noninflamed (green) samples. Each column is a separate sample, and row is a particular pathway. Blue = less than the mean, yellow = greater than mean. B, Principal component analysis of KEGG pathways confirms the segregation of inflamed and noninflamed samples along PC1 and PC2. C, Supervised comparison identifies differential abundance of specific KEGG pathways using LEfSe (LDA score, >3.0). D, Genes in carbohydrate and nucleotide metabolism pathways are more abundant in noninflamed normal tissue from UC patients, with a shift toward more abundance in lipid and amino acid metabolism genes for the mucosal microbiota of inflamed tissues. **P < 0.01, ***P < 0.001.
In UC, 5 of the 8 functional pathways identified (LDA score, >3.0) that were more abundant for bacteria inhabiting areas of inflamed tissue were related to metabolism, particularly amino acid, terpenoid, lipid, and xenobiotic metabolism (Fig. 2C, D). The other pathways were related to cell motility and signaling. Many pathways that were more highly represented in noninflamed tissue were also involved in metabolism; namely, carbohydrate, nucleotide, and energy metabolism (Fig. 2C, D). Hence, the mucosal microbiota of actively inflamed tissue from UC patient may use different metabolic pathways than those in noninflamed tissues.
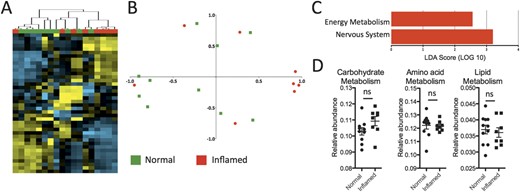
The same analyses were performed on samples from CD patients (Fig. 3). In contrast to UC, hierarchical clustering analysis of samples from CD patients did not segregate well by inflammation state (Fig., Supplemental Digital Content 5, http://links.lww.com/IBD/A452). This was consistent with the principal component analysis (Fig. 3B), which showed that normal and inflamed samples were mostly intermingled. Finally, when we conducted a supervised comparison of inflamed and noninflamed samples by LEfSe analysis, a reduced significance threshold (LDA score, >2.0) was required to identify an increased abundance for energy metabolism in inflamed tissue (Fig. 3C). There was also an increased abundance for genes in the nervous system pathway, but this is difficult to interpret. There were no KEGG pathways that were underrepresented in inflamed CD samples. Amino acid, lipid, and carbohydrate metabolism was unaltered (Fig. 3D). Hence, metabolic pathways of the mucosal microbiota in CD do not vary as much as UC with inflammation state.
Limited differences between the mucosal microbiota of inflamed and noninflamed samples from CD patients. A, Hierarchical clustering analysis of samples from CD patients shows limited segregation of inflamed (red) and noninflamed (green) samples. B, Principal component analysis shows limited segregation of inflamed and noninflamed samples. C, Supervised comparison identifies only 2 differentially abundant KEGG pathways using LEfSe (LDA score, >2.0). D, Genes involved in amino acid, lipid, and nucleotide metabolism pathways are not significantly different between inflamed and normal tissue of CD patients, but there is a increase in the energy metabolism pathway. *P < 0.05. ns, not significant.
Differences in Bacteria Functional Pathways in Inflamed Tissue Compared with Healthy Control Subjects
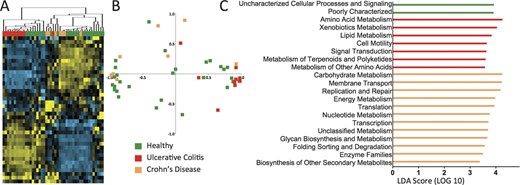
We next compared the functional pathways for the mucosal microbiota of inflamed samples from both UC and CD patients with those from healthy subjects (Fig. 4). Noninflamed samples were not included because they should be more similar to healthy tissue. Unsupervised hierarchical clustering analysis showed that inflamed UC samples and CD samples clustered distinctly in 2 major branches (Fig. 4A; see Fig., Supplemental Digital Content 6, http://links.lww.com/IBD/A453). Some healthy samples clustered with UC samples and others with CD samples. The distinct separation of UC and CD inflamed samples was confirmed by principal coordinate analysis plots where they clustered distinctly (Fig. 4B), intermingled with samples from healthy subjects. This indicates that microbial function of inflamed tissue is more distinct between UC and CD patients than in relation to healthy subjects.
KEGG metabolic pathways are significantly different in the mucosal microbiota of inflamed tissue from UC and CD patients. A, UC and CD patient samples with active inflammation clustered distinctly into separate arms. Some samples from healthy patients cluster with UC samples, whereas other healthy samples cluster with CD samples. B, Principal component analysis confirms the segregation of CD and UC inflamed samples, although there is no clear separation from healthy subjects. C, LEfSe analysis (LDA score, >3.0) shows that amino acid and lipid metabolism was more abundant in the mucosal microbiota of inflamed UC samples, whereas carbohydrate, energy, and nucleotide metabolism pathways were more abundant in inflamed CD samples.
To determine the functional KEGG pathways that are driving these differences, we again performed supervised comparisons by LEfSE (LDA score, >3.0). Different metabolic pathways were enriched in the mucosal microbiota of UC and CD patients (Fig. 4C). Although the KEGG pathways of amino acid and lipid metabolism were enriched in UC samples, CD samples were enriched in carbohydrate, energy, and nucleotide metabolism (Fig. 4C).
Relationship Between Microbiota Metabolic Profiles and Homeostasis of CD4+ T-cell Populations
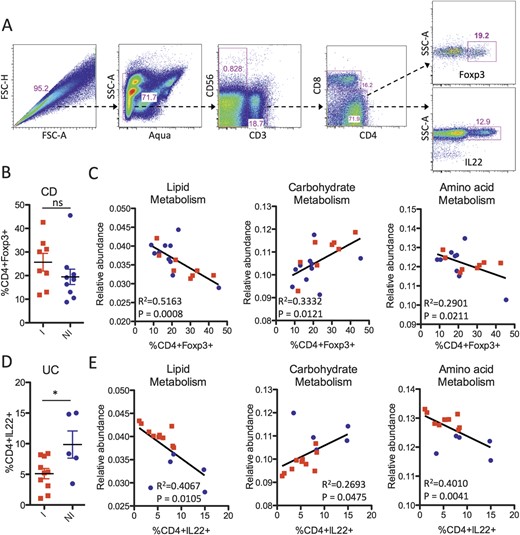
Recently, microbial metabolites were demonstrated to regulate the homeostasis of intestinal regulatory T cells (Tregs).27 We measured the relative frequency of CD4+Foxp3+ Tregs among lamina propria mononuclear cells isolated from pinch biopsies paired with the samples that were sequenced (Fig. 5A, B) and compared them with KEGG microbial metabolic pathways (Fig. 5C). We previously found by flow cytometry that actively inflamed samples from UC patients were depleted of T-helper type-22 (Th22) cells and enriched for mono-IL-17–producing Th17 cells.28 Hence, we also compared the frequency of CD4+IL-22+ cells (Fig. 5A, D) with the relative abundance values of the KEGG metabolic pathways (Fig. 5E).
Correlation of microbial function with the homeostasis of CD4+ effector cells. A, Representative gating strategy for the fluorescence-activated cell sorting analysis of lamina propria mononuclear cells showing live CD3+CD4+ lymphocytes with intracellular staining for IL-22 production and Foxp3 expression. B, Frequency of CD4+Foxp3+ Tregs from lamina propria mononuclear cells of CD patients in inflamed (red) and noninflamed (blue) biopsy sites. ns, not significant. C, Linear regression of KEGG pathways (lipid, carbohydrate, and amino acid metabolism) with the percentage of CD4+Foxp3+ present in the same biopsy location taken from CD patients. D, Frequency of CD4+IL-22+ cells of UC patients in inflamed (red) and noninflamed (blue) biopsy sites. *P < 0.05. E, Linear regression of KEGG pathways with the percentage of CD4+IL-22+ cells present in the same biopsy location taken from UC patients.
For CD patients, the frequency of Foxp3+ Tregs was not significantly different between inflamed and noninflamed biopsy samples (Fig. 5B). However, there was a tight correlation between Treg frequency and microbial lipid metabolism, and to a lesser extent, between carbohydrate and amino acid metabolism (Fig. 5C). These relationships between Treg frequency and microbial metabolism pathways were not significant for UC patients (data not shown). In contrast, UC patients had significantly fewer CD4+IL-22+ cells in inflamed biopsy samples (Fig. 5D), and the frequency of these CD4+IL-22+ cells significantly correlated with microbial metabolism pathways (Fig. 5E). The relationship between CD4+IL-22+ cells and microbial metabolism pathways was not observed in CD samples (data not shown). These results suggest that host–microbial interactions for UC and CD patients may be quite different and that the tissue microenvironment, as reflected by the types of effector CD4+ cells present, may influence the functional phenotype of the mucosal microbiota. Alternatively, metabolites produced by different microbial metabolic pathways may influence the homeostasis of CD4+Foxp3+ and CD4+IL-22+ cells differently for UC and CD patients.
Discussion
In this study, we apply the approach of inferred metagenomics using PICRUSt on colon biopsy samples of known inflammation states from UC and CD patients and control subjects, to determine if there are metabolic alterations to the mucosal microbiota during IBD. We then determined that alterations to microbial metabolism directly correlated with the homeostasis of CD4+Foxp3+ and CD4+IL-22+ cells. Furthermore, the relationship between inflammation status and CD4+ cells with metabolic alterations to the mucosal microbiota is completely different for patients with UC and CD.
Changes to the composition of the microbiome and its interaction with the immune system plays a crucial role in the pathogenesis of CD and UC, but this relationship is still poorly understood.1,–3,29,30 Alterations to the microbiome in IBD include decreased biodiversity, increased numbers of bacteria, and changes in the relative abundance of Firmicutes and Proteobacteria.9,–11 In this study on the mucosal microbiota, we found that only the relative abundance of Bacteroidetes was significantly different from healthy control subjects and only in CD. Contrary to published data, we found no phylum-level significant differences in Firmicutes or Proteobacteria. However, there are many inconsistencies between studies. For example, no significant changes at the phylum level for Bacteroidetes, Firmicutes, or Proteobacteria was found in pediatric IBD patients at the time of diagnosis, compared with healthy patients,31 which raises the possibility that microbial alterations could be a function of previous treatment or disease progression, rather than preceding disease. Because the microbiome profiles are influenced by age, medication history, smoking status, and body mass index,6,32 we are still some distance away from unraveling these complex relationships in the context of IBD.
The tissue microenvironment can influence the nutrient availability for the microbiota, which is critical in defining the composition of the community. The metabolic activities of the microbiota are reflected in the genes encoded into their genomes, which has previously required deep metagenomic sequencing to be analyzed. Deep metagenomic sequencing is not possible on biopsy specimens because the relative content of bacterial DNA to host DNA is extremely small. Recently, a method called PICRUSt that can infer the metagenome through 16S sequence data to extract information from the closest available whole-genome sequences has been developed.17 Using PICRUSt, we investigated functional perturbations to the mucosal microbiota communities of IBD patients. Consistent with findings from other body tissues,13 we found that the overall function was largely conserved across samples, even with widespread differences in bacteria composition. However, important differences exist between bacteria found adhered to inflamed tissue compared with noninflamed tissue.
We find that alterations in microbial function were more significant between inflamed and noninflamed regions of UC patients, in comparison to CD patients. This is somewhat contrary to a previous study that found CD to have larger functional perturbations with inflammation than UC.6 The fact that normal and inflamed tissue is more different in patients with UC than CD is consistent with the hypothesis that UC is a more regionalized disease, whereas CD is a more systemic disease. This observation is also consistent with our previous results comparing the profiles of immune effector CD4+ T cells in normal and inflamed tissues between patients with UC and CD.28 CD4+ T-cell frequencies in normal noninflamed tissue of UC patients were also more similar to those of healthy control subjects than noninflamed tissue from patients with CD.28 Because we also observed that there are greater differences between the mucosal microbiota of inflamed CD samples and inflamed UC and healthy samples, then between inflamed and noninflamed CD samples, there may be a more systemic perturbation of host–bacteria interactions in CD that is independent of the inflammation status of the mucosa.
In inflamed regions of UC patients, the metabolic activities of adherent bacteria was altered to have a reduce abundance of genes used for carbohydrate and nucleotide metabolism and an increased abundance of genes for lipid and amino acid metabolism. Intestinal bacteria compete for the limited supply of nutrients within the intestine, which should be reflected in genes that are encoded into their genomes and their relative abundance.33 Bacteria that are highly dependent on nutrients from the host environment may have a reduced genome lacking in genes for specific metabolic pathways. For example, segmented filamentous bacteria that can promote the differentiation of TH17 cells lack genes involved in nucleotide and amino acid biosynthesis.34 One hypothesis is that inflamed tissue provides less supply of carbohydrates and bacteria that are able to undergo amino acid and lipid metabolism dominate. Hence, in actively inflamed regions of the gut of a UC patient, perhaps nucleotides and carbohydrates might become more readily available, favoring the expansion of carbohydrate auxotrophic bacteria that maintain biosynthetic pathways for amino acids and lipids. Recently, butyrate-producing bacterial taxa were found to be reduced in UC patients, together with a reduction in short-chain fatty acids.35
Interestingly, we find that this functional shift from carbohydrate and nucleotide metabolism toward increased amino acid and lipid metabolism in active inflammation of UC patients was also associated with the frequency of CD4+IL-22+ cells. Alterations in microbial metabolic pathways could result in the production of different metabolites, some of which (e.g., short-chain fatty acid) have been shown to regulate intestinal Foxp3+ Treg populations in mice.27 Although there are few differences for microbial metabolic pathways between inflamed and noninflamed regions of CD patients, there was a very strong association between the frequency of Foxp3+ Treg and microbial metabolism pathways. These observations suggest a link between bacterial metabolism and the intestinal homeostasis of Foxp3+ Tregs in CD patients.
The mucus layer in the intestinal tract is an important barrier to luminal bacteria and a major substrate and adhesion component for bacteria living in this microenvironment. Alterations to the mucus layer may lead to breaches of the epithelial barrier by specific bacteria, resulting in immune responses that may result in chronic inflammation. Changes in the quantity and quality of this mucus layer will influence the constituency of attached microbial populations. Mucin production can be dysfunctional in IBD, and there is a particular paucity of mucin in UC.36 Because mucin is a significant source of carbohydrates for the commensal bacteria,33 mucosal microbiota in inflamed regions of the gut may shift toward bacteria that can metabolize lipids and amino acids for energy. Further support of this principle is that bacteria like Firmicutes33 are unable to metabolize amino acids and are seen with decreased prevalence in IBD.
This study provides new insight into the differences between UC and CD, particularly with regards to microbiota composition and function. We have now found by both flow cytometry analysis of effector T cells and by 16S sequencing of the mucosal microbiota that alterations to the intestinal mucosa in CD are largely colon wide and do not vary as much as UC with inflammation state. Together, this is indicative of a more systemic perturbation of mucosal immunity in CD patients, compared with a more localized dysfunction in UC patients. Therapeutic strategies designed to improve host–bacteria interactions during IBD may have to be selective for UC and CD patients because there is clearly a different interplay between mucosal immune responses and the gut microbiota of these 2 classes of IBD patients.
Acknowledgments
The authors would like to thank the Knight and Huttenhower laboratories for making QIIME and PICRUSt easily available to the scientific community. They also thank the members of the Earth Microbiome Project at Argonne National Laboratories for assistance in microbial sequencing.
References
Author notes
Reprints: P'ng Loke, PhD, Department of Microbiology, New York University School of Medicine, 341 East 25th Street, New York, NY 10010 (e-mail: Png.loke@nyumc.org).
The authors have no conflicts of interest to disclose.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's Web site (www.ibdjournal.org).
M. Davenport, J. Poles and J. M. Leung contributed equally to this study.
Supported by the National Institutes of Health (NIH) grants (5R01AI093811 and 3R21AI094166), the Broad Medical Research Program of The Broad Foundation, and the Kevin and Masha Keating Family Foundation (P.L.); L. Mayer is supported by NIH grant (P01 DK072201); I. Cho is supported in part by NIH grant (UL1 TR000038). J. Poles is a recipient of an AGA-Eli and Edythe Broad Student Research Fellowship.