-
PDF
- Split View
-
Views
-
Cite
Cite
Sophie von der Heyden, Jaco Barendse, Anthony J. Seebregts, Conrad A. Matthee, Misleading the masses: detection of mislabelled and substituted frozen fish products in South Africa, ICES Journal of Marine Science, Volume 67, Issue 1, January 2010, Pages 176–185, https://doi.org/10.1093/icesjms/fsp222
Close - Share Icon Share
Abstract
Mislabelling poses a threat to the sustainability of seafood supply chains and, when frequent, can significantly affect conservation efforts. Here we identify the most popular fish in the South African market through consumer and retailer surveys and data gathered by a sustainable seafood campaign. Of these species, we tested a number of widely available and generally high-market priced fish, utilizing mtDNA 16S rRNA sequencing. Tests of 178 samples revealed that about half of all fillets are mislabelled. Most problematic was kob, Argyrosomus spp., for which some 84% of fillets provided belonged to other species, including mackerel, croaker, and warehou. Phylogenetic analyses provided strong support that the fillets sold as barracuda and wahoo were probably king mackerel and that red snapper fillets included fillets of river snapper, Lutjanus argentimaculatus, which is a species prohibited for sale in South Africa. We also discovered substitution of yellowtail for dorado. From preliminary population genetic comparisons, some 30% of kingklip samples probably had their origin in New Zealand, rather than southern Africa. The research revealed a market conducive to mislabelling through poor consumer and retailer awareness, and highlighted the value of sustainable seafood campaigns to draw attention to this.von der Heyden, S., Barendse, J., Seebregts, A. J., and Matthee, C. A. 2010. Misleading the masses: detection of mislabelled and substituted frozen fish products in South Africa. – ICES Journal of Marine Science, 67: 176–185.
Introduction
In response to the ever-worsening state of the world's commercial fisheries (FAO, 2009), attention has increasingly turned to market-driven incentives, notably eco-labels and seafood awareness campaigns (Roheim and Sutinen, 2006; Jacquet and Pauly, 2007a). Both concepts hope to influence seafood consumers towards making more-informed sustainable choices. Seafood awareness campaigns typically compile seafood lists that make sustainability recommendations about different species (e.g. “best choice” or “avoid”). These lists are dispensed in the form of wallet cards, on websites and mobile telephone wireless application protocol (“WAP”) pages, and more recently as text-message “info-lines”, using short message service (“SMS”) technology (Kinkade and Verclas, 2008). One of the major shortcomings identified with such campaigns is that the recommendations are only as good as the information available to the consumer on the packaging or at the point of sale, i.e. if a species is mislabelled, the “wrong” choice will inadvertently be made regardless of the good intentions of the buyer (Jacquet and Pauly, 2007b; Logan et al., 2008). Until the introduction of molecular testing of seafood and other wildlife products (Baker, 2008, and references therein), detecting the mislabelling of food products was extremely difficult (Woolfe and Primrose, 2004), but currently there is mounting evidence both in popular and scientific literature about its prevalence (Fox, 2008; Wong and Hanner, 2008).
Mislabelling is particularly evident (but not limited to) where fish are processed and pre-packed into fillets (such as restaurant-sized portions) or are components of processed foods, without the buyer seeing the whole specimen (Cocolin et al., 2000). Fraud and consumer deception by mislabelling or “market substitution” of seafood does not only hold implications for the sustainable management and conservation of overexploited marine resources (Jacquet and Pauly, 2007b; Yancy et al., 2008), but also negatively affects food safety and poses potential health-risks to the consumer (Shadbolt et al., 2002; Burger et al., 2004).
Although several techniques have been developed to discriminate between closely related marine species, horse mackerel of the genus Trachurus (Apostolidis et al., 2008), Cape hake (Merluccius capensis and M. paradoxus; von der Heyden et al., 2007), and even broader taxonomic groups such as sharks (Shivji et al., 2002) and sturgeons (Ludwig, 2008), only a small number of studies used molecular methods to estimate the extent of mislabelling in local and global markets. Primarily those studies have been species- or group-specific (Rasmussen and Morrissey, 2009). Using an ELISA- and PCR-based system, Asensio et al. (2008) tested 70 samples sold as grouper (Epinephelus marginatus) and showed that just 12 (∼17%) of these were indeed grouper. Nile perch (Lates niloticus) and wreckfish (Polyprion americanus) constituted the remainder of the samples tested. On a more specific scale, for hake of the genus Merluccius (Machado-Schiaffino et al., 2008), analysis of species-specific mitochondrial single nucleotide polymorphisms (mtSNPs) showed that ∼22% of 40 commercially processed samples were mislabelled. Moreover, a 10-year study by the National Seafood inspection laboratory of the United States showed that ∼37% of fish and 13% of other seafood were labelled incorrectly (see Jacquet and Pauly, 2007b, and references therein). As mislabelling usually holds a financial incentive, rather than management or public health-related ones, it is the high-end market fish that are commonly mislabelled; recent examples include “red” snapper (Lutjanus campechanus; Marko et al., 2004), groupers (Epinephelus spp.; Rasmussen and Morrissey, 2009), and orange roughy (Hoplostethus atlanticus; Jacquet and Pauly, 2007b, and references therein).
The South African Development Community (SADC) includes some of the most important fisheries nations in Africa, with South Africa being the largest fisheries role-player on the continent. The annual African export value of fish during 2002 was US$2.7 billion, and South Africa contributed more than US$890 million to this figure (Teleda, 2004).
To date, no molecular studies have been carried out in South Africa to assess the extent of seafood substitution in the domestic market. Informal visits to restaurants and observations in the market place have suggested a low level of awareness about species identity and origin. Associated with this is often low compliance with national seafood laws and regulations (pers. obs.). Contraventions included purchasing fish from unauthorized sources (such as recreational fishers), and trade in designated “no-sale” or specially protected species. Furthermore, the great variation in regional names for the same species, and the use of vague generic group-names, and terms (such as “linefish”) to market any number of species makes it difficult to accurately assess the species that are on sale (M. Bürgener, pers. comm.). Although quality standards applicable to frozen fish aimed at the prevention of misleading the market do exist in South Africa (Anon., 1996, 2003), they provide no explicit guidelines about which names to use or how to deal with, for example, closely related or imported seafood products. This probably creates a market conducive to misnaming, and Heemstra (2003) remarked that “sometimes dishonest vendors will trim the kob's tail to resemble that of more costly geelbek (Atractoscion aequidens)”.
For this study, we identify the most popular fish species on the South African market using questionnaire surveys of restaurants and consumers carried out in the context of a local sustainable seafood awareness campaign, as well as actual text-message requests by consumers consulting a sustainable seafood list via a mobile text-message service. We then employ a PCR- and sequencing-based technique of the mitochondrial 16S rRNA gene to test the authenticity of a number of these commonly available, highly priced fish acquired from different sources, marketed as “kob” (Argyrosomus spp.), yellowtail (Seriola lalandi), dorado (Coryphaena hippurus), and kingklip (Genypterus capensis). In addition, we opportunistically sourced fillets sold as species that usually are less widely available, such as red snapper (Lutjanus spp.), wahoo (Acanthocybium solandri), and barracuda (Sphyraena spp.). Two fillets sold as “Bassa” and “Cardinal”, for which no other information was available, were also tested.
Material and methods
Market and consumer awareness
The Southern African Sustainable Seafood Initiative (SASSI) was launched in 2005, along similar lines to other international seafood consumer awareness campaigns (see Jacquet and Pauly, 2007a; Logan et al., 2008, for background discussions). A species list containing the most commonly encountered seafood species (mostly from local fisheries) in South Africa was compiled using the familiar “traffic light” system that ranges from green (good choice) through to red (bad choice), based on the conservation, stock, or legal status the species (for the full list visit http://www.wwfsassi.co.za). From December 2007 on, the list was also made available as a mobile telephone text-message service. Known as “FishMS”, the system is customized to interpret and respond to ad hoc queries that should contain acceptable common or market names of the seafood in question. It currently draws its responses from a database of 152 species and 44 groups (e.g. prawns, tuna) constituting 595 separate entries (including aliases for the same species and translations into other languages, such as Afrikaans), matching often wildly misspelt requests using a probabilistic algorithm that takes into account common misspellings and mobile telephone shortcuts. The system itself is based around the Ericsson Erlang OTP platform and records the telephone number, time, date, and content (fish name) of each request. Use of the FishMS service is voluntary, and the cost to the consumer is the standard rate for sending a single text message (approx. ZAR0.5 ≈ US$0.05 in 2009).
The most popular and commonly encountered fish species were identified from three different sources: interviews at seafood restaurants, intercepts of consumers attending food shows, and actual requests received from FishMS users.
Restaurant interviews
Before the launch of the SASSI campaign, several seafood restaurants and fishmongers were visited by arranged meeting, and questionnaire interviews were completed in the two largest coastal cities, Durban (between 5 September and 10 October 2002) and Cape Town (between 11 January and 10 March 2005). Interviewees (owners, managers, or chefs) were asked to state the five most popular (“best seller”) fish species. To ascertain which species were most commonly traded, they were asked to indicate whether they traded species from a predetermined list. A further purpose of these visits was to assess the levels of awareness regarding the legality of species sold, and interviewees were asked whether they were aware of the Marine Living Resources Act (MLRA) of 1998.
Food shows
A major food event is held annually in three major South African cities, the Good Food and Wine Show (GFWS). These were attended on two occasions, once in Cape Town (18–21 May 2006) and once in Johannesburg (1–4 November 2007). Members of the public who showed an indication of interest while strolling past a sustainable seafood stand were intercepted and asked what their favourite seafood was, and whether they were aware of the SASSI initiative.
FishMS
Requests received over a period of 27 months, from 1 December 2006 to 28 February 2009, were extracted from the database. Requests from mobile telephone numbers belonging to known users (staff working directly on either SASSI or FishMS) were removed from the sample. All nonsensical requests, i.e. those that were completely unintelligible, humorous, or considered to be spam, were also removed. Overall monthly and hourly use-patterns were examined. All requests were further ranked according to the frequency of request per species/group.
Molecular analyses
Samples of fish were obtained from a number of seafood wholesalers or restaurants located mostly in the greater Cape Town metropolitan area, and some from Johannesburg, and were primarily purchased as frozen fillets, individually wrapped. In all, 174 tissue samples were analysed, including 54 marketed as kingklip, 25 as dorado, 70 as kob, and 15 as yellowtail, samples being taken from primarily frozen fillets. Opportunistically sampled fillets sold as barracuda (2), wahoo (1), “red snapper” (4), “cardinal” (2), and “bassa” (1) were also obtained.
To determine the regional origin of kingklip, we also analysed 24 samples of kingklip caught off South Africa using a more sensitive molecular marker. As mtDNA 16S rRNA sequences of South African kingklip were not available in any genetic database, and all 16S rRNA sequences were 100% identical, we sequenced a portion of the variable 5' end of the mtDNA control region of all kingklip samples. Four samples from New Zealand were also included to investigate the geographic provenance of kingklip samples purchased in South Africa.
For all samples, total genomic DNA was extracted using the Qiagen DNEasy (Qiagen) kit following the manufacturer's instructions; DNA was eluted in 100 µl of AE buffer. Universal PCR primers 16SAL and 16SBH (Palumbi, 1996) were utilized. For the control region, the primers and cycling conditions of Lee et al. (1995) were used. Products were visualized on 1% agarose gels, and the remainder of the samples were purified using a Nucleofast kit (Macherey–Nagel). PCR products were sequenced using BigDye chemistry and analysed on an ABI 3100 sequencer (Applied Biosystems). As there is no type material available for the fillets examined, sequences were not added to the GenBank database. However, the origin of the South African kingklip sequences is certain, and these were submitted to GenBank with the following accession numbers: GQ324561–GQ324564 (control region) and GQ324565 (16S rRNA). All sequences obtained were blasted against GenBank (http://blast.ncbi.nlm.nih.gov/Blast.cgi) to obtain similarity scores. All sequences with 100% similarity were downloaded for analysis. In instances where no 100% match was found, all sequences of taxa with >95% similarity were downloaded for subsequent comparisons. The 16S rRNA data were manually aligned using MacClade 4.0 (Maddison and Maddison, 2000). Phylogenetic analyses were performed to indicate the similarity among known GenBank sequences, and the sequences obtained from the fillets sampled. We used two techniques (Neighbour Joining and Parsimony) available in PAUP*4.0b10 (Swofford, 2003) software and employed standard procedures (unweighted character transformations, 10 random taxon additions, heuristic searches with TBR branch-swapping, midpoint rooting). Significance for nodes was estimated by 1000 non-parametric bootstrap replicates. All data were analysed (174 fillets), then to reduce cluttering in the trees, just one individual from each clade was retained for further illustrations/analyses. Clades were defined by bootstrap values >90%.
Results
Consumer and market awareness
In all, 28 seafood outlets, 13 in Durban and 15 in Cape Town, were visited and questionnaire interviews completed. In general, there was a positive agreement between the most popular and the most commonly traded species which also rank among the most important local commercial species (Table 1). Some of the species reflected regional distribution of species, e.g. rockcods (groupers, Epinephelus spp.) and king mackerel (Scomberomorus commerson), are found off the east coast. The frequent use of group names prevented species level analysis in some cases; for example, “tuna” presumably refers to yellowfin, bigeye, or longfin tuna, and kob (Argyrosomus spp.) could include all three species (dusky, silver, and squaretail). Two obviously imported species were commercially important in Cape Town, Atlantic salmon (Salmo salar, farmed) and bluenose (Hyperoglyphe antarctica). In Durban and Cape Town, respectively, just 23 and 20% of the outlets were familiar with legislation (the MLRA).
Most commonly encountered and popular fish species sold by 28 Durban and Cape Town restaurants and fishmongers.
| Most commonly traded species selected from a predetermined list (% of dealers trading) . | Most popular fish species as perceived by interviewees (overall rank) . | ||
|---|---|---|---|
| Durban . | Cape Town . | Durban . | Cape Town . |
| Rockcodsa, geelbek (62) | Kingklip, koba (100) | Geelbek | Koba |
| Kingklip, king mackerel (54) | Geelbek (93) | Kingklip | Kingklip |
| Yellowtail, tunaa, hake, dorado (46) | Dorado, solea, tunaa (87) | King mackerel | Geelbek |
| Red steenbras, snoek, solea, sharka (38) | Atlantic pomfret (angelfish), bluenose, Roman, Atlantic salmon (80) | Rockcodsa | Hake |
| Musselcrackera, skatea, koba (31) | Hake (73) | Hake | Tunaa, yellowtail |
| Most commonly traded species selected from a predetermined list (% of dealers trading) . | Most popular fish species as perceived by interviewees (overall rank) . | ||
|---|---|---|---|
| Durban . | Cape Town . | Durban . | Cape Town . |
| Rockcodsa, geelbek (62) | Kingklip, koba (100) | Geelbek | Koba |
| Kingklip, king mackerel (54) | Geelbek (93) | Kingklip | Kingklip |
| Yellowtail, tunaa, hake, dorado (46) | Dorado, solea, tunaa (87) | King mackerel | Geelbek |
| Red steenbras, snoek, solea, sharka (38) | Atlantic pomfret (angelfish), bluenose, Roman, Atlantic salmon (80) | Rockcodsa | Hake |
| Musselcrackera, skatea, koba (31) | Hake (73) | Hake | Tunaa, yellowtail |
aGroups that may include more than one species.
Most commonly encountered and popular fish species sold by 28 Durban and Cape Town restaurants and fishmongers.
| Most commonly traded species selected from a predetermined list (% of dealers trading) . | Most popular fish species as perceived by interviewees (overall rank) . | ||
|---|---|---|---|
| Durban . | Cape Town . | Durban . | Cape Town . |
| Rockcodsa, geelbek (62) | Kingklip, koba (100) | Geelbek | Koba |
| Kingklip, king mackerel (54) | Geelbek (93) | Kingklip | Kingklip |
| Yellowtail, tunaa, hake, dorado (46) | Dorado, solea, tunaa (87) | King mackerel | Geelbek |
| Red steenbras, snoek, solea, sharka (38) | Atlantic pomfret (angelfish), bluenose, Roman, Atlantic salmon (80) | Rockcodsa | Hake |
| Musselcrackera, skatea, koba (31) | Hake (73) | Hake | Tunaa, yellowtail |
| Most commonly traded species selected from a predetermined list (% of dealers trading) . | Most popular fish species as perceived by interviewees (overall rank) . | ||
|---|---|---|---|
| Durban . | Cape Town . | Durban . | Cape Town . |
| Rockcodsa, geelbek (62) | Kingklip, koba (100) | Geelbek | Koba |
| Kingklip, king mackerel (54) | Geelbek (93) | Kingklip | Kingklip |
| Yellowtail, tunaa, hake, dorado (46) | Dorado, solea, tunaa (87) | King mackerel | Geelbek |
| Red steenbras, snoek, solea, sharka (38) | Atlantic pomfret (angelfish), bluenose, Roman, Atlantic salmon (80) | Rockcodsa | Hake |
| Musselcrackera, skatea, koba (31) | Hake (73) | Hake | Tunaa, yellowtail |
aGroups that may include more than one species.
In all, 1120 people were questioned at the GFWS in Cape Town, of which 8% were aware of SASSI, and 18 months later in Johannesburg, 45% of 666 people were aware of the initiative. The most popular seafood (top 15 choices) included non-finfish items such as prawns (shrimps), which were the top overall choice, and calamari and oysters (Table 2).
Most frequently recorded responses by consumers attending two food shows in Cape Town and Johannesburg to the question “What is your favourite seafood choice?”
| Rank . | Cape Town (May 2006) . | Johannesburg (November 2007) . |
|---|---|---|
| 1 | Prawns | Prawns |
| 2 | Kob | Kingklip |
| 3 | Kingklip | Calamari (squid) |
| 4 | Tuna | Sole |
| 5 | Calamari (squid) | Norwegian salmon |
| 6 | Sole | Tuna |
| 7 | Crayfish (rock lobster) | Crayfish (rock lobster) |
| 8 | Musselcracker, yellowtail | “Do not eat seafood” |
| 9 | Geelbek | Hake |
| 10 | Norwegian salmon | Yellowtail |
| 11 | Hake | Kob |
| 12 | Oysters | Dorado, oysters |
| 13 | Rockcod | Roman |
| 14 | Red steenbras, snoek | Rockcod |
| 15 | Dorado | Butterfish, sushi |
| Rank . | Cape Town (May 2006) . | Johannesburg (November 2007) . |
|---|---|---|
| 1 | Prawns | Prawns |
| 2 | Kob | Kingklip |
| 3 | Kingklip | Calamari (squid) |
| 4 | Tuna | Sole |
| 5 | Calamari (squid) | Norwegian salmon |
| 6 | Sole | Tuna |
| 7 | Crayfish (rock lobster) | Crayfish (rock lobster) |
| 8 | Musselcracker, yellowtail | “Do not eat seafood” |
| 9 | Geelbek | Hake |
| 10 | Norwegian salmon | Yellowtail |
| 11 | Hake | Kob |
| 12 | Oysters | Dorado, oysters |
| 13 | Rockcod | Roman |
| 14 | Red steenbras, snoek | Rockcod |
| 15 | Dorado | Butterfish, sushi |
Most frequently recorded responses by consumers attending two food shows in Cape Town and Johannesburg to the question “What is your favourite seafood choice?”
| Rank . | Cape Town (May 2006) . | Johannesburg (November 2007) . |
|---|---|---|
| 1 | Prawns | Prawns |
| 2 | Kob | Kingklip |
| 3 | Kingklip | Calamari (squid) |
| 4 | Tuna | Sole |
| 5 | Calamari (squid) | Norwegian salmon |
| 6 | Sole | Tuna |
| 7 | Crayfish (rock lobster) | Crayfish (rock lobster) |
| 8 | Musselcracker, yellowtail | “Do not eat seafood” |
| 9 | Geelbek | Hake |
| 10 | Norwegian salmon | Yellowtail |
| 11 | Hake | Kob |
| 12 | Oysters | Dorado, oysters |
| 13 | Rockcod | Roman |
| 14 | Red steenbras, snoek | Rockcod |
| 15 | Dorado | Butterfish, sushi |
| Rank . | Cape Town (May 2006) . | Johannesburg (November 2007) . |
|---|---|---|
| 1 | Prawns | Prawns |
| 2 | Kob | Kingklip |
| 3 | Kingklip | Calamari (squid) |
| 4 | Tuna | Sole |
| 5 | Calamari (squid) | Norwegian salmon |
| 6 | Sole | Tuna |
| 7 | Crayfish (rock lobster) | Crayfish (rock lobster) |
| 8 | Musselcracker, yellowtail | “Do not eat seafood” |
| 9 | Geelbek | Hake |
| 10 | Norwegian salmon | Yellowtail |
| 11 | Hake | Kob |
| 12 | Oysters | Dorado, oysters |
| 13 | Rockcod | Roman |
| 14 | Red steenbras, snoek | Rockcod |
| 15 | Dorado | Butterfish, sushi |
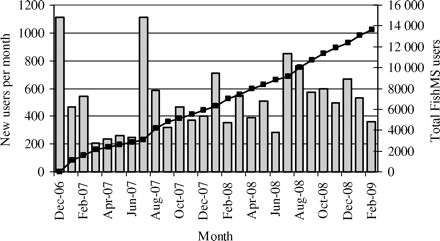
FishMS received a total of 54 294 requests from 13 968 individual telephone numbers (users) over the 27 months, each sending 3.89 requests on average. The number of new users joining every month ranged from 206 to 1112 (Figure 1). The highest monthly total of requests (3494) was in July 2007, but on average, 2011 requests were received each month. Most requests were made between 13:00 and 15:00, and between 19:00 and 22:00, suggesting that consumers were making use of the service during peak mealtimes. Table 3 provides a breakdown of requested information, which can be inferred as the most frequently requested species during meals. Kingklip (excluding ling, which was requested 136 times), and the kob groups ranked first and second, respectively. There were 24 requests for “Gastora” (see below), the first received only in July 2008. Red snapper was ranked 13th (972 requests), whereas river snapper received 123 requests. Other variations in snapper included the “generic” snapper (132) and ruby snapper (4). “Basa” received just two requests, although Pangasius received 422. Wahoo was requested 67, and Barracuda 327 times. “Cardinal” was first recorded in May 2007 with a total of 237 requests.
Monthly number of new (grey bars) and total (black squares) number of users of the FishMS service between December 2006 and February 2009.
Ten most requested seafood species or groups via the FishMS text message service between 1 December 2006 and 28 February 2009.
| Rank . | Fish name . | Species included in group (number of requests) . | Total requests . |
|---|---|---|---|
| 1 | Kingklip | n.a. | 4 102 |
| 2 | Kob group | Silver kob (2 108), “generic” kob (1 540), dusky kob (49), squaretail kob (10) | 3 707 |
| 3 | Tuna group | “Generic” tuna (1 954), yellowfin (307), bluefin (179), longfin (46), bigeye (17) | 2 503 |
| 4 | Dorado | n.a. | 2 498 |
| 5 | Sole group | “Generic” sole (2 120), east coast sole (83), west coast sole (29) | 2 232 |
| 6 | Hake | Shallow-water and deep-water hake, no market distinction | 2 087 |
| 7 | Yellowtail | n.a. | 1 964 |
| 8 | Geelbek | n.a. | 1 641 |
| 9 | Salmon group | “Generic” salmon (1 136), Norwegian (287), Atlantic (22), Pink (25), Scottish (71) | 1 544 |
| 10 | Roman | n.a. | 1 438 |
| Rank . | Fish name . | Species included in group (number of requests) . | Total requests . |
|---|---|---|---|
| 1 | Kingklip | n.a. | 4 102 |
| 2 | Kob group | Silver kob (2 108), “generic” kob (1 540), dusky kob (49), squaretail kob (10) | 3 707 |
| 3 | Tuna group | “Generic” tuna (1 954), yellowfin (307), bluefin (179), longfin (46), bigeye (17) | 2 503 |
| 4 | Dorado | n.a. | 2 498 |
| 5 | Sole group | “Generic” sole (2 120), east coast sole (83), west coast sole (29) | 2 232 |
| 6 | Hake | Shallow-water and deep-water hake, no market distinction | 2 087 |
| 7 | Yellowtail | n.a. | 1 964 |
| 8 | Geelbek | n.a. | 1 641 |
| 9 | Salmon group | “Generic” salmon (1 136), Norwegian (287), Atlantic (22), Pink (25), Scottish (71) | 1 544 |
| 10 | Roman | n.a. | 1 438 |
Ten most requested seafood species or groups via the FishMS text message service between 1 December 2006 and 28 February 2009.
| Rank . | Fish name . | Species included in group (number of requests) . | Total requests . |
|---|---|---|---|
| 1 | Kingklip | n.a. | 4 102 |
| 2 | Kob group | Silver kob (2 108), “generic” kob (1 540), dusky kob (49), squaretail kob (10) | 3 707 |
| 3 | Tuna group | “Generic” tuna (1 954), yellowfin (307), bluefin (179), longfin (46), bigeye (17) | 2 503 |
| 4 | Dorado | n.a. | 2 498 |
| 5 | Sole group | “Generic” sole (2 120), east coast sole (83), west coast sole (29) | 2 232 |
| 6 | Hake | Shallow-water and deep-water hake, no market distinction | 2 087 |
| 7 | Yellowtail | n.a. | 1 964 |
| 8 | Geelbek | n.a. | 1 641 |
| 9 | Salmon group | “Generic” salmon (1 136), Norwegian (287), Atlantic (22), Pink (25), Scottish (71) | 1 544 |
| 10 | Roman | n.a. | 1 438 |
| Rank . | Fish name . | Species included in group (number of requests) . | Total requests . |
|---|---|---|---|
| 1 | Kingklip | n.a. | 4 102 |
| 2 | Kob group | Silver kob (2 108), “generic” kob (1 540), dusky kob (49), squaretail kob (10) | 3 707 |
| 3 | Tuna group | “Generic” tuna (1 954), yellowfin (307), bluefin (179), longfin (46), bigeye (17) | 2 503 |
| 4 | Dorado | n.a. | 2 498 |
| 5 | Sole group | “Generic” sole (2 120), east coast sole (83), west coast sole (29) | 2 232 |
| 6 | Hake | Shallow-water and deep-water hake, no market distinction | 2 087 |
| 7 | Yellowtail | n.a. | 1 964 |
| 8 | Geelbek | n.a. | 1 641 |
| 9 | Salmon group | “Generic” salmon (1 136), Norwegian (287), Atlantic (22), Pink (25), Scottish (71) | 1 544 |
| 10 | Roman | n.a. | 1 438 |
Molecular analyses
Of the 174 fillet analyses, 50% were mislabelled. The results are summarized in Table 4.
Results for 16S rRNA matches from GenBank.
| Species sold as . | Sample size . | Percentage correctly labelled . | Identified substitutes . | BLAST % match . |
|---|---|---|---|---|
| Kingklip, Genypterus capensis | 54 | 100a | Pink cuskeel, Genypterus blacodes (EU848470) | 100 |
| Kob, Argyrosomus spp. | 70 | 16 | Warehou, Seriolella (AB205418) | 100 |
| Bigscale mackerel, Gasterochisma melampus (29) | n.a.b | |||
| Blackspotted croaker, Protonibea diacanthus (EF528202) | 100 | |||
| Sciaenops ocellatus (AY857951)/Leiostomus xanthurus (EU239813) | – | |||
| Yellowtail, Seriola lalandi | 15 | 100 | n.a. | – |
| Dorado, Coryphaena hippurus | 25 | 79 | Seriola lalandi (DQ521033) | 98 |
| Barracuda, Sphyraena spp. | 2 | 0 | King mackerel, Scomberomorus cavalla (DQ536428) | 96 |
| Red Snapper, Lutjanus spp. | 4 | 0 | River snapper, Lutjanus argentimaculatus (DQ784728) | 99 |
| Common bluestripe snapper, Lutjanus kasmira (FJ416614) | 95 | |||
| “Bassa” (no FishBase common name) | 1 | – | Sutchi catfish, Pangasianodon hypothalamus (DQ334285) | 99 |
| Wahoo, Acanthocybium solandri | 1 | 0 | King mackerel, Scomberomorus cavalla (DQ536428) | 96 |
| “Cardinal” | 2 | 0 | Bull's eye, Epigonus telescopus (EU848458) | 100 |
| Species sold as . | Sample size . | Percentage correctly labelled . | Identified substitutes . | BLAST % match . |
|---|---|---|---|---|
| Kingklip, Genypterus capensis | 54 | 100a | Pink cuskeel, Genypterus blacodes (EU848470) | 100 |
| Kob, Argyrosomus spp. | 70 | 16 | Warehou, Seriolella (AB205418) | 100 |
| Bigscale mackerel, Gasterochisma melampus (29) | n.a.b | |||
| Blackspotted croaker, Protonibea diacanthus (EF528202) | 100 | |||
| Sciaenops ocellatus (AY857951)/Leiostomus xanthurus (EU239813) | – | |||
| Yellowtail, Seriola lalandi | 15 | 100 | n.a. | – |
| Dorado, Coryphaena hippurus | 25 | 79 | Seriola lalandi (DQ521033) | 98 |
| Barracuda, Sphyraena spp. | 2 | 0 | King mackerel, Scomberomorus cavalla (DQ536428) | 96 |
| Red Snapper, Lutjanus spp. | 4 | 0 | River snapper, Lutjanus argentimaculatus (DQ784728) | 99 |
| Common bluestripe snapper, Lutjanus kasmira (FJ416614) | 95 | |||
| “Bassa” (no FishBase common name) | 1 | – | Sutchi catfish, Pangasianodon hypothalamus (DQ334285) | 99 |
| Wahoo, Acanthocybium solandri | 1 | 0 | King mackerel, Scomberomorus cavalla (DQ536428) | 96 |
| “Cardinal” | 2 | 0 | Bull's eye, Epigonus telescopus (EU848458) | 100 |
Sample sizes for each fish species are given, along with the percentage correctly labelled. Identified substitutes with their corresponding GenBank numbers are also listed.
aOrigin of these may not be southern Africa, but they could represent fish imported from New Zealand; to date, there is no information available on whether South African G. capensis and New Zealand G. blacodes represent different species or whether there is population genetic structuring within the distributional range.
b100% match obtained from sequencing Gasterochisma melampus.
Results for 16S rRNA matches from GenBank.
| Species sold as . | Sample size . | Percentage correctly labelled . | Identified substitutes . | BLAST % match . |
|---|---|---|---|---|
| Kingklip, Genypterus capensis | 54 | 100a | Pink cuskeel, Genypterus blacodes (EU848470) | 100 |
| Kob, Argyrosomus spp. | 70 | 16 | Warehou, Seriolella (AB205418) | 100 |
| Bigscale mackerel, Gasterochisma melampus (29) | n.a.b | |||
| Blackspotted croaker, Protonibea diacanthus (EF528202) | 100 | |||
| Sciaenops ocellatus (AY857951)/Leiostomus xanthurus (EU239813) | – | |||
| Yellowtail, Seriola lalandi | 15 | 100 | n.a. | – |
| Dorado, Coryphaena hippurus | 25 | 79 | Seriola lalandi (DQ521033) | 98 |
| Barracuda, Sphyraena spp. | 2 | 0 | King mackerel, Scomberomorus cavalla (DQ536428) | 96 |
| Red Snapper, Lutjanus spp. | 4 | 0 | River snapper, Lutjanus argentimaculatus (DQ784728) | 99 |
| Common bluestripe snapper, Lutjanus kasmira (FJ416614) | 95 | |||
| “Bassa” (no FishBase common name) | 1 | – | Sutchi catfish, Pangasianodon hypothalamus (DQ334285) | 99 |
| Wahoo, Acanthocybium solandri | 1 | 0 | King mackerel, Scomberomorus cavalla (DQ536428) | 96 |
| “Cardinal” | 2 | 0 | Bull's eye, Epigonus telescopus (EU848458) | 100 |
| Species sold as . | Sample size . | Percentage correctly labelled . | Identified substitutes . | BLAST % match . |
|---|---|---|---|---|
| Kingklip, Genypterus capensis | 54 | 100a | Pink cuskeel, Genypterus blacodes (EU848470) | 100 |
| Kob, Argyrosomus spp. | 70 | 16 | Warehou, Seriolella (AB205418) | 100 |
| Bigscale mackerel, Gasterochisma melampus (29) | n.a.b | |||
| Blackspotted croaker, Protonibea diacanthus (EF528202) | 100 | |||
| Sciaenops ocellatus (AY857951)/Leiostomus xanthurus (EU239813) | – | |||
| Yellowtail, Seriola lalandi | 15 | 100 | n.a. | – |
| Dorado, Coryphaena hippurus | 25 | 79 | Seriola lalandi (DQ521033) | 98 |
| Barracuda, Sphyraena spp. | 2 | 0 | King mackerel, Scomberomorus cavalla (DQ536428) | 96 |
| Red Snapper, Lutjanus spp. | 4 | 0 | River snapper, Lutjanus argentimaculatus (DQ784728) | 99 |
| Common bluestripe snapper, Lutjanus kasmira (FJ416614) | 95 | |||
| “Bassa” (no FishBase common name) | 1 | – | Sutchi catfish, Pangasianodon hypothalamus (DQ334285) | 99 |
| Wahoo, Acanthocybium solandri | 1 | 0 | King mackerel, Scomberomorus cavalla (DQ536428) | 96 |
| “Cardinal” | 2 | 0 | Bull's eye, Epigonus telescopus (EU848458) | 100 |
Sample sizes for each fish species are given, along with the percentage correctly labelled. Identified substitutes with their corresponding GenBank numbers are also listed.
aOrigin of these may not be southern Africa, but they could represent fish imported from New Zealand; to date, there is no information available on whether South African G. capensis and New Zealand G. blacodes represent different species or whether there is population genetic structuring within the distributional range.
b100% match obtained from sequencing Gasterochisma melampus.
Kingklip
At the mtDNA 16S rRNA level, all fillets examined showed a 100% match with Genypterus blacodes (pink ling/pink cuskeel), as also indicated by the zero branch length in the phylogenetic trees (Figure 2). As there is no published population genetic or phylogenetic data for southern African G. capensis, it was not possible to infer the country of origin from these comparisons. Therefore, the variable 5' prime part of the mtDNA control region was sequenced for 24 kingklip caught off South Africa, four fillets labelled with the origin “New Zealand”, and all commercially obtained fillets. The two geographic populations are differentiated by an average uncorrected sequence divergence of 8.44% (s.d. = 1.2%). Intra-population variation was interesting in that New Zealand samples probably attributed to G. blacodes showed a high sequence diversity of 4.8% (±1.8%), whereas South African kingklip shows a high level of similarity among individual fish, with a sequence diversity of 0.8% (±0.07%). This suggests that G. blacodes/capensis consists of at least two well-separated stocks. Given this approach, we identified 15 samples of kingklip fillets that probably originated from New Zealand.
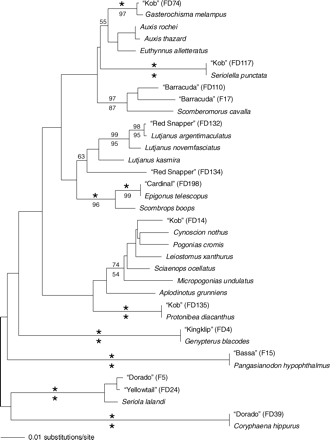
Neighbour-joining tree, showing phylogenetic associations of samples tested in this study (labelled as FDXXX or FXXX), with voucher sequences obtained from GenBank. Only bootstrap values for the clades of interest are indicated. Asterisks indicate 100% bootstrap support. Values above branches are those obtained from 1000 NJ replicates, values below are from 1000 parsimony replicates.
Kob
Kob, also commonly known locally as kabeljou, was by far the most substituted fish in this study. Of the 70 fillets sampled, only 11 matched Argyrosomus; 8 fillets were identified as silver kob (A. inodorus), and three as dusky kob (A. japonicus). Of the total, 29 matched a sample sold as “gastora” from a different restaurant (presumably the bigscale mackerel, Gasterochisma melampus) with 100% similarity, although no voucher specimen was available. Two fillets matched warehou (Seriolella spp.) with 100%, and another two matched 100% to blackspotted croaker (Protonibea diacanthus; Table 4). The remaining samples matched the family Sciaenidae (kob family), but no satisfactory species match could be found for them in GenBank (Table 4, Figure 2).
Yellowtail
The 15 fillets purchased as yellowtail showed 0.03% uncorrected sequence divergence from Seriola lalandi in the GenBank database (Table 4); this clustering was supported with 100% bootstrap in the phylogenetic analyses (Figure 2).
Dorado
Of the 24 samples tested for this study, 19 samples were correctly identified as dorado, with a 100% similarity to the GenBank sequence (Table 4, Figure 2). Five “dorado” fillets showed a 99% sequence similarity (0.1% sequence divergence) to yellowtail, Seriola lalandi.
Barracuda
Neither of the two fillets sequenced showed a match to the barracuda genus Sphyraena, but they did show a 98% sequence similarity to Scomberomorus cavalla (king mackerel) of the western Atlantic, despite Sphyraena 16S rRNA sequences being present in GenBank (EU099477, DQ874739, DQ532964, and others). Interestingly, in South Africa, the name king mackerel is used for the related S. commerson.
Red snapper
Three of the fillets examined showed a 99.9% sequence similarity to L. argentimaculatus in GenBank, and one a 96% similarity to Lutjanus kasmira (Table 4, Figure 2).
Wahoo
The fillet sold as wahoo showed a 97% sequence similarity to S. cavalla (see barracuda; Figure 2, Table 2), despite wahoo (Acanthocybium sp.) being represented in GenBank by 16S rRNA sequences (EU099493, DQ874727).
“Bassa”
Basa (Bassa) and Pangasius are common market names that refer to mostly two species, tra (Pangasianodon hypophthalmus) and basa (Pangasius bocourti). The fillet matched 99% with the Sutchi catfish (P. hypophthalmus; Table 4, Figure 2). There are known importers of farmed “Pangasius” from Vietnam, and it is sold by a number of seafood restaurant franchises, with a growing local market.
“Cardinal”
The two fillets of cardinal obtained from a restaurant showed a 100% match to Bull's eye (Epigonus telescopus), a fish belonging to the family Epigonidae (deep-water cardinals; Figure 2).
Discussion
The consequences of mislabelling and seafood substitution are numerous and include financial losses in both the public and governmental sectors, management implications, especially for endangered and already overfished species, undermining of consumer confidence in the seafood trade and in conservation-driven campaigns, as well as potential health concerns (Jacquet and Pauly, 2007b; GAO, 2009). Focusing on the South African markets and consumers, this study showed that substitution of seafood in the supply chain of the outlets tested is frequent, with relevance to all the above.
Seafood mislabelling in South Africa
The results clearly show that the rate of mislabelling seafood in South Africa determined in this study (50%) compares with mislabelling levels found in other studies (Logan et al., 2008). The most misrepresented fish species studied was kob, which for South African consumers comprise three species all belonging to the genus Argyrosomus (Griffiths and Heemstra, 1995). Of 70 frozen fillets analysed, almost 41% belonged to fish from the family Scombridae. A likely candidate is the bigscale mackerel (G. melampus), a species presumably caught as bycatch by tuna longliners throughout the Southern Ocean, including off the west coasts of South Africa and Nambia (Collette and Nauen, 1983). The origin of this fish on the South African market is not clear, although the name “gastora” is frequently used by the wholesale sector (M. Bürgener, pers. comm.). Other substitutions are not indigenous to South African waters; two of the fillets were warehou (Seriolella spp.), and another two fillets were probably blackspot croaker (P. diacanthus). The substitution of exotic fish for kob has significant financial implications, because both Protonibea and Seriolella are fish commanding lower prices than the premium-priced kob.
The detection of river snapper (mangrove red snapper; Lutjanus argentimaculatus), sold in two seafood retail outlets, has major management and legal implications. Fillets purchased as “red snapper” showed a 99% match to river snapper (Table 4, Figure 2). The estuary-dependent species is a designated recreational species in South Africa, based on its limited distribution on the east coast, and its sale is prohibited. The sale of river snapper may indicate deliberate marketing of a no-sale species as something closely related. The fillets were sold as frozen portions, suggesting that they were imported rather than caught in South Africa. Whole fresh river snappers are sometimes seen in informal markets in Durban and Johannesburg, but the colloquial name “rock salmon” is mostly used there (pers. obs.). It is unlikely that an import permit would be granted for a local no-sale species, so it has to be assumed that the fish was imported illegally under another name, or misidentified at source where similar fish were all sold as red snapper, and imported as a batch of mixed species. The fact remains that by labelling river snapper as red snapper, consumers are unable to avoid illegally purchasing a locally protected fish.
A second example is the trade in E. telescopus, or Bull's eye (a first record in SA). It highlights another emerging conservation issue in fisheries management: that of the growing trade in deep-water species (Roberts, 2002). FishMS data first showed this species to infiltrate the South African market in May 2007, with a growing number of requests for information. This species, like other deep-water species such as orange roughy (H. atlanticus), is extremely slow-growing and long-lived, reaching an estimated maximum age of more than 100 years (www.fishbase.org). Concerns relating to the trade of such species do not only pertain to vulnerable life histories and susceptibility to overfishing, but also to the sensitivity of the deep-water habitats where these species often occur (Roberts, 2002). It is therefore imperative that reliable information, including life-history data, as well as labelling for each species, is more accessible to consumers.
Health issues are frequently cited as a possible problem when substituting fish, and some of the most well-documented cases involve two members of the family Gempylidae, the escolar (Lepidocybium flavobrunneum) and the oilfish (Ruvettus pretiosus; Shadbolt et al., 2002). These species are routinely sold under the market name “butterfish” in South Africa (the 14th most requested name in FishMS), but despite the potential side effects of their consumption or their obscure identity (escolar received only 13, and oilfish 10 specific requests), they seem reasonably popular and widely available. Our findings did highlight other potential risks, e.g. substitution with scombroid fish (e.g. king mackerel) that have been implicated in cases of ciguatera poisoning (Lehane and Lewis, 2000).
Overall, perhaps one of the most startling findings was that so many of the species sold in the South African market seemed to originate from foreign sources, with more than half the species examined not even occurring in the South African Exclusive Economic Zone. This value excludes the widely distributed yellowtail and dorado that are found around the southern African coast, but may also be imported, especially with the growing aquaculture production for the former. As Baker (2008) notes, the next step from species identification would be the identification of geographical provenance or population of marine species. This, however, is still some way in the future, because population genetic studies that require such detail have been carried out for just a handful of exploited marine resources, and as such, their use in seafood traceability remains limited.
Consumer and seafood supply chain awareness in South Africa
Survey results confirmed a generally low awareness towards legislation (the MLRA) and species identity among restaurants and consumers. However, the results from the two food shows suggested a growing awareness (from 8 to 45%) among consumers about sustainable seafood, supported by the growth in the number of FishMS users (Figure 1). Yet, despite overall greater awareness, overexploited or “orange listed” species were still among the most popular. Both consumers and restaurants showed poor species knowledge or distinction when it came to certain groups such as sole and tuna, a trend that was further supported by FishMS requests where “generic” species were the most requested for these two groups. A notable exception is found in the kob group, where specific requests for silver kob were more frequent than for generic kob. Interestingly, most of the true kob fillets identified in this study were silver kob. There was also dominance of the use of vague group names to describe certain species, e.g. musselcracker, steenbras, rockcod, and in some cases market names such as “butterfish” (see above).
Substitution clearly undermines any messaging about sustainability of specific local species where a limited supply is available, or where a species is overexploited (Logan et al., 2008). For example, the annual catch limit for kingklip in South Africa is currently 3500 t, although for 2006/2007 the catch of kingklip was ∼2800 t per annum (Brandão and Butterworth, 2008). However, kingklip features on virtually every restaurant menu and retail seafood counters (pers. obs.), and it was recognized as early as the 1960s that “as it is nowhere abundant, far more fish named ‘kingklip’ on menus is eaten than is ever caught” (Smith and Smith, 1966). This is probably only possible with pink ling (also known as pink cuskeel) imports from New Zealand, although there may be other species substitutions. Consumers experience this as a contradiction, because they are on the one hand cautioned about conservation issues surrounding kingklip stocks, yet see it on offer wherever they go. Our results for the control region analysed show that ∼31% of the 49 kingklip fillets analysed were G. blacodes from New Zealand, although all these came from one wholesaler. Smith and Paulin (2003) found 2.8% nucleotide substitutions between G. blacodes and the local G. capensis in the mtDNA control region, so further suggesting differentiation between South African and New Zealand stocks.
Kob is a further example, with the stocks of all three species of kob in South Africa considered to be overexploited (Griffiths, 2000). However, based on its wide availability, consumers may gain the impression that kob remains plentiful, despite massive conservation concerns.
The examples above provide a number of reasons why there is mislabelling in South African markets. These may include masking illegal sourcing, and a limited interest or obligation in naming fish correctly because of generally poor compliance and legal awareness. It could further include a direct financial incentive (i.e. higher prices for low-value species), capitalizing on the popularity of certain well-known species, or perhaps the fear of trying to sell unknown species to a generally ignorant consumer-base. It is important to distinguish between deliberate and “accidental” mislabelling. For example, king mackerel in South Africa (S. commerson) is often called baracouta, cuda, couta, or barracouda, especially in KwaZulu–Natal (Collette, 2003). Two barracuda (Sphyraena) fillets tested were probably Scomberomorus, but in that instance the mislabelling may have been accidental, given the multitude of false common and local names in southern Africa. However, the renaming of this species to wahoo was apparently deliberately carried out at one restaurant investigated (perhaps to make it sound more exotic), because the invoices examined contained the correct name (L. Fish, pers. comm. Flying Fish Productions, Cape Town, South Africa).
For river snapper, it is not inconceivable that some of the material was processed as part of a mixed catch of snappers that was separated at source. However, it is likely that most mislabelling in South Africa is not accidental, but rather constitutes a deliberate act.
Seafood labelling in South Africa: a way forward
We have shown mislabelling to take place intentionally or unintentionally at various levels in the South African seafood value chain, and for a variety of reasons. Similar to the North American situation (Jacquet and Pauly, 2007b), it seems that most of this deception is perpetrated at a wholesale level, especially on imported products. We have also shown that South African consumers are undoubtedly encountering seafood fraud regularly when shopping for their favourite species. As a result, their ability to make informed choices, whether based on sustainability or other considerations, is severely curtailed. Local quality standards that require a true description of the variety of fish that should not be misleading do exist (Anon., 1996, 2003). However, neither these nor international guidelines such as the Codex Alimentarius (to which South Africa is a signatory) seem able to achieve their similar goals of protecting consumer health and ensuring fair trade practice in the food trade, through promoting (international) standards. This situation is complicated further by the extent of international trade, with >800 fish species being traded internationally, and some 45% of the global catch marketed away from the country of origin (http://www.fao.org/fishery/topic/2004/en). Disparate labelling and naming requirements, policies, and enforcement that exist in different countries further aggravate an already complex situation.
Local authorities in South Africa recognize that seafood is often mislabelled, whether as a result of inadequate policies and enforcement, or inefficient interagency collaboration, as is true in the United States (Jacquet and Pauly, 2007b). In fact, in the United States, it seems that seafood fraud prevails despite the presence of more guidelines, e.g. the standardized “Seafood List” or more heavily regulated trade (GAO, 2009).
For correct labelling in South Africa to begin to work, the fishing and processing industries, regulatory and management authorities, and other stakeholders involved (such as restaurateurs and wholesalers) must be willing to adopt and implement tools for sound seafood trade, with routine monitoring and auditing an integral part of the process. The key points listed below need to be considered. Our study has shown that sustainable seafood campaigns are useful in gaining information, or identifying and interpreting trends in the seafood trade. However, as Jacquet and Pauly (2007a, b) point out, their efficacy is undermined by the lack of traceability and misnaming, such as discovered in this study. The high prevalence of imported and exotic species in South Africa has clearly highlighted the limited relevance of consumer seafood lists that contain only local species. Further, it is likely that with the growing international seafood trade, such lists would require regular updating as new species become available to the market. Finally, the use of a consistent methodology to compile these lists becomes critical (Armsby and Roheim, 2008), because all evidence suggests that consumers will continue to consult them to guide their seafood choices.
Recognition of the issue: Authorities need to recognize that seafood fraud occurs regularly and that current standards, legislation, or policies (or their application and enforcement) are inadequate to address it. Certainly, the process currently in place for issuing import permits seems incapable of regulating or keeping record of which species enter the country. Dealers of seafood in the wholesale and retail sector need to appreciate that seafood fraud may cause irreparable damage to consumer trust and the reputation of seafood companies and industry as a whole.
Establishment of a standardized market and trade name list and naming protocol: As has been done in many countries (the United States, the UK, Australia/New Zealand), South Africa should compile a list of all locally traded species, taking into consideration variations between provinces and within the subregion. The list should include species from local fisheries and provide guidance on how to name imported species with local equivalents (e.g. would New Zealand kingklip be an acceptable trade name for ling?) or species unknown to the local market. The list and protocol should be developed in consultation with the industry, and ultimately become a national or regional trade policy.
Legislation on labelling requirements for seafood products: A minimum standard for compulsory information to be contained on any seafood label should be established. This should include the acceptable market name, scientific name, country of origin, and capture area (FAO fishing area), and the production method (wild-caught or farmed), as has been the case in the EU since 2002 (www.fao.org/fishery/topic/13293/en). Additional information that is strongly encouraged to be included is the capture gear type, and information relating to traceability.
Identify agencies responsible for monitoring and compliance and establish monitoring regime: As is true in the United States (GAO, 2009), there may be significant overlap in agencies that monitor different aspects of the seafood trade, e.g. quality (National Regulator for Compulsory Specifications, NRCS; formerly the South African Bureau of Standards, SABS), health (Department of Health), import and trade-related matters (South African Revenue Services, SARS), and compliance with local marine laws (Department of Environment Affairs and Tourism, DEAT). These agencies need to establish their respective roles in monitoring and detecting seafood fraud. An obligatory or voluntary monitoring framework should be developed to aid this.
Improved sustainability information: The inclusion of information on sustainability such as eco-labels should be supported.
Acknowledgements
We are indebted to L. Fish of Flying Fish Productions, who approached us with the idea of sampling local suppliers to test the validity of fish offered in South Africa and who also provided samples. We also thank P. Bloomer and A. Klopper for providing Argyrosomus sequences, and M. R. Lipiński (Marine and Coastal Management) and D. Griffiths (Sea Harvest, Saldanha Bay) for providing kingklip samples. F. Smith from Island Seafood assisted L. Fish in sourcing a number of samples. JB thanks K. Sink for co-conducting the restaurant surveys, and T. Siebert and L. Atkinson for tireless assistance with the consumer surveys. An investigation on linefish trade (unpublished) carried out by M. Bürgener (TRAFFIC) in 2006 was invaluable to our work. The project was partially supported by WWF–South Africa/Southern African Sustainable Seafood Initiative (SASSI), with funding from The Green Trust and Pick n Pay. SvdH was funded by a Stellenbosch University postdoctoral fellowship and the Claude Leon Foundation postdoctoral fellowship.