-
PDF
- Split View
-
Views
-
Cite
Cite
Phillip C. Watts, Suzanne M. Kay, Drew Wolfenden, Clive J. Fox, Audrey J. Geffen, Stephen J. Kemp, Richard D. M. Nash, Temporal patterns of spatial genetic structure and effective population size in European plaice (Pleuronectes platessa) along the west coast of Scotland and in the Irish Sea, ICES Journal of Marine Science, Volume 67, Issue 4, May 2010, Pages 607–616, https://doi.org/10.1093/icesjms/fsp274
Close - Share Icon Share
Abstract
The European plaice (Pleuronectes platessa) is a relatively mobile flatfish species, and previous studies have reported broad-scale genetic homogeneity among samples distributed throughout much of its northern European range, with no evidence for isolation-by-distance (IBD) population structure. Using microsatellite loci, the pattern of spatial genetic structure and effective population size is characterized for >800 plaice collected from locations off the west coast of Great Britain over a 3-year period (2001–2003). The plaice populations are characterized by weak spatial genetic structure, consistent with tagging data, and relatively low effective population sizes. In contrast to previous work, a pattern of isolation by distance is present among pairs of plaice from within each sampling period. However, IBD spatial structure was not observed for comparisons of plaice from different sampling years or using the entire dataset, indicating a patchy temporal genetic structure. Therefore, pooling the data from several years can mask subtle patterns of population structure and potentially confound estimation of other important demographic parameters, such as effective population size.Watts, P. C., Kay, S. M., Wolfenden, D., Fox, C. J., Geffen, A. J., Kemp, S. J., and Nash, R. D. M. 2010. Temporal patterns of spatial genetic structure and effective population size in European plaice (Pleuronectes platessa) along the west coast of Scotland and in the Irish Sea. – ICES Journal of Marine Science, 67: 607–616.
Introduction
Marine finfish management is underpinned by population dynamics models that are based on the concept of ideal stocks which are, in effect, isolated and self-sustaining populations (Walters and Martell, 2004). With the rapid advances in molecular biology techniques, it has become routine to use high-resolution genetic markers to determine patterns of gene flow and, therefore, to define biological stocks. Indeed, several studies have uncovered some degree of genetic structure among allopatric populations of marine fish that are indicative of separate stocks (O'Connell et al., 1998; Castillo et al., 2005), but typically there is little evidence of strong genetic differences (e.g. Hoarau et al., 2002b; Watts et al., 2004; Mariani et al., 2005; Carlsson et al., 2006; Pampoulie et al., 2007). Although the studies just listed may be in accordance with the view that the marine environment lacks significant barriers to organisms with dispersive life histories, this idea is challenged by marine ecologists, who point out that many oceanographic features can limit dispersal (Jørgensen et al., 2005a; Gallindo et al., 2006; Hansen and Hemmer-Hansen, 2007).
Even in the absence of specific features that restrict dispersal, spatial genetic structure is expected to develop where there is gene flow between neighbouring areas—the isolation-by-distance (IBD) model of population structure described by Wright (1943). Under IBD, the magnitude of genetic differences between pairs of populations (or individuals along a continuum) increases with spatial separation. As the dispersal capabilities of most species are less than their geographic ranges, IBD is expected to be at an appropriate spatial scale, but actually is rarely detected among marine teleost populations (Hoarau et al., 2002b; Knutsen et al., 2003; McPherson et al., 2004; Jørgensen et al., 2005a). An obvious reason for this is that the species are mobile, so a significant proportion of the fish stray to non-neighbouring populations. Alternatively, there may have been insufficient time for populations to reach genetic equilibrium, for instance, because the species has undergone a recent range expansion (Slatkin, 1993) or exists as a metapopulation (Hanski, 2003). Moreover, IBD can be overlooked simply because too few samples were analysed to attain statistical rigour (Watts et al., 2007a). One strategy to extricate the actual signal of gene flow, particularly for species with weak population structure, is to quantify the observed pattern of spatial genetic structure between temporally replicated samples (Waples, 1998; McPherson et al., 2004), although this practice is not employed routinely. Clearly, caution is needed when inferring patterns of gene flow in cases where few genetic differences have been uncovered.
The European plaice (Pleuronectes platessa) is a widely distributed flatfish found in waters shallower than 200 m from the English Channel to as far north as Iceland and Norway. Around the UK, it is heavily exploited, and some stocks are considered to be below safe biological limits (www.ices.dk). Movements of adult plaice around the UK are reasonably well understood, based on extensive tagging programmes (Dunn and Pawson, 2002; Hunter et al., 2003, 2004), and the dispersal of eggs and larvae has been studied using particle-tracking models (de Graaf et al., 2004; Fox et al., 2006, 2009). These data indicate that although the majority (e.g. 100% of tagged female plaice in one study; Hunter et al., 2003) of adults remain faithful to spawning areas, a significant proportion of adult plaice nonetheless move between management units (Dunn and Pawson, 2002; Kell et al., 2004), with consequent management implications (Kell et al., 2004). Moreover, eggs and larvae from different spawning grounds may be transported to the same nursery grounds, where the juveniles mix and may join other populations (Dunn and Pawson, 2002; Fox et al., 2009).
High levels of prerecruit dispersal combined with significant adult straying would suggest that genetic structuring in plaice should be weak. This statement seems to be confirmed by Hoarau et al. (2002b), who failed to find significant genetic differences throughout much of the northern European range of plaice, except for the Icelandic stock which is isolated by deep water, leading to the hypothesis that the species constitutes panmictic (or nearly panmictic) units at large spatial scales (the North Sea Basin). Subsequently, Hoarau et al. (2005) reported effective population sizes (Ne) of the North Sea and Icelandic plaice stocks that were several orders of magnitude less than the adult census abundance (N). In essence, Ne is a measure of the reproductively successful population rather than of all the fish present. This is a critical distinction, because it is the former and not the latter that determines the rate of loss of genetic diversity and therefore the associated risk of extinction (Saccheri et al., 1998) and level of evolutionary potential (Franklin, 1980). Despite this, fishery management operates on N, which may put even greater strain on the reproductive fraction of the population. A crucial assumption of any genetic method used to estimate Ne is that the sample of genotypes is representative. It is significant, therefore, that our current understanding of the population structure of North Atlantic European plaice has several limitations. First, thorough sampling of many potential plaice populations within a relatively limited area has not been undertaken. Second, the analysis of populations has not been repeated for successive years to assess the level of temporal stability to population structure. Third, relatively little is known about the population structure of stocks to the west of Great Britain: Hoarau et al. (2002b) collected one sample from the Irish Sea and one from Oban (west coast of Scotland). The study by Watts et al. (2004) did not uncover significant spatial genetic structure within the Irish Sea, but sample sizes were too limited to draw robust conclusions.
The objectives of this paper are (i) to quantify the pattern of localized spatial genetic differentiation in European plaice and to contrast this with the apparent homogeneity throughout much of northern Europe, (ii) to assess the degree of temporal stability in any population structure, and (iii) to estimate the effective population size of plaice stocks in the Irish Sea and west of Scotland.
Material and methods
Fieldwork and genotyping
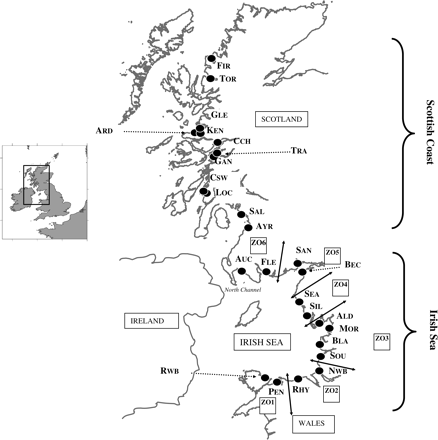
Juvenile plaice were sampled from inshore nursery grounds along the west coast of Great Britain between 2001 and 2003 (Figure 1); the numbers collected are listed in the Appendix. Samples along the Scottish coast were collected with a 1.5-m beam trawl towed 0.5 m deep, parallel with the shore, and samples from the Irish Sea were collected using 1.5- or 2-m beam trawls towed from 100 m offshore (maximum 1.5 m deep) on to the beach (Fox et al., 2007). The position of each sample was recorded using GPS, except the collections from the Irish Sea (2001) that were taken from several sites within one of six distinct zones (Figure 1). For spatial statistics purposes, these samples were allocated coordinates at the centre of the sample zone. Whole fish were stored in absolute ethanol until processing.
Sample locations for juvenile plaice, P. platessa, collected during the years 2001–2003. The three-letter code for each location is used to identify samples in the Appendix, with zones around the Irish Sea identified for 2001 samples by ZO1–ZO6.
Genomic DNA was extracted using a high salt protocol (Sambrook and Russell, 2000) from muscle tissue. Plaice were genotyped at nine microsatellite loci (LIST1-001, LIST1-003, LIST1-007, LIST1-009, LIST1-010, Pl06, Pl09, Pl92, Pl115) characterized by Watts et al. (1999, 2001) and Hoarau et al. (2002a). Between 10 and 50 ng of DNA was used in a 10 µl PCR containing 75 mM Tris–HCl pH 8.9, 20 mM (NH4)2SO4, 0.01% v/v Tween-20, 0.2 mM each dNTP, 1.5–3.0 mM MgCl2, 2 pmol of each primer, and 0.25 U Taq polymerase (ABgene). Forward primers were 5′-labelled with either 6-fam, ned, pet, or vic flourophores (Applied Biosystems). Thermal cycling conditions are described by Watts et al. (1999, 2001) and Hoarau et al. (2002a). PCR products were pooled into one of two genotyping panels along with a genescan-500 liz size standard (Applied Biosystems), then separated by capillary electrophoresis through a denaturing polymer on an ABI3100 automated sequencer (Applied Biosystems). Allele sizes were determined using the cubic model of analysis in genemapper software (Applied Biosystems).
Statistical analysis of genetic data
Genotypes were examined for the presence of null alleles, potential mis-scoring attributable to stutter bands and/or PCR bias against amplification of larger alleles using microchecker (Van Oosterhout et al., 2003). fstat ver. 2.9.3 (Goudet, 1995) software was used to calculate indices of genetic diversity: the number of alleles (Na), observed heterozygosity (Ho), expected heterozygosity (He), and Wright's (1951) inbreeding coefficient (f), which was estimated using the method of Weir and Cockerham (1984). The significance of any departures from expected Hardy–Weinberg equilibrium conditions was assessed by permuting alleles among individuals within samples (1000 permutations) using fstat. Genotypic linkage equilibrium among all locus–pair combinations was determined for the pooled dataset within each sampling period using the log-likelihood ratio G-statistic in fstat. Sequential Bonferroni corrections for k multiple tests were applied where appropriate (Rice, 1989).
Population subdivision was characterized for each locus and as an average over all loci by calculating θ, Weir and Cockerham's (1984) unbiased estimator of Wright's (1951),FST, using fstat. The 95% confidence intervals about estimates of θ were generated by jackknifing over loci (Goudet, 1995).
Genetic differentiation under IBD is expected to increase linearly with spatial distance in a one-dimensional habitat, and linearly with the logarithm of spatial separation in two-dimensional space (Rousset, 1997). Although our sample sites are (roughly) linearly distributed, the appropriate treatment to account for the surrounding matrix of other (unsampled) plaice populations from west of our sample sites, i.e. off the Irish coast, is that of a two-dimensional population matrix. The strength of any IBD was examined by regression of estimates of genetic differentiation among pairs of samples, defined by FST/(1 – FST), against the corresponding distances (ln m) separating them (Rousset, 1997); values of FST/(1 – FST) were calculated using the online version (3.1c, http://wbiomed.curtin.edu.au/genepop/) of genepop (Raymond and Rousset, 1995), with the significance of any correlation between genetic differentiation and geographic distance assessed by the Mantel test (1000 permutations of population locations among all locations). Distances between individuals and samples were the shortest marine route between sample locations.
IBD was also investigated by spatial autocorrelation, a technique that yields information about the pattern and scale of spatial structure and has proved to be sensitive at recovering fine-scale genetic structure (Vekemans and Hardy, 2004). To determine whether any genetic structure was temporally stable, we restricted comparisons among individuals that were collected either within or between each sampling period (year). For each treatment, the variation in average kinship among individuals (Fij; Loiselle et al., 1995) was estimated across a range of spatial scales using spagedi version 1.2 (Hardy and Vekemans, 2002). Average (over all loci) correlograms are presented to avoid variation in correlogram profiles based on the frequencies of individual alleles subject to stochastic processes. Standard errors for average Fij were generated by jackknifing over loci, and the significance of average estimates of Fij was calculated by making 1000 permutations of spatial group locations among spatial groups within category.
A point estimate of short-term effective population size (Ne) was calculated from the strength of linkage disequilibrium (LD; Hill, 1981) using Waples’ (2006) correction for a downward bias that occurs when the sample size is small relative to Ne. Briefly, the LD method of estimating Ne is based on the premise that if there is no immigration, population substructure or selection and the genetic sample is representative, then genetic drift in a finite population generates a measurable correlation between alleles among different loci that informs on the Ne. Estimates of Ne were computed using LDNe ver. 1.31 (Waples and Do, 2008) for pooled samples (within each year) from two distinct areas, the Irish Sea and the Scottish coast, that are separated by the narrow North channel (Figure 1). For this analysis, we assumed random mating populations and calculated parametric 95% confidence intervals using Equation (12) of Waples (2006). Estimates of Ne were calculated using the subset of six loci that are unlinked and do not suffer from null alleles (see below).
Estimates of plaice population size in the Irish Sea and west coast of Scotland
ICES carries out annual analytical stock assessments of the Irish Sea plaice stock (Division VIIa; ICES, 2006) based upon commercial catch data and a dedicated beam trawl survey. Estimates of spawning–stock biomass (t) and the numbers of fish at age for years 2001–2005 were taken from the latest available assessment. There is no formal assessment of the status of plaice for the west of Scotland, but a groundfish survey using a Grande Overture Verticale (GOV) trawl is conducted annually in the first quarter of each year. That survey also includes a number of stations in the Irish Sea (VIIa). Although not an ideal gear for sampling flatfish, the GOV trawl does catch plaice. Catch data were obtained from Marine Scotland, Aberdeen, and an approximate estimate of the status of plaice west of Scotland obtained by scaling the data from the formal VIIa assessment by the ratio of mean catches in Division VIa for stations east of 8°W and between 55 and 58.5°N to the mean catch in Division VIIa.
Results
Genetic diversity
We genotyped 864 plaice at nine microsatellite loci. One pair of loci (LIST1-009 and LIST1-010) demonstrated significant (p < 0.05, k = 36) linkage disequilibrium in all three temporal samples; because LIST1-009 was weakly polymorphic, it was omitted from subsequent analyses. There was no evidence for mis-scoring of genotypes as a consequence of stutter bands or large allele dropout, but several loci apparently suffer from null alleles: for the two loci Pl06 and Pl09, there was evidence of nulls in almost all (97 and 89%, respectively) of the 38 tests, whereas at two other loci (Pl92 and Pl115), roughly one-quarter of the samples may have been affected by null alleles. Null alleles were not a problem for the remaining loci. Therefore, the subset of six loci, LIST1-001, LIST1-003, LIST1-007, LIST1-010, Pl92, and Pl115, was selected as a panel of unlinked genetic markers that are unaffected by non-amplifying alleles. Only these genetic markers were used for genetic analyses.
Raw data on basic measures of genetic diversity at LIST1-001, LIST1-003, LIST1-007, LIST1-010, Pl92, and Pl115 are provided in the Appendix. Briefly, the number of alleles over all samples varied from a moderate 8 (LIST1-007 and LIST1-010) to a maximum of 61 (Pl92). Within samples, the number of alleles ranged between 2 and 10 at four loci (LIST1-001, LIST1-003, LIST1-007, and LIST1-010), and between 3 and 21 at two loci (Pl92 and Pl115); therefore, average levels of gene diversity He (over all samples) were greater in the last two loci (He = 0.89 and 0.69) than in the other four (He = 0.62, 0.44, 0.41, and 0.53). The f values of most locus–sample combinations were not significantly different from zero (p > 0.05, k = 38 tests per locus), but the most polymorphic locus (Pl92) had significant heterozygote deficits in four samples (Appendix). The average f over all loci varied from –0.098 (SAL 2001) up to 0.423 (FIR 2001), but was <0.1 for most (79%) samples.
Spatio-temporal genetic structure
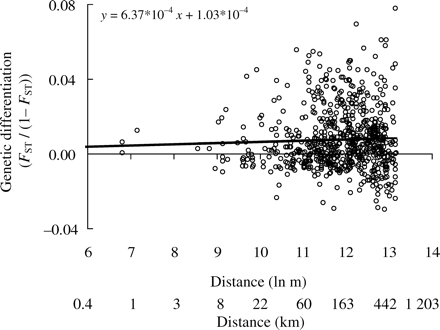
Overall level of genetic differentiation was weak, with all fixation indices below 0.007. The variance of FST/(1 – FST) increased with the distance separating populations (Figure 2), which is a qualitative pattern of IBD genetic structure. Although regressions of FST/(1 – FST) against spatial separation produced weak positive slopes, neither the 2001 samples (there were too few samples to analyse the 2002 and 2003 datasets individually) nor the entire dataset demonstrated a statistically significant relationship (p = 0.50 and 0.34, respectively). The results were the same when all loci were included in the analyses (data not shown).
Relationship between the level of genetic differentiation (FST/(1 – FST)) between pairs of samples and their distance of separation.
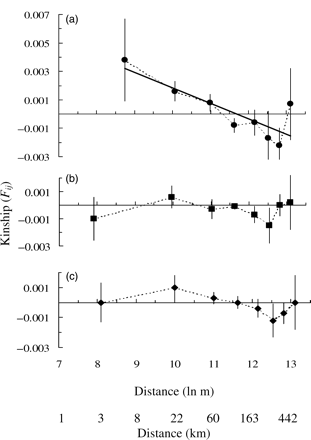
Neighbouring plaice were genetically more alike than were pairs of more distantly separated individuals. Average kinship (Fij) between pairs of fish from the same sample period was significantly (p < 0.05) greater than zero in the first distance category (up to 10 km separation). There was a significant (p = 0.0021, r2 = 0.814) linear decline in average kinship (Fij) with increasing (logarithm of) distance among individuals, with the first three distance classes (up to ∼60 km) having positive values of Fij (Figure 3a). In contrast, no significant spatial trend in the level of Fij (i.e. no pattern of IBD) was observed when the analysis was restricted to pairs of plaice collected from different sample periods (Figure 3b) or when the entire dataset was analysed (Figure 3c).
Correlogram of the combined genetic correlation in kinship (Fij; Loiselle et al., 1995) as a function of average spatial separation for pairs of juvenile plaice, from inshore areas off the western coast of Great Britain. Analysis using (a) pairs of individuals within the same sampling period only, (b) pairs from different sampling periods, and (c) the entire dataset.
Effective population size
Estimates of Ne for the samples of Irish Sea plaice were ∼700 (during 2001 and 2003, but could not be determined during 2002) and were noticeably lower (∼40) for plaice from the Scottish coast (2001 only). Pooling all samples (in 2001) from the Irish Sea and Scottish coast provided an estimate of Ne ∼ 90, which was substantially lower than the estimates of Ne for the Irish Sea alone. From approximate adult censuses (N), the Ne/N ratio for all samples is well below 1, typically in the order of 10−5 (Table 1).
Estimates of effective population size (Ne) (±95% CI) and approximate total numbers of adults (N) of plaice, P. platessa.
| Sample . | Ne . | –95% CI . | +95% CI . | N . | Ne/N . |
|---|---|---|---|---|---|
| 2001 All | 89.1 | 75.6 | 105.3 | 18 925 684 | 0.47 × 10−5 |
| 2001 Scotland | 36.9 | 31.7 | 42.9 | 1 683 464 | 2.19 × 10−5 |
| 2001 Irish Sea | 648.9 | 289.7 | Inf. | 17 242 220 | 3.76 × 10−5 |
| 2002 Irish Sea | – | – | – | 20 924 870 | – |
| 2003 Irish Sea | 774.4 | 289.0 | Inf. | 25 840 220 | 3.00 × 10−5 |
| Sample . | Ne . | –95% CI . | +95% CI . | N . | Ne/N . |
|---|---|---|---|---|---|
| 2001 All | 89.1 | 75.6 | 105.3 | 18 925 684 | 0.47 × 10−5 |
| 2001 Scotland | 36.9 | 31.7 | 42.9 | 1 683 464 | 2.19 × 10−5 |
| 2001 Irish Sea | 648.9 | 289.7 | Inf. | 17 242 220 | 3.76 × 10−5 |
| 2002 Irish Sea | – | – | – | 20 924 870 | – |
| 2003 Irish Sea | 774.4 | 289.0 | Inf. | 25 840 220 | 3.00 × 10−5 |
Ne was estimated from single generation samples using the corrected linkage disequilibrium method of Waples (2006).
Estimates of effective population size (Ne) (±95% CI) and approximate total numbers of adults (N) of plaice, P. platessa.
| Sample . | Ne . | –95% CI . | +95% CI . | N . | Ne/N . |
|---|---|---|---|---|---|
| 2001 All | 89.1 | 75.6 | 105.3 | 18 925 684 | 0.47 × 10−5 |
| 2001 Scotland | 36.9 | 31.7 | 42.9 | 1 683 464 | 2.19 × 10−5 |
| 2001 Irish Sea | 648.9 | 289.7 | Inf. | 17 242 220 | 3.76 × 10−5 |
| 2002 Irish Sea | – | – | – | 20 924 870 | – |
| 2003 Irish Sea | 774.4 | 289.0 | Inf. | 25 840 220 | 3.00 × 10−5 |
| Sample . | Ne . | –95% CI . | +95% CI . | N . | Ne/N . |
|---|---|---|---|---|---|
| 2001 All | 89.1 | 75.6 | 105.3 | 18 925 684 | 0.47 × 10−5 |
| 2001 Scotland | 36.9 | 31.7 | 42.9 | 1 683 464 | 2.19 × 10−5 |
| 2001 Irish Sea | 648.9 | 289.7 | Inf. | 17 242 220 | 3.76 × 10−5 |
| 2002 Irish Sea | – | – | – | 20 924 870 | – |
| 2003 Irish Sea | 774.4 | 289.0 | Inf. | 25 840 220 | 3.00 × 10−5 |
Ne was estimated from single generation samples using the corrected linkage disequilibrium method of Waples (2006).
Discussion
We found generally weak or non-significant spatial genetic structure among populations of plaice from the western coast of Great Britain, similar to that reported for populations of this species from the continental shelf of northern Europe (Hoarau et al., 2002b; Watts et al., 2004). However, beneath this weak spatial genetic structure lies a subtle annual pattern of IBD that has not been reported for the species and which may impact upon the assumption that the species comprises a single, broad panmictic unit in northern European seas (excluding the Icelandic stock).
Spatial genetic structure
Adult plaice are capable of long-distance movement (Metcalfe and Arnold, 1997; Hunter et al., 2004), but generally return to the spawning site where they were tagged (Hunter et al., 2003). The mechanisms by which juvenile plaice locate spawning grounds are not known, but immature fish probably learn the migration routes from adults (Arnold and Metcalfe, 1996), rather than possessing an innate natal homing instinct. Stochastic attachment of juveniles to adults from different spawning areas would lead to gene flow among spawning stocks even if mature fish subsequently used one spawning ground only. In addition to a possible juvenile dispersal phase, the opportunity for straying between spawning grounds, despite evidence for homing, has been identified by conventional tagging (Dunn and Pawson, 2002), and with rates sufficiently high to prevent genetic divergence through genetic drift alone. Therefore, the overall weak spatial genetic structure reported here is consistent with the known dispersal characteristics of this species, particularly the level of mixing between Irish Sea plaice stocks reported by Dunn and Pawson (2002). Moreover, the gradual decline in Fij points to IBD rather than any specific barrier to gene flow in the study region; as such, there is no evidence that the narrow North Channel (Figure 1) acts as a substantive barrier to movement by plaice from the Irish Sea and the west coast of Scotland.
That juvenile plaice are characterized by IBD within a sampling year points to restricted gene flow in this sample area. Extremely localized retention, and therefore sampling, of fish from just a few families would generate high genetic similarities at small distance classes, but this phenomenon may be expected to exhibit a patchy genetic structure at broader scales (owing to chance sampling effects) rather than the consistent linear decline in relatedness observed here (Figure 3a). IBD has been reported for some marine teleost populations (Pogson et al., 2001; O'Reilly et al., 2004), but, as stated earlier, IBD is not usually detected and it was not reported for plaice stocks sampled across much of its northern European range (Hoarau et al., 2002b). On the one hand, this difference in observed spatial genetic structure may reflect our sampling of juveniles that have travelled a limited distance from spawning grounds (see below), whereas Hoarau et al.'s (2002b) sample of adults comprised mainly non-spawning fish from feeding grounds and therefore potentially sampled mixed stocks [note that Hoarau et al. (2002b) analysed both adults and juveniles]. However, a similar contrast in the pattern of spatial genetic structure at relatively limited and broader geographic scales has been uncovered during extensive research into two of the most important commercially exploited teleosts in the North Atlantic, cod Gadus morhua (Bentzen et al., 1996; Ruzzante et al., 1996, 1997; Hutchinson et al., 2001; Pogson et al., 2001) and herring Clupea harengus (Shaw et al., 1999; Mariani et al., 2005; Jørgensen et al. 2005b). That the pattern of spatial structure depends on spatial scale likely reflects greater, and possibly more predictable, rates of gene flow at local scales and a more rapid approach to genetic equilibrium conditions (Crow and Aoki, 1984; Slatkin, 1993). Together, these studies demonstrate that a single genetic survey at a broad spatial scale can be misleading and underscores the need for repeated temporal sampling of marine teleost populations to provide meaningful appraisals of their stock structure.
To the west of the British Isles, plaice are formally assessed and managed as discrete stocks in the Western Channel, Celtic Sea, and Irish Sea (www.ices.dk). Plaice to the west of Scotland are not an important component of commercial catches, so there is no formal management of the stock in that area, and there are no data on the level of mixing between the Irish Sea and west coast of Scotland. Within the eastern Irish Sea and west of Scotland, we failed to find discreet genetic boundaries corresponding to distinct stocks, but our results demonstrate that the management unit is also not a single, panmictic stock. Aside from potential dispersal by juveniles and/or adults, the pattern of IBD is consistent with results from a recently developed particle-tracking model for the Irish Sea (van der Molen et al., 2007), where the modal distance eggs and larvae are transported is ∼80 km. On the west coast of the British Isles and in the Irish Sea in particular, the duration of the pelagic egg and larva phase is of the order of 50–70 d (Nash, 1998; Fox et al., 2009), so given the prevailing oceanography during early spring, the distances between spawning and settlement on the nursery grounds are, in general, relatively short (Fox et al., 2007, 2009). Therefore, our sampling reflects genetic differences between recruits from spawning grounds off the western British Isles; the pattern of IBD indicates dispersal mainly between neighbouring areas. The results from these particle models also demonstrate transport of eggs and larvae from the western Irish Sea to the east (see also Watts et al., 2004; Fox et al., 2009), and that eggs and larvae could be moved north through the North Channel under certain meteorological conditions, consistent with our genetic data and providing a prerecruit mechanism for dispersal (Fox et al., 2009). Given improved knowledge of mixing rates between fish populations, it is relatively easy to set up management simulation models to investigate the impacts on management advice (see Kell et al., 2009, for issues relating to herring stocks west of the British Isles). Although this has been undertaken for plaice in the North Sea and Eastern Channel (Kell et al., 2004), this approach has not yet been applied to plaice stocks to the west of the UK.
The distinction between methods used to detect IBD (cf. Figures 2 and 3a) is interesting because most studies determine the strength of IBD from the pattern of genetic differentiation among pairs of populations. One likely reason for the wide variance in estimates of FST/(1 – FST) is when the distance between sample pairs does not reflect the distance travelled by dispersing individuals. To some extent, this imprecision may have less effect when using autocorrelation because data are placed into distance classes. Nonetheless, as we estimated the shortest marine route between sample sites, perhaps the patterns of movement among plaice stocks are more complex. Moreover, IBD may not be detected when too few populations are sampled to provide sufficient statistical power. In this study, we found a qualitative pattern of IBD (i.e. an increase in the variance) among estimates of differentiation among pairs of populations (Figure 2), but the regression slope was non-significant. Certainly, other studies have found greater statistical power to detect IBD associated with analyses based on genetic relationships among individuals rather than populations (Castric and Bernatchez, 2004; Watts et al., 2007a).
Temporal genetic structure
An absence of IBD between samples from different sample periods (Figure 3b and c) is indicative of annual variation in the genetic composition of juvenile plaice; such temporal genetic patchiness appears to be a relatively common feature of populations of marine species (Larson and Julian, 1999). Crucially, when samples were combined across years, the subtle IBD spatial genetic structure was masked. Hoarau et al. (2002b) analysed samples that were collected from several years which, in addition to the issues discussed above, may account for the failure to detect IBD.
Reasons for temporal variation in genetic structure generally are not known, but typically are believed to reflect the chance survival of just a small fraction of individuals from a much larger pool of offspring (reviewed by Larson and Julian, 1999). Other explanations lie in social structure and breeding behaviour. For example, spawning aggregations of cod are not simple, random mating units, but involve complex lekking behaviour that allows dominant males to fertilize most eggs (Morgan and Trippel, 1996; Rowe et al., 2004). Similarly, some Atlantic herring populations apparently form genetically discreet spawning waves (McPherson et al., 2003; Jørgensen et al., 2005b). In addition, a recent work has implicated selective mortality as an example of a process that can alter gene frequencies between annual cohorts (Vigliola et al., 2007). Whatever the mechanism, young plaice at a particular nursery ground are the progeny of a relatively small fraction (compared with the number of mature adults) of the available breeding individuals (see the discussion on effective population size below), and the transient IBD genetic structure indicates that the success of these individuals varies annually. Determining the relative importance of potential stochastic or deterministic factors that cause variance in the composition of the breeding population, and hence prevent temporal stability in gene frequencies, clearly would help better understand demographic processes relevant to the management of European plaice.
Effective population size
That our estimates of Ne varied between years and could not be resolved for one sample (Table 1) may reflect some of the biases and imprecision associated with single generation estimates of Ne (Frankham, 1995), although the use of large sample sizes and at least six loci may provide reasonable estimates of Ne with the linkage disequilibrium method (Bartley et al., 1992). Alternatively, the apparent variation in Ne may reflect temporal changes in the size of the successful breeding population attributable to variable survivorship of planktonic eggs and larvae (possibly reflecting, for example, annual variations in weather conditions or food availability), but it has to be stressed that further work is essential to corroborate this, because the variation could reflect different sampling strategies. Even if the estimates of Ne are reasonable, it is important to make the contrast between this parameter (an indication of the successful breeding population) and the total adult population size that may not fluctuate in the same way.
Bearing in mind the well-documented limitations of single-generation methods of estimating Ne, the estimated Nes of Irish Sea and Scottish populations of plaice are low. The Ne/N ratios in the order of 10−5 (Table 1) are comparable with values reported for other marine teleosts (Hauser et al., 2002; Turner et al., 2002; Hutchinson et al., 2003), including the Icelandic and North Sea plaice stocks (Hoarau et al., 2005). A reduction in Ne is typically attributed to some combination of three factors, (i) uneven sex ratio, (ii) population fluctuations, or (iii) variance in reproductive success (Frankham, 1995), with the latter, in particular, thought to be typical of many marine teleosts that are highly fecund, have complex mating behaviours, and experience high mortality and stochastic survival of planktonic eggs and larvae (i.e. a type III survivorship curve).
Although a low Ne may make biological sense for plaice, it is essential to note that one assumption of the LD (like many other) method of estimating Ne is that the population is closed and, given putative mobility of eggs, larvae, juveniles, and adult plaice, this does not seem likely for many plaice stocks. The impact of immigration on these estimates of Ne depends on the genetic composition of immigrants (Fraser et al., 2007; Chevolot et al., 2008); indeed, a comparison of the LD method with other methods of estimating Ne that account for dispersal failed to find a systematic bias (Watts et al., 2007b). The clear difference in the estimates of Ne for the Irish Sea samples and that of the entire study region (cf. Table 1) highlights the impact of the scale of sampling upon estimates of Ne (and the ratio Ne/N), and the issue of spatial structure within a large population that lacks obvious genetic boundaries. Therefore, another crucial implication of subtle population structure is that an apparently homogeneous population may not be strictly panmictic. This is important because estimates of Ne require a sample of representative genotypes, which is straightforward to collect when a population is truly mixed. Failure to recognize subtle population structure will lead to the signal of genetic drift that is used to estimate Ne, i.e. the temporal variation in allele frequencies, being confounded with differences arising from spatial genetic variation, so generating inaccurate estimates of Ne. It is relevant therefore that the contemporary “North Sea” sample used by Hoarau et al. (2005) was collected from a limited area (two nursery grounds in the Dutch Wadden Sea), whereas the historical samples were sourced more generally from “the North Sea”. To what extent the temporal variance in allele frequencies among samples reflects divergence because of IBD spatial structure rather than genetic drift, i.e. the Ne, requires further study before management plans are adopted to account for an apparently severely reduced Ne.
To summarize, plaice populations along the west coast of Great Britain are characterized by IBD genetic structure against a background of weak spatial genetic differentiation. This pattern of spatial genetic structure agrees with our current understanding of the dispersal characteristics of plaice, although we cannot yet partition the relative importance of different stages in its life history. Our data contrast with previous work that reported broad-scale genetic homogeneity among samples of plaice throughout much of its northern European distribution, and an absence of IBD. Our results also raise questions about the validity of present estimates of Ne for populations of mobile marine species that lack clear genetic boundaries. Clearly, it is essential to determine whether representative genetic samples have been taken before such data are used for genetic management and conservation. Despite this, it appears likely that the Ne values of plaice populations are several orders of magnitude lower than the adult censuses, which is typical of highly fecund species.
Acknowledgements
The work was funded by the University of Liverpool research development fund, and collection of samples within the Irish Sea was done under Defra project MF0423. Samples from Scotland were kindly provided by Tim Targett (University of Maryland), Mike Burrows, and Bob Batty (SAMS, Oban, Scotland).
References
Appendix
Sample sizes and basic measure of genetic variation at six unlinked microsatellite loci for samples of plaice (P. platessa) from the west coast of Great Britain
| . | . | . | LIST1-001 . | LIST1-003 . | LIST1-007 . | LIST1-010 . | Pl92 . | Pl115 . | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | Sample . | n . | He . | Na . | f . | He . | Na . | f . | He . | Na . | f . | He . | Na . | f . | He . | Na . | f . | He . | Na . | f . |
| 2001 | ARD | 15 | 0.591 | 6 | −0.016 | 0.260 | 6 | −0.077 | 0.404 | 4 | −0.188 | 0.524 | 3 | −0.374 | 0.894 | 18 | 0.150* | 0.714 | 10 | 0.160 |
| AYR | 20 | 0.625 | 4 | −0.280 | 0.625 | 4 | −0.280 | 0.200 | 2 | 0.000 | 0.550 | 2 | 0.273 | 0.975 | 6 | 0.590* | 0.375 | 3 | −0.067 | |
| CCH | 11 | 0.723 | 4 | −0.132 | 0.405 | 4 | 0.326 | 0.391 | 3 | −0.163 | 0.518 | 2 | −0.228 | 0.959 | 12 | 0.336* | 0.523 | 5 | −0.217 | |
| CSW | 12 | 0.695 | 4 | −0.055 | 0.576 | 6 | 0.306 | 0.500 | 5 | 0.200 | 0.510 | 2 | 0.738* | 0.876 | 10 | 0.239* | 0.643 | 10 | 0.170 | |
| FIR | 25 | 0.617 | 5 | 0.027 | 0.583 | 5 | 0.314 | 0.372 | 4 | 0.463 | 0.633 | 3 | 0.684* | 0.917 | 8 | 0.515* | 0.594 | 6 | 0.495* | |
| GAN | 26 | 0.673 | 4 | −0.029 | 0.077 | 2 | 0.000 | 0.538 | 4 | −0.143 | 0.494 | 2 | 0.065 | 0.917 | 13 | −0.007 | 0.619 | 7 | 0.130 | |
| GLE | 10 | 0.709 | 6 | −0.026 | 0.595 | 6 | 0.084 | 0.464 | 3 | 0.608* | 0.636 | 3 | 0.286 | 0.933 | 11 | 0.250* | 0.786 | 7 | −0.040 | |
| GRU | 5 | 0.581 | 5 | 0.174 | 0.432 | 7 | −0.070 | 0.289 | 3 | −0.154 | 0.525 | 3 | −0.246 | 0.909 | 17 | 0.154* | 0.613 | 13 | −0.045 | |
| KEN | 15 | 0.638 | 6 | 0.035 | 0.535 | 8 | −0.007 | 0.535 | 4 | −0.078 | 0.506 | 2 | −0.064 | 0.856 | 15 | 0.236* | 0.719 | 12 | 0.144 | |
| LOC | 5 | 0.492 | 3 | 0.154 | 0.436 | 5 | 0.043 | 0.485 | 4 | 0.141 | 0.500 | 2 | −0.333 | 0.860 | 9 | 0.225 | 0.765 | 8 | 0.020 | |
| SAL | 11 | 0.675 | 4 | −0.481 | 0.375 | 3 | −0.067 | 0.550 | 2 | 0.273 | 0.550 | 2 | −0.091 | 0.950 | 7 | 0.158 | 0.725 | 5 | −0.379 | |
| TOR | 11 | 0.743 | 6 | 0.462* | 0.450 | 4 | 0.667* | 0.629 | 4 | 0.364* | 0.667 | 3 | 0.368* | 0.829 | 14 | 0.111 | 0.751 | 11 | 0.468* | |
| TRA | 26 | 0.441 | 3 | 0.175 | 0.255 | 3 | −0.071 | 0.473 | 4 | 0.231 | 0.527 | 2 | 0.310 | 0.782 | 9 | −0.047 | 0.845 | 10 | 0.032 | |
| 2001 | ZO1 | 16 | 0.680 | 4 | 0.095 | 0.489 | 7 | 0.056 | 0.425 | 4 | −0.205 | 0.516 | 3 | −0.142 | 0.877 | 18 | 0.064 | 0.607 | 12 | 0.177* |
| ZO2 | 33 | 0.657 | 5 | 0.060 | 0.429 | 6 | −0.093 | 0.504 | 4 | 0.008 | 0.506 | 2 | −0.019 | 0.880 | 19 | 0.219* | 0.795 | 13 | 0.136 | |
| ZO3 | 24 | 0.694 | 5 | −0.002 | 0.414 | 7 | 0.056 | 0.438 | 5 | 0.106 | 0.510 | 3 | 0.233 | 0.877 | 21 | 0.133* | 0.689 | 14 | 0.148* | |
| ZO5 | 46 | 0.512 | 5 | 0.186 | 0.395 | 6 | −0.055 | 0.408 | 4 | −0.124 | 0.511 | 2 | 0.021 | 0.876 | 13 | 0.057 | 0.743 | 13 | 0.159 | |
| ZO5 | 34 | 0.657 | 6 | −0.015 | 0.524 | 9 | 0.016 | 0.405 | 4 | 0.028 | 0.536 | 4 | −0.188 | 0.891 | 18 | 0.116* | 0.655 | 9 | 0.306* | |
| ZO6 | 39 | 0.792 | 8 | −0.026 | 0.660 | 8 | 0.338* | 0.385 | 5 | −0.135 | 0.696 | 5 | 0.281 | 0.815 | 9 | 0.079 | 0.744 | 12 | 0.076 | |
| 2002 | ALD | 21 | 0.607 | 5 | −0.301 | 0.332 | 5 | −0.110 | 0.329 | 4 | −0.120 | 0.507 | 3 | −0.245 | 0.886 | 13 | 0.050 | 0.752 | 11 | 0.113 |
| AUC | 16 | 0.547 | 5 | −0.023 | 0.385 | 6 | 0.004 | 0.403 | 4 | 0.059 | 0.506 | 3 | −0.004 | 0.884 | 18 | 0.181* | 0.558 | 13 | 0.134* | |
| BEC | 22 | 0.591 | 5 | −0.073 | 0.491 | 8 | 0.055 | 0.325 | 4 | 0.099 | 0.530 | 4 | 0.080 | 0.890 | 18 | 0.150* | 0.720 | 15 | 0.052 | |
| BLA | 23 | 0.574 | 5 | −0.054 | 0.429 | 6 | 0.080 | 0.396 | 4 | 0.069 | 0.508 | 2 | 0.223 | 0.880 | 18 | 0.222* | 0.712 | 12 | 0.002 | |
| FLE | 22 | 0.603 | 4 | −0.245 | 0.314 | 5 | −0.113 | 0.388 | 4 | 0.098 | 0.538 | 3 | 0.071 | 0.846 | 13 | 0.232* | 0.650 | 9 | 0.077 | |
| MOR | 24 | 0.740 | 5 | 0.070 | 0.387 | 4 | 0.631* | 0.335 | 4 | −0.118 | 0.505 | 2 | −0.585 | 0.892 | 14 | 0.019 | 0.724 | 7 | 0.447* | |
| NWB | 21 | 0.654 | 4 | −0.113 | 0.576 | 7 | −0.026 | 0.404 | 5 | 0.550* | 0.530 | 3 | −0.114 | 0.935 | 20 | −0.069 | 0.588 | 9 | 0.072 | |
| PEN | 24 | 0.576 | 5 | −0.026 | 0.255 | 6 | −0.068 | 0.449 | 4 | 0.494* | 0.459 | 2 | −0.090 | 0.890 | 13 | 0.029 | 0.735 | 11 | 0.072 | |
| RWB | 29 | 0.598 | 4 | −0.018 | 0.468 | 5 | −0.167 | 0.344 | 5 | 0.241 | 0.488 | 2 | 0.109 | 0.921 | 15 | 0.112 | 0.629 | 12 | 0.240* | |
| RHY | 19 | 0.546 | 5 | 0.176 | 0.429 | 5 | −0.049 | 0.429 | 5 | 0.184 | 0.517 | 3 | 0.420* | 0.916 | 18 | 0.072 | 0.689 | 11 | 0.275* | |
| SAN | 23 | 0.699 | 5 | −0.120 | 0.477 | 6 | −0.002 | 0.439 | 4 | 0.108 | 0.551 | 4 | 0.133 | 0.863 | 18 | −0.008 | 0.649 | 9 | 0.397* | |
| SEA | 20 | 0.645 | 5 | −0.008 | 0.415 | 7 | −0.032 | 0.191 | 4 | −0.048 | 0.505 | 2 | −0.188 | 0.892 | 15 | −0.065 | 0.710 | 8 | 0.262* | |
| SIL | 23 | 0.603 | 4 | 0.062 | 0.405 | 4 | 0.034 | 0.399 | 5 | 0.129 | 0.555 | 4 | 0.139 | 0.923 | 20 | 0.011 | 0.687 | 9 | 0.177 | |
| SOU | 17 | 0.640 | 5 | −0.171 | 0.540 | 6 | −0.003 | 0.415 | 4 | 0.096 | 0.548 | 3 | 0.088 | 0.929 | 21 | 0.058 | 0.624 | 11 | −0.001 | |
| 2003 | BEC | 41 | 0.531 | 3 | −0.107 | 0.272 | 4 | −0.081 | 0.399 | 3 | −0.032 | 0.518 | 2 | 0.319 | 0.926 | 15 | −0.079 | 0.833 | 12 | 0.294* |
| FLE | 19 | 0.651 | 4 | −0.024 | 0.344 | 7 | −0.107 | 0.538 | 4 | −0.062 | 0.525 | 3 | −0.270 | 0.911 | 17 | 0.059 | 0.765 | 9 | 0.005 | |
| NWB | 60 | 0.472 | 4 | −0.115 | 0.439 | 4 | −0.080 | 0.364 | 4 | −0.157 | 0.515 | 2 | 0.182 | 0.876 | 11 | 0.038 | 0.808 | 11 | 0.219* | |
| SAN | 38 | 0.664 | 4 | −0.004 | 0.572 | 7 | −0.094 | 0.383 | 4 | −0.087 | 0.509 | 2 | −0.146 | 0.862 | 17 | 0.082 | 0.791 | 13 | 0.262* | |
| SIL | 20 | 0.591 | 4 | −0.051 | 0.557 | 8 | 0.196 | 0.355 | 4 | 0.029 | 0.501 | 2 | −0.238 | 0.923 | 20 | −0.009 | 0.769 | 16 | 0.118 | |
| All | 866 | 13 | 11 | 8 | 8 | 61 | 23 | |||||||||||||
| . | . | . | LIST1-001 . | LIST1-003 . | LIST1-007 . | LIST1-010 . | Pl92 . | Pl115 . | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | Sample . | n . | He . | Na . | f . | He . | Na . | f . | He . | Na . | f . | He . | Na . | f . | He . | Na . | f . | He . | Na . | f . |
| 2001 | ARD | 15 | 0.591 | 6 | −0.016 | 0.260 | 6 | −0.077 | 0.404 | 4 | −0.188 | 0.524 | 3 | −0.374 | 0.894 | 18 | 0.150* | 0.714 | 10 | 0.160 |
| AYR | 20 | 0.625 | 4 | −0.280 | 0.625 | 4 | −0.280 | 0.200 | 2 | 0.000 | 0.550 | 2 | 0.273 | 0.975 | 6 | 0.590* | 0.375 | 3 | −0.067 | |
| CCH | 11 | 0.723 | 4 | −0.132 | 0.405 | 4 | 0.326 | 0.391 | 3 | −0.163 | 0.518 | 2 | −0.228 | 0.959 | 12 | 0.336* | 0.523 | 5 | −0.217 | |
| CSW | 12 | 0.695 | 4 | −0.055 | 0.576 | 6 | 0.306 | 0.500 | 5 | 0.200 | 0.510 | 2 | 0.738* | 0.876 | 10 | 0.239* | 0.643 | 10 | 0.170 | |
| FIR | 25 | 0.617 | 5 | 0.027 | 0.583 | 5 | 0.314 | 0.372 | 4 | 0.463 | 0.633 | 3 | 0.684* | 0.917 | 8 | 0.515* | 0.594 | 6 | 0.495* | |
| GAN | 26 | 0.673 | 4 | −0.029 | 0.077 | 2 | 0.000 | 0.538 | 4 | −0.143 | 0.494 | 2 | 0.065 | 0.917 | 13 | −0.007 | 0.619 | 7 | 0.130 | |
| GLE | 10 | 0.709 | 6 | −0.026 | 0.595 | 6 | 0.084 | 0.464 | 3 | 0.608* | 0.636 | 3 | 0.286 | 0.933 | 11 | 0.250* | 0.786 | 7 | −0.040 | |
| GRU | 5 | 0.581 | 5 | 0.174 | 0.432 | 7 | −0.070 | 0.289 | 3 | −0.154 | 0.525 | 3 | −0.246 | 0.909 | 17 | 0.154* | 0.613 | 13 | −0.045 | |
| KEN | 15 | 0.638 | 6 | 0.035 | 0.535 | 8 | −0.007 | 0.535 | 4 | −0.078 | 0.506 | 2 | −0.064 | 0.856 | 15 | 0.236* | 0.719 | 12 | 0.144 | |
| LOC | 5 | 0.492 | 3 | 0.154 | 0.436 | 5 | 0.043 | 0.485 | 4 | 0.141 | 0.500 | 2 | −0.333 | 0.860 | 9 | 0.225 | 0.765 | 8 | 0.020 | |
| SAL | 11 | 0.675 | 4 | −0.481 | 0.375 | 3 | −0.067 | 0.550 | 2 | 0.273 | 0.550 | 2 | −0.091 | 0.950 | 7 | 0.158 | 0.725 | 5 | −0.379 | |
| TOR | 11 | 0.743 | 6 | 0.462* | 0.450 | 4 | 0.667* | 0.629 | 4 | 0.364* | 0.667 | 3 | 0.368* | 0.829 | 14 | 0.111 | 0.751 | 11 | 0.468* | |
| TRA | 26 | 0.441 | 3 | 0.175 | 0.255 | 3 | −0.071 | 0.473 | 4 | 0.231 | 0.527 | 2 | 0.310 | 0.782 | 9 | −0.047 | 0.845 | 10 | 0.032 | |
| 2001 | ZO1 | 16 | 0.680 | 4 | 0.095 | 0.489 | 7 | 0.056 | 0.425 | 4 | −0.205 | 0.516 | 3 | −0.142 | 0.877 | 18 | 0.064 | 0.607 | 12 | 0.177* |
| ZO2 | 33 | 0.657 | 5 | 0.060 | 0.429 | 6 | −0.093 | 0.504 | 4 | 0.008 | 0.506 | 2 | −0.019 | 0.880 | 19 | 0.219* | 0.795 | 13 | 0.136 | |
| ZO3 | 24 | 0.694 | 5 | −0.002 | 0.414 | 7 | 0.056 | 0.438 | 5 | 0.106 | 0.510 | 3 | 0.233 | 0.877 | 21 | 0.133* | 0.689 | 14 | 0.148* | |
| ZO5 | 46 | 0.512 | 5 | 0.186 | 0.395 | 6 | −0.055 | 0.408 | 4 | −0.124 | 0.511 | 2 | 0.021 | 0.876 | 13 | 0.057 | 0.743 | 13 | 0.159 | |
| ZO5 | 34 | 0.657 | 6 | −0.015 | 0.524 | 9 | 0.016 | 0.405 | 4 | 0.028 | 0.536 | 4 | −0.188 | 0.891 | 18 | 0.116* | 0.655 | 9 | 0.306* | |
| ZO6 | 39 | 0.792 | 8 | −0.026 | 0.660 | 8 | 0.338* | 0.385 | 5 | −0.135 | 0.696 | 5 | 0.281 | 0.815 | 9 | 0.079 | 0.744 | 12 | 0.076 | |
| 2002 | ALD | 21 | 0.607 | 5 | −0.301 | 0.332 | 5 | −0.110 | 0.329 | 4 | −0.120 | 0.507 | 3 | −0.245 | 0.886 | 13 | 0.050 | 0.752 | 11 | 0.113 |
| AUC | 16 | 0.547 | 5 | −0.023 | 0.385 | 6 | 0.004 | 0.403 | 4 | 0.059 | 0.506 | 3 | −0.004 | 0.884 | 18 | 0.181* | 0.558 | 13 | 0.134* | |
| BEC | 22 | 0.591 | 5 | −0.073 | 0.491 | 8 | 0.055 | 0.325 | 4 | 0.099 | 0.530 | 4 | 0.080 | 0.890 | 18 | 0.150* | 0.720 | 15 | 0.052 | |
| BLA | 23 | 0.574 | 5 | −0.054 | 0.429 | 6 | 0.080 | 0.396 | 4 | 0.069 | 0.508 | 2 | 0.223 | 0.880 | 18 | 0.222* | 0.712 | 12 | 0.002 | |
| FLE | 22 | 0.603 | 4 | −0.245 | 0.314 | 5 | −0.113 | 0.388 | 4 | 0.098 | 0.538 | 3 | 0.071 | 0.846 | 13 | 0.232* | 0.650 | 9 | 0.077 | |
| MOR | 24 | 0.740 | 5 | 0.070 | 0.387 | 4 | 0.631* | 0.335 | 4 | −0.118 | 0.505 | 2 | −0.585 | 0.892 | 14 | 0.019 | 0.724 | 7 | 0.447* | |
| NWB | 21 | 0.654 | 4 | −0.113 | 0.576 | 7 | −0.026 | 0.404 | 5 | 0.550* | 0.530 | 3 | −0.114 | 0.935 | 20 | −0.069 | 0.588 | 9 | 0.072 | |
| PEN | 24 | 0.576 | 5 | −0.026 | 0.255 | 6 | −0.068 | 0.449 | 4 | 0.494* | 0.459 | 2 | −0.090 | 0.890 | 13 | 0.029 | 0.735 | 11 | 0.072 | |
| RWB | 29 | 0.598 | 4 | −0.018 | 0.468 | 5 | −0.167 | 0.344 | 5 | 0.241 | 0.488 | 2 | 0.109 | 0.921 | 15 | 0.112 | 0.629 | 12 | 0.240* | |
| RHY | 19 | 0.546 | 5 | 0.176 | 0.429 | 5 | −0.049 | 0.429 | 5 | 0.184 | 0.517 | 3 | 0.420* | 0.916 | 18 | 0.072 | 0.689 | 11 | 0.275* | |
| SAN | 23 | 0.699 | 5 | −0.120 | 0.477 | 6 | −0.002 | 0.439 | 4 | 0.108 | 0.551 | 4 | 0.133 | 0.863 | 18 | −0.008 | 0.649 | 9 | 0.397* | |
| SEA | 20 | 0.645 | 5 | −0.008 | 0.415 | 7 | −0.032 | 0.191 | 4 | −0.048 | 0.505 | 2 | −0.188 | 0.892 | 15 | −0.065 | 0.710 | 8 | 0.262* | |
| SIL | 23 | 0.603 | 4 | 0.062 | 0.405 | 4 | 0.034 | 0.399 | 5 | 0.129 | 0.555 | 4 | 0.139 | 0.923 | 20 | 0.011 | 0.687 | 9 | 0.177 | |
| SOU | 17 | 0.640 | 5 | −0.171 | 0.540 | 6 | −0.003 | 0.415 | 4 | 0.096 | 0.548 | 3 | 0.088 | 0.929 | 21 | 0.058 | 0.624 | 11 | −0.001 | |
| 2003 | BEC | 41 | 0.531 | 3 | −0.107 | 0.272 | 4 | −0.081 | 0.399 | 3 | −0.032 | 0.518 | 2 | 0.319 | 0.926 | 15 | −0.079 | 0.833 | 12 | 0.294* |
| FLE | 19 | 0.651 | 4 | −0.024 | 0.344 | 7 | −0.107 | 0.538 | 4 | −0.062 | 0.525 | 3 | −0.270 | 0.911 | 17 | 0.059 | 0.765 | 9 | 0.005 | |
| NWB | 60 | 0.472 | 4 | −0.115 | 0.439 | 4 | −0.080 | 0.364 | 4 | −0.157 | 0.515 | 2 | 0.182 | 0.876 | 11 | 0.038 | 0.808 | 11 | 0.219* | |
| SAN | 38 | 0.664 | 4 | −0.004 | 0.572 | 7 | −0.094 | 0.383 | 4 | −0.087 | 0.509 | 2 | −0.146 | 0.862 | 17 | 0.082 | 0.791 | 13 | 0.262* | |
| SIL | 20 | 0.591 | 4 | −0.051 | 0.557 | 8 | 0.196 | 0.355 | 4 | 0.029 | 0.501 | 2 | −0.238 | 0.923 | 20 | −0.009 | 0.769 | 16 | 0.118 | |
| All | 866 | 13 | 11 | 8 | 8 | 61 | 23 | |||||||||||||
n, sample size; He, expected heterozygosity; Na, number of alleles; f, inbreeding coefficient (Wright, 1951). ZO, Zone (Figure 1).
* Significant (p < 0.05) deviations from expected Hardy–Weinberg conditions, with tests that remain significant after Bonferroni correction highlighted emboldened.