-
PDF
- Split View
-
Views
-
Cite
Cite
Christophe Pampoulie, Sigurlaug Skirnisdottir, Sigurbjorg Hauksdottir, Kristinn Olafsson, Hrafnkell Eiríksson, Valérie Chosson, Gudmundur O. Hreggvidsson, Gudmundur H. Gunnarsson, Sigridur Hjorleifsdottir, A pilot genetic study reveals the absence of spatial genetic structure in Norway lobster (Nephrops norvegicus) on fishing grounds in Icelandic waters, ICES Journal of Marine Science, Volume 68, Issue 1, January 2011, Pages 20–25, https://doi.org/10.1093/icesjms/fsq165
Close - Share Icon Share
Abstract
Stock structure of Norway lobster off southern Iceland was investigated using 12 microsatellite loci. No genetic method detected significant genetic differentiation among the locations sampled, even among Icelandic samples and an out-group from Scotland. Testing the power of resolution of microsatellite loci, the loci and sample sizes used were sufficient to detect significant genetic differentiation with confidence. The lack of genetic structure is discussed in terms of the level of gene flow, recent isolation of populations, and the statistical power of the experimental design.Pampoulie, C., Skirnisdottir, S., Hauksdottir, S., Olafsson, K., Eiríksson, H., Chosson, V., Hreggvidsson, G. O., Gunnarsson, G. H., and Hjorleifsdottir, S. 2011. A pilot genetic study reveals the absence of spatial genetic structure in Norway lobster (Nephrops norvegicus) on fishing grounds in Icelandic waters. – ICES Journal of Marine Science, 68: 20–25.
Introduction
The identification of stock structure has been recognized widely as a prerequisite for sustainable management of marine fisheries (Reiss et al., 2009), and different methods have been tested for this purpose recently (see Cadrin et al., 2005, for a review). One of the currently most popular approaches is the use of highly variable genetic markers such as microsatellite loci. Although marine organisms have been thought to constitute homogenous entities, microsatellite studies have revealed the presence of subtle genetic structure at small and large geographic scales (Knutsen et al., 2003; Nielsen et al., 2003; Jørgensen et al., 2005; D'Amato, 2006). Moreover, the recent application of microsatellite loci to stock identification has revealed a number of cases with notable discrepancy between biological and fisheries management units (Lundy et al., 1999; Hoarau et al., 2002; Pampoulie et al., 2006; Charrier et al., 2007; Was et al., 2008; see Reiss et al., 2009, for a review). Such results are likely to have a major effect on fisheries management. Microsatellite studies have hence been playing a more significant role in stock discrimination of marine resources and on their potential connectivity, through indirect assessment of gene flow (Viñas et al., 2004; Jørgensen et al., 2005; Fritsch et al., 2007; Was et al., 2008; Pampoulie et al., 2009; Stefánsson et al., 2009a).
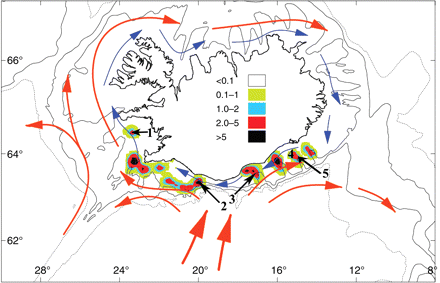
Despite the many species studied using microsatellite loci, some important commercial species remain to be considered. This is the case for Norway lobster, Nephrops norvegicus, which is distributed widely in the Northeast Atlantic from Iceland to Mauritania, and in the Mediterranean Sea (Figueiredo and Thomas, 1967). Adult N. norvegicus are fairly sedentary, usually occupying burrows at depths of 10–1200 m and, according to tagging data, with movements rarely exceeding 100 m from their burrow (Chapman and Rice, 1971). In contrast, movements among populations can take place during the 4–8-week planktonic larval phase, i.e. through passive dispersal of larvae by oceanic currents. In Icelandic waters, Norway lobster occupy burrows at depths of 100–300 m and are exclusively in the south (Figure 1; Eiríksson, 1999). Within that area, the continental shelf is characterized by a series of straight troughs up to 250 m deep inhabited by various densities of Norway lobster at depths of 150–250 m, with the intervening banks (100–120 m) inappropriate for the species owing to the nature of bottom substrata. However, in some areas off the southwest coast, dense bank populations of Norway lobster are found at 130–180 m (Eiríksson, 1999). Populations contained within troughs and banks have been suggested to be self-contained unit stocks owing to the characteristics of the seafloor and the biological cycle of the species (Eiríksson, 1999). Therefore, present fishing units correspond to potentially isolated areas (populations within troughs), which are also considered as breeding units.
Sampling locations for Norway lobster off Iceland. The numbers refer to the samples depicted in Table 1, and fishing areas (which are also breeding areas) are indicated by colours. The scale indicates the density of catches from 2005 to 2009 (t nautical mile−2), and the arrows indicate the main currents around Iceland: red, branch of the North Atlantic Current; blue, coastal current.
The strong spatial variability of catch per unit effort (cpue) and mean size between isolated areas (e.g. the most eastbound vs. the most westbound; see Eiríksson, 1999) fuelled interest in testing whether population genetic approaches would reveal reproductive isolation between separate fishing units.
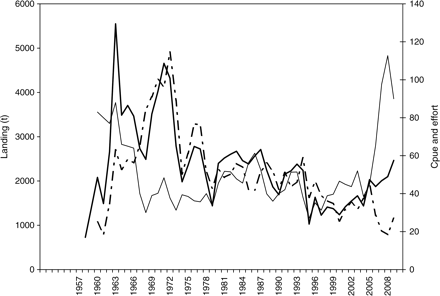
The Icelandic N. norvegicus fishery dates back to the late 1950s, with landings increasing rapidly to a historical high of 5550 t in 1963 (Figure 2). However, heavy fishing caused a decline in the catches and just 2500 t were taken in 1968, with cpue falling by 65%. Then, with improved recruitment and greater fishing effort, annual landings rose again to >4000 t in 1970–1972. As a consequence of the declining cpue (Eiríksson, 1968, 1970a, b), a total allowable catch was recommended for N. norvegicus by the Marine Research Institute, Reykjavík, and put into practice in 1973. In 1990, a system of individual transferable quotas was established for Icelandic fisheries. The history of the fishery from the mid-1970s to the early 2000s can be generally described by fluctuating landings, from ∼2000–2500 t in the late 1970s and the 1980s, with cpue increasing in the 1980s and the early 1990s, and a decline to 1500 t or less by the mid-1990s accompanied by a historically low cpue, associated with a record low recruitment to the fishable stock. However, recruitment and landings have increased gradually in recent years, accompanied by a notable decrease in effort and a historically high cpue of 90–110 kg h−1 from 2007 to 2009 (Figure 2).
Fisheries data for Norway lobster in Icelandic waters. Bold line, landings (t); line, catch per unit effort (kg h−1); dashed–dotted line, effort (‘000 h).
The aims of the present study were to provide preliminary information on the possible genetic structure of N. norvegicus in Icelandic waters, to assess whether actual fishing units correspond to genetically distinct populations, and to assess whether or not the variability in cpue and mean size between isolated areas could be explained by reproductive isolation of the fishing units.
Material and methods
Sampling areas and protocol
In all, 549 Norway lobster were collected at several locations in an annual Nephrops survey in Icelandic waters in May 2007 (Figure 1). The research vessel was rigged with a conventional Nephrops trawl of 45 m headline and mesh size 80 mm. An additional sample of 94 animals collected at North Clyde, west of the Isle of Cumbrae, Scotland, was analysed genetically as an out-group.
Genetic samples were collected from tails preserved in 99% ethanol. Samples were genotyped at 12 microsatellite loci, namely B11, C12, E4, and G2 (Streiff et al., 2001) and PLH4, PLH5, PLH12, PLH15, PLH21, PLH33, PLH35, and PLH46 (Skirnisdottir et al., 2010). DNA extraction, PCR, and genotyping were performed as described in Skirnisdottir et al. (2010). The Streiff et al. (2001) loci were amplified with annealing temperatures 58°C (for G2) and 60°C (for B11, C12, and E4), and the PCR conditions were as described in Skirnisdottir et al. (2010).
Genetic analyses
Genetic diversity was evaluated using allele frequencies, observed (Ho) and unbiased expected heterozygosity (He) calculated in GENEPOP’007 (Rousset, 2008). Deviations from the Hardy–Weinberg expectation (HWE) were tested using the inbreeding coefficient FIS (Weir and Cockerham, 1984) implemented in GENEPOP, and significance was assessed with exact tests. Genetic differentiation was estimated using theta estimates (θ; Weir and Cockerham, 1984) implemented in GENEPOP, and significance was assessed with allelic and genotypic frequency homogeneity tests (5000 permutations). The significance levels were adjusted by a simple Bonferroni correction (Rice, 1989) when multiple tests were applied.
The statistical power of the microsatellite loci was estimated using the program POWSIM (Ryman and Palm, 2006), which assesses the α (type I) error (the probability of rejecting Ho when it is true) and the β (type II) error, which is the probability of rejecting (Ho: genetic homogeneity) when it is false. The program estimates the power of the genetic design performed using information on sample size, number of samples, number of loci, and allele frequencies for any hypothetical degree of true differentiation quantified as FST (Ryman and Palm, 2006). The significance of the tests is assessed with Fisher's exact tests as well as with χ2 tests.
STRUCTURE 2.3.2 (Pritchard et al., 2000) was used to enumerate the potential number of populations within our samples. Owing to the very low genetic differentiation level detected, we used the admixture model with the LOCPRIOR setting, which considers location information. This recently developed method (Hubisz et al., 2009) has been suggested to perform better than the traditional STRUCTURE methods when genetic structure is weak or when the number of loci is low (<20). The model was run with a burn-in period of 300 000 iterations and 600 000 Markov chain Monte Carlo iterations. The potential number of populations (K) varied from 1 to 15 and was tested with five independent analyses for each K.
Results
Biological information retrieved from the samples is listed in Table 1. Genetic diversity assessed as the number of alleles per locus was high, ranging from 8 (C12) to 53 (PLH33; data not shown). The value of He per sample ranged from 0.821 (Sample 4) to 0.859 (Sample 2; Table 2). Genotypic proportions were out of HWE in 2 of 96 exact tests after the Bonferroni correction for multiple tests and were not attributable to any specific loci or samples. Only Sample 4 deviated from HWE (Table 2). The overall genetic estimates did not reveal significant FST (FST = −0.0003, p> 0.05, 95% CI: −0.0009–0.0002) or FIS values (FIS = 0.0232, p> 0.05, 95% CI: 0.0092–0.0360). This genetic pattern was reflected in the pairwise FST comparisons of samples, because none of the comparisons were significant after the Bonferroni correction (Table 3).
Sampling area and information (tow number, coordinates, and depth range), sample size, sex ratio (females vs. males, F:M), and size distribution (mean carapace length in mm, standard deviation s.d., and range) for six samples of N. norvegicus.
| Information . | Sample 1 . | Sample 2 . | Sample 3 . | Sample 4 . | Sample 5 . | Sample 6 . |
|---|---|---|---|---|---|---|
| Sampling area | Faxaflói | Háfadjúp | Meðallands | Breiðamerkurdjúp | Breiðamerkurdjúp | Scotland |
| Tow number | 56 | 32 | 6 | 18 | 12 | Not known |
| Coordinates | 64°24.75′N 23°16.91′W | 63°17.25′N 19°59.81′W | 63°32.95′N 17°40.00′W | 63°46.08′N 15°51.56′W | 63°40.37′N 15°48.36′W | 55°44.89′N 04°59.37′W |
| Depth range (m) | 156–166 | 219–238 | 141–143 | 199–210 | 193–196 | Not known |
| Sample size | 185 | 92 | 75 | 97 | 100 | 94 |
| F:M | 4:181 | 7:85 | 0:75 | 3:94 | 0:100 | 15:30a |
| Length (mm) | ||||||
| Mean | 54.15 | 41.14 | 57.62 | 45.96 | 49.13 | 36.35 |
| s.d. | 8.75 | 8.20 | 7.87 | 7.60 | 7.08 | 5.56 |
| Range | 32–73 | 23–62 | 36–80 | 29–64 | 28–73 | 25– 51 |
| Information . | Sample 1 . | Sample 2 . | Sample 3 . | Sample 4 . | Sample 5 . | Sample 6 . |
|---|---|---|---|---|---|---|
| Sampling area | Faxaflói | Háfadjúp | Meðallands | Breiðamerkurdjúp | Breiðamerkurdjúp | Scotland |
| Tow number | 56 | 32 | 6 | 18 | 12 | Not known |
| Coordinates | 64°24.75′N 23°16.91′W | 63°17.25′N 19°59.81′W | 63°32.95′N 17°40.00′W | 63°46.08′N 15°51.56′W | 63°40.37′N 15°48.36′W | 55°44.89′N 04°59.37′W |
| Depth range (m) | 156–166 | 219–238 | 141–143 | 199–210 | 193–196 | Not known |
| Sample size | 185 | 92 | 75 | 97 | 100 | 94 |
| F:M | 4:181 | 7:85 | 0:75 | 3:94 | 0:100 | 15:30a |
| Length (mm) | ||||||
| Mean | 54.15 | 41.14 | 57.62 | 45.96 | 49.13 | 36.35 |
| s.d. | 8.75 | 8.20 | 7.87 | 7.60 | 7.08 | 5.56 |
| Range | 32–73 | 23–62 | 36–80 | 29–64 | 28–73 | 25– 51 |
aThe sex ratio was based on 45 animals.
Sampling area and information (tow number, coordinates, and depth range), sample size, sex ratio (females vs. males, F:M), and size distribution (mean carapace length in mm, standard deviation s.d., and range) for six samples of N. norvegicus.
| Information . | Sample 1 . | Sample 2 . | Sample 3 . | Sample 4 . | Sample 5 . | Sample 6 . |
|---|---|---|---|---|---|---|
| Sampling area | Faxaflói | Háfadjúp | Meðallands | Breiðamerkurdjúp | Breiðamerkurdjúp | Scotland |
| Tow number | 56 | 32 | 6 | 18 | 12 | Not known |
| Coordinates | 64°24.75′N 23°16.91′W | 63°17.25′N 19°59.81′W | 63°32.95′N 17°40.00′W | 63°46.08′N 15°51.56′W | 63°40.37′N 15°48.36′W | 55°44.89′N 04°59.37′W |
| Depth range (m) | 156–166 | 219–238 | 141–143 | 199–210 | 193–196 | Not known |
| Sample size | 185 | 92 | 75 | 97 | 100 | 94 |
| F:M | 4:181 | 7:85 | 0:75 | 3:94 | 0:100 | 15:30a |
| Length (mm) | ||||||
| Mean | 54.15 | 41.14 | 57.62 | 45.96 | 49.13 | 36.35 |
| s.d. | 8.75 | 8.20 | 7.87 | 7.60 | 7.08 | 5.56 |
| Range | 32–73 | 23–62 | 36–80 | 29–64 | 28–73 | 25– 51 |
| Information . | Sample 1 . | Sample 2 . | Sample 3 . | Sample 4 . | Sample 5 . | Sample 6 . |
|---|---|---|---|---|---|---|
| Sampling area | Faxaflói | Háfadjúp | Meðallands | Breiðamerkurdjúp | Breiðamerkurdjúp | Scotland |
| Tow number | 56 | 32 | 6 | 18 | 12 | Not known |
| Coordinates | 64°24.75′N 23°16.91′W | 63°17.25′N 19°59.81′W | 63°32.95′N 17°40.00′W | 63°46.08′N 15°51.56′W | 63°40.37′N 15°48.36′W | 55°44.89′N 04°59.37′W |
| Depth range (m) | 156–166 | 219–238 | 141–143 | 199–210 | 193–196 | Not known |
| Sample size | 185 | 92 | 75 | 97 | 100 | 94 |
| F:M | 4:181 | 7:85 | 0:75 | 3:94 | 0:100 | 15:30a |
| Length (mm) | ||||||
| Mean | 54.15 | 41.14 | 57.62 | 45.96 | 49.13 | 36.35 |
| s.d. | 8.75 | 8.20 | 7.87 | 7.60 | 7.08 | 5.56 |
| Range | 32–73 | 23–62 | 36–80 | 29–64 | 28–73 | 25– 51 |
aThe sex ratio was based on 45 animals.
Expected heterozygosity (He) and deviation from HWE (FIS) for 12 microsatellite loci in six samples of N. norvegicus.
| . | Sample 1 . | Sample 2 . | Sample 3 . | Sample 4 . | Sample 5 . | Sample 6 . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus . | He . | FIS . | He . | FIS . | He . | FIS . | He . | FIS . | He . | FIS . | He . | FIS . |
| B11 | 0.822 | 0.032 | 0.841 | 0.043 | 0.847 | 0.027 | 0.833 | −0.014 | 0.835 | 0.030 | 0.837 | −0.002 |
| C12 | 0.464 | 0.081 | 0.507 | 0.035 | 0.542 | −0.008 | 0.481 | 0.185 | 0.442 | 0.028 | 0.459 | −0.005 |
| E4 | 0.876 | 0.038 | 0.887 | −0.030 | 0.882 | 0.002 | 0.877 | 0.094 | 0.895 | 0.040 | 0.876 | 0.065 |
| G2 | 0.966 | −0.007 | 0.971 | 0.037 | 0.971 | 0.011 | 0.966 | 0.040 | 0.969 | 0.019 | 0.968 | 0.099 |
| PLH4 | 0.797 | 0.019 | 0.787 | −0.008 | 0.767 | 0.062 | 0.772 | 0.052 | 0.766 | −0.045 | 0.772 | 0.022 |
| PLH5 | 0.845 | 0.010 | 0.818 | −0.037 | 0.857 | −0.011 | 0.821 | −0.093 | 0.829 | −0.001 | 0.866 | −0.016 |
| PLH12 | 0.842 | 0.076 | 0.847 | 0.014 | 0.824 | 0.013 | 0.836 | 0.018 | 0.826 | 0.022 | 0.836 | −0.047 |
| PLH15 | 0.943 | −0.014 | 0.948 | −0.020 | 0.940 | −0.021 | 0.939 | −0.011 | 0.944 | −0.015 | 0.949 | 0.003 |
| PLH31 | 0.875 | 0.068 | 0.886 | 0.033 | 0.867 | 0.018 | 0.868 | 0.054 | 0.873 | 0.017 | 0.895 | −0.054 |
| PLH33 | 0.963 | 0.029 | 0.958 | 0.051 | 0.957 | 0.096 | 0.959 | 0.164a | 0.963 | 0.036 | 0.954 | 0.076 |
| PLH35 | 0.964 | 0.010 | 0.966 | 0.083 | 0.969 | 0.027 | 0.967 | 0.021 | 0.964 | 0.098a | 0.959 | 0.025 |
| PLH46 | 0.942 | −0.019 | 0.951 | 0.031 | 0.938 | −0.008 | 0.949 | 0.001 | 0.946 | 0.031 | 0.928 | −0.017 |
| Overall loci | 0.856 | 0.024 | 0.859 | 0.020 | 0.858 | 0.018 | 0.851 | 0.038a | 0.850 | 0.023 | 0.854 | 0.014 |
| . | Sample 1 . | Sample 2 . | Sample 3 . | Sample 4 . | Sample 5 . | Sample 6 . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus . | He . | FIS . | He . | FIS . | He . | FIS . | He . | FIS . | He . | FIS . | He . | FIS . |
| B11 | 0.822 | 0.032 | 0.841 | 0.043 | 0.847 | 0.027 | 0.833 | −0.014 | 0.835 | 0.030 | 0.837 | −0.002 |
| C12 | 0.464 | 0.081 | 0.507 | 0.035 | 0.542 | −0.008 | 0.481 | 0.185 | 0.442 | 0.028 | 0.459 | −0.005 |
| E4 | 0.876 | 0.038 | 0.887 | −0.030 | 0.882 | 0.002 | 0.877 | 0.094 | 0.895 | 0.040 | 0.876 | 0.065 |
| G2 | 0.966 | −0.007 | 0.971 | 0.037 | 0.971 | 0.011 | 0.966 | 0.040 | 0.969 | 0.019 | 0.968 | 0.099 |
| PLH4 | 0.797 | 0.019 | 0.787 | −0.008 | 0.767 | 0.062 | 0.772 | 0.052 | 0.766 | −0.045 | 0.772 | 0.022 |
| PLH5 | 0.845 | 0.010 | 0.818 | −0.037 | 0.857 | −0.011 | 0.821 | −0.093 | 0.829 | −0.001 | 0.866 | −0.016 |
| PLH12 | 0.842 | 0.076 | 0.847 | 0.014 | 0.824 | 0.013 | 0.836 | 0.018 | 0.826 | 0.022 | 0.836 | −0.047 |
| PLH15 | 0.943 | −0.014 | 0.948 | −0.020 | 0.940 | −0.021 | 0.939 | −0.011 | 0.944 | −0.015 | 0.949 | 0.003 |
| PLH31 | 0.875 | 0.068 | 0.886 | 0.033 | 0.867 | 0.018 | 0.868 | 0.054 | 0.873 | 0.017 | 0.895 | −0.054 |
| PLH33 | 0.963 | 0.029 | 0.958 | 0.051 | 0.957 | 0.096 | 0.959 | 0.164a | 0.963 | 0.036 | 0.954 | 0.076 |
| PLH35 | 0.964 | 0.010 | 0.966 | 0.083 | 0.969 | 0.027 | 0.967 | 0.021 | 0.964 | 0.098a | 0.959 | 0.025 |
| PLH46 | 0.942 | −0.019 | 0.951 | 0.031 | 0.938 | −0.008 | 0.949 | 0.001 | 0.946 | 0.031 | 0.928 | −0.017 |
| Overall loci | 0.856 | 0.024 | 0.859 | 0.020 | 0.858 | 0.018 | 0.851 | 0.038a | 0.850 | 0.023 | 0.854 | 0.014 |
Emboldened values differ significantly from zero (Fisher's exact test, p < 0.05).
aValues remaining significant after Bonferroni correction (α = 0.05/72 = 0.00069).
Expected heterozygosity (He) and deviation from HWE (FIS) for 12 microsatellite loci in six samples of N. norvegicus.
| . | Sample 1 . | Sample 2 . | Sample 3 . | Sample 4 . | Sample 5 . | Sample 6 . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus . | He . | FIS . | He . | FIS . | He . | FIS . | He . | FIS . | He . | FIS . | He . | FIS . |
| B11 | 0.822 | 0.032 | 0.841 | 0.043 | 0.847 | 0.027 | 0.833 | −0.014 | 0.835 | 0.030 | 0.837 | −0.002 |
| C12 | 0.464 | 0.081 | 0.507 | 0.035 | 0.542 | −0.008 | 0.481 | 0.185 | 0.442 | 0.028 | 0.459 | −0.005 |
| E4 | 0.876 | 0.038 | 0.887 | −0.030 | 0.882 | 0.002 | 0.877 | 0.094 | 0.895 | 0.040 | 0.876 | 0.065 |
| G2 | 0.966 | −0.007 | 0.971 | 0.037 | 0.971 | 0.011 | 0.966 | 0.040 | 0.969 | 0.019 | 0.968 | 0.099 |
| PLH4 | 0.797 | 0.019 | 0.787 | −0.008 | 0.767 | 0.062 | 0.772 | 0.052 | 0.766 | −0.045 | 0.772 | 0.022 |
| PLH5 | 0.845 | 0.010 | 0.818 | −0.037 | 0.857 | −0.011 | 0.821 | −0.093 | 0.829 | −0.001 | 0.866 | −0.016 |
| PLH12 | 0.842 | 0.076 | 0.847 | 0.014 | 0.824 | 0.013 | 0.836 | 0.018 | 0.826 | 0.022 | 0.836 | −0.047 |
| PLH15 | 0.943 | −0.014 | 0.948 | −0.020 | 0.940 | −0.021 | 0.939 | −0.011 | 0.944 | −0.015 | 0.949 | 0.003 |
| PLH31 | 0.875 | 0.068 | 0.886 | 0.033 | 0.867 | 0.018 | 0.868 | 0.054 | 0.873 | 0.017 | 0.895 | −0.054 |
| PLH33 | 0.963 | 0.029 | 0.958 | 0.051 | 0.957 | 0.096 | 0.959 | 0.164a | 0.963 | 0.036 | 0.954 | 0.076 |
| PLH35 | 0.964 | 0.010 | 0.966 | 0.083 | 0.969 | 0.027 | 0.967 | 0.021 | 0.964 | 0.098a | 0.959 | 0.025 |
| PLH46 | 0.942 | −0.019 | 0.951 | 0.031 | 0.938 | −0.008 | 0.949 | 0.001 | 0.946 | 0.031 | 0.928 | −0.017 |
| Overall loci | 0.856 | 0.024 | 0.859 | 0.020 | 0.858 | 0.018 | 0.851 | 0.038a | 0.850 | 0.023 | 0.854 | 0.014 |
| . | Sample 1 . | Sample 2 . | Sample 3 . | Sample 4 . | Sample 5 . | Sample 6 . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus . | He . | FIS . | He . | FIS . | He . | FIS . | He . | FIS . | He . | FIS . | He . | FIS . |
| B11 | 0.822 | 0.032 | 0.841 | 0.043 | 0.847 | 0.027 | 0.833 | −0.014 | 0.835 | 0.030 | 0.837 | −0.002 |
| C12 | 0.464 | 0.081 | 0.507 | 0.035 | 0.542 | −0.008 | 0.481 | 0.185 | 0.442 | 0.028 | 0.459 | −0.005 |
| E4 | 0.876 | 0.038 | 0.887 | −0.030 | 0.882 | 0.002 | 0.877 | 0.094 | 0.895 | 0.040 | 0.876 | 0.065 |
| G2 | 0.966 | −0.007 | 0.971 | 0.037 | 0.971 | 0.011 | 0.966 | 0.040 | 0.969 | 0.019 | 0.968 | 0.099 |
| PLH4 | 0.797 | 0.019 | 0.787 | −0.008 | 0.767 | 0.062 | 0.772 | 0.052 | 0.766 | −0.045 | 0.772 | 0.022 |
| PLH5 | 0.845 | 0.010 | 0.818 | −0.037 | 0.857 | −0.011 | 0.821 | −0.093 | 0.829 | −0.001 | 0.866 | −0.016 |
| PLH12 | 0.842 | 0.076 | 0.847 | 0.014 | 0.824 | 0.013 | 0.836 | 0.018 | 0.826 | 0.022 | 0.836 | −0.047 |
| PLH15 | 0.943 | −0.014 | 0.948 | −0.020 | 0.940 | −0.021 | 0.939 | −0.011 | 0.944 | −0.015 | 0.949 | 0.003 |
| PLH31 | 0.875 | 0.068 | 0.886 | 0.033 | 0.867 | 0.018 | 0.868 | 0.054 | 0.873 | 0.017 | 0.895 | −0.054 |
| PLH33 | 0.963 | 0.029 | 0.958 | 0.051 | 0.957 | 0.096 | 0.959 | 0.164a | 0.963 | 0.036 | 0.954 | 0.076 |
| PLH35 | 0.964 | 0.010 | 0.966 | 0.083 | 0.969 | 0.027 | 0.967 | 0.021 | 0.964 | 0.098a | 0.959 | 0.025 |
| PLH46 | 0.942 | −0.019 | 0.951 | 0.031 | 0.938 | −0.008 | 0.949 | 0.001 | 0.946 | 0.031 | 0.928 | −0.017 |
| Overall loci | 0.856 | 0.024 | 0.859 | 0.020 | 0.858 | 0.018 | 0.851 | 0.038a | 0.850 | 0.023 | 0.854 | 0.014 |
Emboldened values differ significantly from zero (Fisher's exact test, p < 0.05).
aValues remaining significant after Bonferroni correction (α = 0.05/72 = 0.00069).
Pairwise FST (above diagonal) and values of p (below diagonal) among six samples of N. norvegicus based on allelic frequencies.
| . | 1 . | 2 . | 3 . | 4 . | 5 . | 6 . |
|---|---|---|---|---|---|---|
| 1 | 0.000 | −0.001 | −0.001 | 0.000 | 0.001 | |
| 2 | 0.156 | 0.001 | −0.002 | 0.000 | 0.001 | |
| 3 | 0.308 | 0.085 | 0.001 | 0.000 | 0.001 | |
| 4 | 0.405 | 0.228 | 0.032 | −0.001 | 0.001 | |
| 5 | 0.437 | 0.442 | 0.507 | 0.668 | 0.001 | |
| 6 | 0.011 | 0.011 | 0.048 | 0.139 | 0.016 |
| . | 1 . | 2 . | 3 . | 4 . | 5 . | 6 . |
|---|---|---|---|---|---|---|
| 1 | 0.000 | −0.001 | −0.001 | 0.000 | 0.001 | |
| 2 | 0.156 | 0.001 | −0.002 | 0.000 | 0.001 | |
| 3 | 0.308 | 0.085 | 0.001 | 0.000 | 0.001 | |
| 4 | 0.405 | 0.228 | 0.032 | −0.001 | 0.001 | |
| 5 | 0.437 | 0.442 | 0.507 | 0.668 | 0.001 | |
| 6 | 0.011 | 0.011 | 0.048 | 0.139 | 0.016 |
Emboldened values differ significantly from zero (Fisher's exact test, p < 0.05), none remained significant after Bonferroni correction (α = 0.05/15 = 0.003).
Pairwise FST (above diagonal) and values of p (below diagonal) among six samples of N. norvegicus based on allelic frequencies.
| . | 1 . | 2 . | 3 . | 4 . | 5 . | 6 . |
|---|---|---|---|---|---|---|
| 1 | 0.000 | −0.001 | −0.001 | 0.000 | 0.001 | |
| 2 | 0.156 | 0.001 | −0.002 | 0.000 | 0.001 | |
| 3 | 0.308 | 0.085 | 0.001 | 0.000 | 0.001 | |
| 4 | 0.405 | 0.228 | 0.032 | −0.001 | 0.001 | |
| 5 | 0.437 | 0.442 | 0.507 | 0.668 | 0.001 | |
| 6 | 0.011 | 0.011 | 0.048 | 0.139 | 0.016 |
| . | 1 . | 2 . | 3 . | 4 . | 5 . | 6 . |
|---|---|---|---|---|---|---|
| 1 | 0.000 | −0.001 | −0.001 | 0.000 | 0.001 | |
| 2 | 0.156 | 0.001 | −0.002 | 0.000 | 0.001 | |
| 3 | 0.308 | 0.085 | 0.001 | 0.000 | 0.001 | |
| 4 | 0.405 | 0.228 | 0.032 | −0.001 | 0.001 | |
| 5 | 0.437 | 0.442 | 0.507 | 0.668 | 0.001 | |
| 6 | 0.011 | 0.011 | 0.048 | 0.139 | 0.016 |
Emboldened values differ significantly from zero (Fisher's exact test, p < 0.05), none remained significant after Bonferroni correction (α = 0.05/15 = 0.003).
The estimate of the statistical α (type I) error, e.g. the probability of rejecting the null hypothesis (Ho; genetic homogeneity) when it is true varied from 0.068 with Fisher's exact tests to 0.097 with χ2 tests (Table 4), much higher than the 5% limit for significance. Moreover, simulation analyses on the power analysis of the microsatellite loci revealed that the combination of microsatellite loci and sample sizes used conferred a statistical power sufficient to detect a very low (FST = 0.0001) level of differentiation (Table 4).
Estimate of the resolution power of the microsatellite loci using POWSIM (Ryman and Palm, 2006).
| Expected FST . | Average FST . | χ2-test . | Fisher's test . | Ne . | Generation (t) . | Runs . |
|---|---|---|---|---|---|---|
| 0.0000 | 0.0000 | 0.097 | 0.068 | 1 000 | 0 | 10 000 |
| 0.0001 | 0.0001 | 0.996 | 0.997 | 1 000 | 2 | 10 000 |
| 0.0010 | 0.0010 | 0.997 | 0.998 | 2 000 | 4 | 10 000 |
| 0.0020 | 0.0020 | 1.000 | 1.000 | 1 000 | 4 | 10 000 |
| 0.0100 | 0.0100 | 1.000 | 1.000 | 1 000 | 20 | 10 000 |
| 0.0198 | 0.0198 | 1.000 | 1.000 | 1 000 | 40 | 10 000 |
| Expected FST . | Average FST . | χ2-test . | Fisher's test . | Ne . | Generation (t) . | Runs . |
|---|---|---|---|---|---|---|
| 0.0000 | 0.0000 | 0.097 | 0.068 | 1 000 | 0 | 10 000 |
| 0.0001 | 0.0001 | 0.996 | 0.997 | 1 000 | 2 | 10 000 |
| 0.0010 | 0.0010 | 0.997 | 0.998 | 2 000 | 4 | 10 000 |
| 0.0020 | 0.0020 | 1.000 | 1.000 | 1 000 | 4 | 10 000 |
| 0.0100 | 0.0100 | 1.000 | 1.000 | 1 000 | 20 | 10 000 |
| 0.0198 | 0.0198 | 1.000 | 1.000 | 1 000 | 40 | 10 000 |
Resolution power is assessed by simulating different expected level of FST according to the effective population size (Ne) and generations (t) and to the Nei (1987) formula FST = 1 − (1 – 1/2Ne)t. The significance, evaluated using Fisher's exact tests and χ2 tests, reflects the power to detect any given level of differentiation (average FST) with the sampling design developed during our study. Values of Ne used during the test are based on estimates calculated from fisheries data. The column headed “Runs” denotes the number of simulations performed. The setting FST = 0 and t= 0 estimates α (type I error, in the absence of genetic drift).
Estimate of the resolution power of the microsatellite loci using POWSIM (Ryman and Palm, 2006).
| Expected FST . | Average FST . | χ2-test . | Fisher's test . | Ne . | Generation (t) . | Runs . |
|---|---|---|---|---|---|---|
| 0.0000 | 0.0000 | 0.097 | 0.068 | 1 000 | 0 | 10 000 |
| 0.0001 | 0.0001 | 0.996 | 0.997 | 1 000 | 2 | 10 000 |
| 0.0010 | 0.0010 | 0.997 | 0.998 | 2 000 | 4 | 10 000 |
| 0.0020 | 0.0020 | 1.000 | 1.000 | 1 000 | 4 | 10 000 |
| 0.0100 | 0.0100 | 1.000 | 1.000 | 1 000 | 20 | 10 000 |
| 0.0198 | 0.0198 | 1.000 | 1.000 | 1 000 | 40 | 10 000 |
| Expected FST . | Average FST . | χ2-test . | Fisher's test . | Ne . | Generation (t) . | Runs . |
|---|---|---|---|---|---|---|
| 0.0000 | 0.0000 | 0.097 | 0.068 | 1 000 | 0 | 10 000 |
| 0.0001 | 0.0001 | 0.996 | 0.997 | 1 000 | 2 | 10 000 |
| 0.0010 | 0.0010 | 0.997 | 0.998 | 2 000 | 4 | 10 000 |
| 0.0020 | 0.0020 | 1.000 | 1.000 | 1 000 | 4 | 10 000 |
| 0.0100 | 0.0100 | 1.000 | 1.000 | 1 000 | 20 | 10 000 |
| 0.0198 | 0.0198 | 1.000 | 1.000 | 1 000 | 40 | 10 000 |
Resolution power is assessed by simulating different expected level of FST according to the effective population size (Ne) and generations (t) and to the Nei (1987) formula FST = 1 − (1 – 1/2Ne)t. The significance, evaluated using Fisher's exact tests and χ2 tests, reflects the power to detect any given level of differentiation (average FST) with the sampling design developed during our study. Values of Ne used during the test are based on estimates calculated from fisheries data. The column headed “Runs” denotes the number of simulations performed. The setting FST = 0 and t= 0 estimates α (type I error, in the absence of genetic drift).
The Bayesian cluster analysis (using location information) confirmed the observed pattern with the pairwise FST comparisons and showed that the most likely number of K was 1 (mean ln P(D)±s.d.: K= 1, −41 348 ± 196; K= 2, −41 812 ± 1968; K = 3, −42 096 ± 3689; K= 4, −41 518 ± 3377; K= 5, −41 400 ± 3654; K= 6, −42 636 ± 4283; K= 7, −41 436 ± 4589; K= 8, −41 789 ± 5924; K= 9, −42 062 ± 6321; K= 10, −41 344 ± 6745; K= 15, −42 459 ± 9130).
Discussion
Genetic markers have been used extensively to assess the stock structure of commercial species, yet several species remain to be investigated, including Norway lobster. This study aimed to assess the genetic structure of this species in Icelandic waters using microsatellite loci. The results of this pilot study revealed that N. norvegicus do not exhibit any significant genetic differentiation around Iceland, results supported by a non-significant overall FST, non-significant pairwise FST comparisons among samples collected, and an absence of genetically distinguishable groups during the Bayesian cluster analyses. The lack of spatial genetic divergence observed can be explained by several non-exclusive hypotheses among which are high gene flow, historical genetic signal, and type II error. A detailed discussion on these three hypotheses is provided below.
Although this view has been largely challenged (Lundy et al., 1999; Hoarau et al., 2002; Pampoulie et al., 2006; Charrier et al., 2007; Was et al., 2008), marine species have been thought to be genetically homogeneous owing to their ability to disperse as both eggs and larvae and adults (Waples, 1998). Adult Norway lobster are sedentary, occupying burrows between 10 and 1200 m deep and, according to tagging data, rarely move more than 100 m from their burrow (Chapman and Rice, 1971). Further, females carry the eggs under their tail and stay largely in their burrows until the larvae hatch, so only passive dispersal of larvae by local oceanic currents can explain the lack of genetic structure observed. In Icelandic waters, two main currents might promote larval dispersal in the south (Figure 1). A branch of the North Atlantic current flows towards the mid-south Icelandic coast, with the main warm water branching towards the southwest, and another branch turns east towards the southeast of the country. Closer to shore, the coastal current originates from the southeast coast and flows clockwise around the island. Modelling the effect of these oceanic currents (particle-tracking) on the drift probabilities of Icelandic cod (Gadus morhua) larvae, Brickman et al. (2007) showed that larval drift in the south area of Iceland can be important within a 4–8-week pelagic period (see Figure 7 and 8 of Brickman et al., 2007). The dispersal of larvae has already been mentioned as a possible cause for the absence of genetic structure in crustacean species over vast distances (Sotelo et al., 2008; Ungfors et al., 2009).
An alternative hypothesis would be the presence of a historical signal in the genetic data. Recent microsatellite studies have demonstrated that the contemporary genetic patterns might have originated from the isolation of populations in glacial refugia during Pleistocene ice ages (Hardie et al., 2006; Hoarau et al., 2007; Pampoulie et al., 2008; Stefánsson et al., 2009b). In the area studied, the last glacial maximum (LGM) some 21 000 years ago was suggested to be one of the possible explanations for the observed genetic structure of commercial marine species such as cod (Pampoulie et al., 2008) and deep-sea redfish (Stefánsson et al., 2009b). During LGM, the northern part of the North Atlantic was covered with ice and average reconstructed temperatures ranged from −2 to −4°C (Siegert and Dowdeswell, 2004), conditions likely too extreme for Norway lobster. After the LGM, colonization of the newly opened environment from refuge areas probably resulted in a typical lack of mutation-drift equilibrium as a result of recent population expansion, and hence the lack of genetic differentiation. This phenomenon has already been suggested to explain the genetic pattern of N. norvegicus (Stamatis et al., 2004).
Finally, the lack of significant genetic differentiation is unlikely to result from a type II error, because the statistical test indicated that our genetic design should be able to detect any significant level of differentiation equal to or above FST = 0.0001 with a power >99% if it were present.
Although temporal approaches are proposed to ascertain the genetic structure of marine organisms owing to the generally low level of differentiation, this preliminary study has revealed that (i) the microsatellite loci employed are powerful enough to detect any significant differentiation, (ii) the N. norvegicus population is unlikely to be structured on the geographic scale investigated, but also that (iii) further analyses are needed to elucidate fully the structure of this species in Icelandic waters. Because of the low level of differentiation observed, future genetic investigation needs to be based on larger sample size, potentially more microsatellite loci (if available), and a temporal design. Sample collection might also be better during several months within a year to avoid a potentially biased sex ratio to assess its effect on the genetic pattern detected.
Norway lobster are currently managed as a single fishing unit in Icelandic waters, and although this preliminary study does not suggest any discrepancy between biological and fisheries management units, other biological parameters such as variability in cpue, mean size, and growth pattern might be taken into account for future management advice, as already stated (Anon., 2002).
Acknowledgements
This research was supported by the Icelandic Fisheries Research Fund in Iceland (Stofnerfðafræði leturhumars á Íslandsmiðum, Grant R013). We thank Stefán H. Brynjólfsson, Sæunn K. Erlingsdóttir, and G. Skúli Bragason for collecting the Icelandic samples, and Hasnita Charun and Douglas Neil for providing the Scottish samples and commenting on an early draft of the manuscript. We also acknowledge A. K. Sigmarsdóttir and S. Magnúsdóttir for their laboratory work, and K. Kristinsson and J. Sólmundsson for providing Figure 1.