-
PDF
- Split View
-
Views
-
Cite
Cite
Christophe Pampoulie, Sigurlaug Skirnisdóttir, Anna Kristín Daníelsdóttir, Ásgeir Gunnarsson, Genetic structure of the Atlantic wolffish (Anarhichas lupus L.) at Icelandic fishing grounds: another evidence of panmixia in Iceland?, ICES Journal of Marine Science, Volume 69, Issue 4, May 2012, Pages 508–515, https://doi.org/10.1093/icesjms/fss017
Close - Share Icon Share
Abstract
The stock structure of the Atlantic wolffish was investigated at Icelandic fishing grounds, using 16 microsatellite loci. Despite the potential of the Atlantic wolffish to exhibit genetic structure (lack of eggs/larval dispersal and adults are sedentary), none of the genetic tests applied in this study detected significant genetic differentiation among the contemporary samples as well as among the contemporary and archived samples. The results of this study therefore suggested a lack of genetic structure among the populations of Atlantic wolffish in Icelandic waters and temporal stability over a period of ∼10 years. These results are discussed in terms of biological characteristics of Atlantic wolffish, recent isolation of populations, and their application to sustainable fisheries management issues.Pampoulie, C., Skirnisdóttir, S., Daníelsdóttir, A. K., and Gunnarsson, Á. 2012. Genetic structure of the Atlantic wolffish (Anarhichas lupus L.) at Icelandic fishing grounds: another evidence of panmixia in Iceland? – ICES Journal of Marine Science, 69: 508–515.
Introduction
The Atlantic wolffish Anarhichas lupus (Linnaeus, 1758) is widely distributed across the North Atlantic and is an important commercial fish species. Like many other marine fishery resources, the abundance of the Atlantic wolffish has drastically declined over the last decades, especially in the Northwest Atlantic where it has been listed by the Canadian Species at Risk Act (SARA) as a species of “special concern” (McCusker et al., 2008). Habitat destruction by bottom trawl and bycatch and recreational fishing activities have been mentioned as possible reasons for the decline in the species (Collie et al., 2000). However, despite its status of endangered species, very few biological studies exist for the Atlantic wolffish, although length and age at maturity as well as growth and fecundity have been studied in various environments (Barsukov, 1959; Jónsson, 1982; Templeman, 1986; Nelson and Ross, 1992; Pavlov and Novikov, 1993; Liao and Lucas, 2000).
Available biological data suggest life-history trait (LHT) differences between Atlantic wolffish collected at the east and the west of Iceland (Gunnarsson et al., 2006). Fish collected in the west in warmer water tend to grow faster and to mature at an earlier age and at a smaller size, than fish collected in the cold water in the east (Gunnarsson et al., 2006). In addition, tagging experiments using anchor and alcathene tags have been performed from 1966 to 1975 to investigate the migration pattern of the species and showed that the Atlantic wolffish exhibits spawning-site fidelity and migration to feeding grounds of <100 miles in Icelandic waters (Jónsson, 1982).
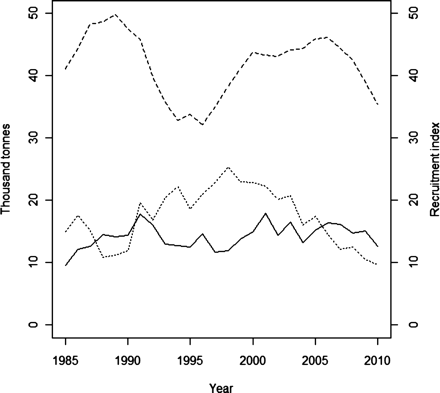
Data from the fisheries of Atlantic wolffish in the North Atlantic date back to the 1950s with landings averaging 20 000 to 40 000 t. In the year 2000, the catches averaged 40 000 t within the whole North Atlantic, but since then it has been constantly decreasing, with catches dropping to 23 000 t in 2008 (FAO, 2011). In Icelandic waters, annual landings averaged 19 000 t from the 1950s to the 1960s, and subsequently declined to 11 000 to 12 000 t in the 1970s–1980s (Figure 1). In 1977, foreign ships stopped fishing in Icelandic grounds, which resulted in a slight decline in catches until 1985 (∼10 000 t). From 1986 to 1992, the long-line effort increased again and the catches averaged 15 000 t year−1. Since 1999, the catches have been steady and were on average ∼15 500 t (Anon., 2010). However, during this period, increased catch efforts using bottom trawls at one of the main spawning locations (west of Iceland) have raised concern about the recruitment of Atlantic wolffish in Icelandic waters. In 2010, the area of this location, which was closed during spawning time (from 15 September to the end of April), was therefore increased from 500 to 1000 km2.
Annual landings of A. lupus (solid line) and fishable stock size (dashed line) in Iceland. The fishable stock size is based on the Bormicon model. Recruitment index (dotted line) represents the number of Atlantic wolffish at the length between 19 and 41 cm (in millions) and is based on data collected from the annual groundfish survey performed in March by the Marine Research Institute of Iceland.
Although genetic structure has been intensively studied in several important commercial species, e.g. because of the drastic decline in their stock, there is an obvious lack of genetic structure studies in the Atlantic wolffish. A recent study based on microsatellite loci revealed a weak genetic structure of the Atlantic wolffish from Western Greenland and Iceland to the Barents Sea (McCusker and Bentzen, 2010a). The apparent lack of genetic structure within these geographical locations was explained by a possible recent recolonization of these waters after the last glacial maximum from a refugia area located in the Rockall Bank (McCusker and Bentzen, 2010a).
For decades, marine species have been thought to be genetically homogenous due to their extensive eggs/larval dispersal and the obvious lack of barriers in the marine environment; thoughts that have been largely challenged in recent years (Ruzzante et al., 1996; Lage et al., 2004; Bekkevold et al., 2005; Jørgensen et al., 2005; Pampoulie et al., 2008b). For the Atlantic wolffish, one would expect to find reproductively isolated populations due to its peculiar life cycle and to the absence of long-distance migrations of adults. In addition, the reproductive tactics and the peculiarities of the eggs and larvae are likely to prevent passive gene flow among populations through dispersal of young stages. The fertilization of the eggs is internal (Pavlov and Moksness, 1995) and eggs are deposited in a nest that the male guards until hatching (Pavlov and Novikov, 1993). The larvae will typically hatch at a size around 20 mm and almost exclusively stay around the nest area until the juveniles become bottom dwelling owing to their large size and negative buoyancy (Bigelow and Schroeder, 1953; Moksness and Pavlov, 1996).
In Icelandic waters, the strong variability of LHTs previously observed between samples collected at the east and the west Icelandic regions (Gunnarsson et al., 2006), as well as the biological characteristics of the species described above fuelled our interest in testing whether or not population genetic approaches would reveal: To achieve our goals, contemporary as well as archived genetic samples were genotyped for 16 microsatellite loci, shedding light into contemporary genetic structure as well as possible temporal variation.
reproductive isolation of these potentially separated fishing units (east vs. west) and
a loss of genetic variability due to the decline in the population and recruitment.
Material and methods
Sampling areas and protocol
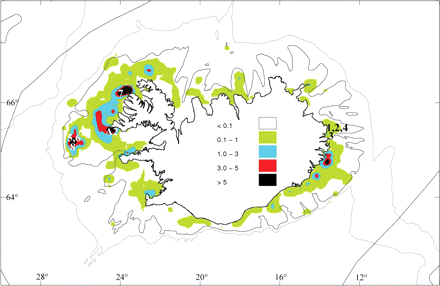
In all, 770 individuals were collected at two spawning grounds of the Atlantic wolffish, located the east and the west of Iceland, during fisheries (commercial catches) in autumn of 2010 (Figure 2; Table 1). Three of eight samples were collected at the main spawning ground (west of Iceland, samples 5, 6, and 8), which is usually closed for fisheries during spawning time (from 15 September to the end of April), except in 2010 (the area was closed from 8 October to the end of April because of the extension of the area). Since biological data suggest that the arrival of mature Atlantic wolffish at spawning grounds occurred in successive waves, a temporal sampling scheme was developed at the two spawning grounds, e.g. samples were taken in August and later in September 2010 (Table 1; Figure 2). In addition, three archived samples collected in 2002 and 2004 were also genotyped for temporal stability analyses (Table 1, archived samples A1, A2, and A3; n = 240). Two of these samples (A2 and A3) were already used in a previous large-scale analysis of Atlantic wolffish genetic structure across the North Atlantic (McCusker and Bentzen, 2010a).
Sampling area and information (coordinates, depth range, sampling dates), sample size, and size distribution (mean standard length in mm, standard deviation s.d., and range) for 11 samples of A. lupus.
| Sampling identifier . | 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | 7 . | 8 . | A1 . | A2 . | A3 . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Coordinates | 65°28.00′N 13°19.00′W | 65°27.00′N 13°21.00′W | 65°12.03′N 13°21.00′W | 65°26.50′N 13°14.00′W | 65°10.00′N 26°00.00′W | 65°10.00′N 26°30.00′W | 66°11.00′N 23°28.00′W | 65°11.00′N 26°00.00′W | n.a. | 65°25.00′N 24°33.00′W | n.a. |
| Depth (m) | 75 | 66 | 82 | 82 | 156 | 156 | 46 | 156 | n.a. | 52 | n.a. |
| Sampling dates | 30 August 2010 | 31 August 2010 | 13 September 2010 | 13 September 2010 | 18 August 2010 | 25 August 2010 | 13 September 2010 | 21 September 2010 | 18 October 2004 | 19 April 2002 | 11 March 2004 |
| Sample size | 100 | 100 | 70 | 100 | 100 | 100 | 100 | 100 | 67 | 95 | 78a |
| Length (mm) | |||||||||||
| Mean | 68.72 | 69.97 | 66.39 | 67.98 | 76.78 | 74.84 | 61.33 | 74.46 | 44.86 | 66.91 | 50.05 |
| s.d. | 5.78 | 6.38 | 6.42 | 6.46 | 8.95 | 8.81 | 7.55 | 8.30 | 18.71 | 9.12 | 19.61 |
| Range | 53–82 | 53–83 | 50–88 | 53–82 | 48–94 | 57–95 | 47–87 | 58–100 | 16–85 | 46–91 | 11–110 |
| Sampling identifier . | 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | 7 . | 8 . | A1 . | A2 . | A3 . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Coordinates | 65°28.00′N 13°19.00′W | 65°27.00′N 13°21.00′W | 65°12.03′N 13°21.00′W | 65°26.50′N 13°14.00′W | 65°10.00′N 26°00.00′W | 65°10.00′N 26°30.00′W | 66°11.00′N 23°28.00′W | 65°11.00′N 26°00.00′W | n.a. | 65°25.00′N 24°33.00′W | n.a. |
| Depth (m) | 75 | 66 | 82 | 82 | 156 | 156 | 46 | 156 | n.a. | 52 | n.a. |
| Sampling dates | 30 August 2010 | 31 August 2010 | 13 September 2010 | 13 September 2010 | 18 August 2010 | 25 August 2010 | 13 September 2010 | 21 September 2010 | 18 October 2004 | 19 April 2002 | 11 March 2004 |
| Sample size | 100 | 100 | 70 | 100 | 100 | 100 | 100 | 100 | 67 | 95 | 78a |
| Length (mm) | |||||||||||
| Mean | 68.72 | 69.97 | 66.39 | 67.98 | 76.78 | 74.84 | 61.33 | 74.46 | 44.86 | 66.91 | 50.05 |
| s.d. | 5.78 | 6.38 | 6.42 | 6.46 | 8.95 | 8.81 | 7.55 | 8.30 | 18.71 | 9.12 | 19.61 |
| Range | 53–82 | 53–83 | 50–88 | 53–82 | 48–94 | 57–95 | 47–87 | 58–100 | 16–85 | 46–91 | 11–110 |
n.a. depicts information that could not be collected from the archived samples. Dates are depicted in day/month/year.
aIn all, 96 individuals were genotyped and only 78 were successfully screened for more than three-fourths of the loci investigated.
Sampling area and information (coordinates, depth range, sampling dates), sample size, and size distribution (mean standard length in mm, standard deviation s.d., and range) for 11 samples of A. lupus.
| Sampling identifier . | 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | 7 . | 8 . | A1 . | A2 . | A3 . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Coordinates | 65°28.00′N 13°19.00′W | 65°27.00′N 13°21.00′W | 65°12.03′N 13°21.00′W | 65°26.50′N 13°14.00′W | 65°10.00′N 26°00.00′W | 65°10.00′N 26°30.00′W | 66°11.00′N 23°28.00′W | 65°11.00′N 26°00.00′W | n.a. | 65°25.00′N 24°33.00′W | n.a. |
| Depth (m) | 75 | 66 | 82 | 82 | 156 | 156 | 46 | 156 | n.a. | 52 | n.a. |
| Sampling dates | 30 August 2010 | 31 August 2010 | 13 September 2010 | 13 September 2010 | 18 August 2010 | 25 August 2010 | 13 September 2010 | 21 September 2010 | 18 October 2004 | 19 April 2002 | 11 March 2004 |
| Sample size | 100 | 100 | 70 | 100 | 100 | 100 | 100 | 100 | 67 | 95 | 78a |
| Length (mm) | |||||||||||
| Mean | 68.72 | 69.97 | 66.39 | 67.98 | 76.78 | 74.84 | 61.33 | 74.46 | 44.86 | 66.91 | 50.05 |
| s.d. | 5.78 | 6.38 | 6.42 | 6.46 | 8.95 | 8.81 | 7.55 | 8.30 | 18.71 | 9.12 | 19.61 |
| Range | 53–82 | 53–83 | 50–88 | 53–82 | 48–94 | 57–95 | 47–87 | 58–100 | 16–85 | 46–91 | 11–110 |
| Sampling identifier . | 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | 7 . | 8 . | A1 . | A2 . | A3 . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Coordinates | 65°28.00′N 13°19.00′W | 65°27.00′N 13°21.00′W | 65°12.03′N 13°21.00′W | 65°26.50′N 13°14.00′W | 65°10.00′N 26°00.00′W | 65°10.00′N 26°30.00′W | 66°11.00′N 23°28.00′W | 65°11.00′N 26°00.00′W | n.a. | 65°25.00′N 24°33.00′W | n.a. |
| Depth (m) | 75 | 66 | 82 | 82 | 156 | 156 | 46 | 156 | n.a. | 52 | n.a. |
| Sampling dates | 30 August 2010 | 31 August 2010 | 13 September 2010 | 13 September 2010 | 18 August 2010 | 25 August 2010 | 13 September 2010 | 21 September 2010 | 18 October 2004 | 19 April 2002 | 11 March 2004 |
| Sample size | 100 | 100 | 70 | 100 | 100 | 100 | 100 | 100 | 67 | 95 | 78a |
| Length (mm) | |||||||||||
| Mean | 68.72 | 69.97 | 66.39 | 67.98 | 76.78 | 74.84 | 61.33 | 74.46 | 44.86 | 66.91 | 50.05 |
| s.d. | 5.78 | 6.38 | 6.42 | 6.46 | 8.95 | 8.81 | 7.55 | 8.30 | 18.71 | 9.12 | 19.61 |
| Range | 53–82 | 53–83 | 50–88 | 53–82 | 48–94 | 57–95 | 47–87 | 58–100 | 16–85 | 46–91 | 11–110 |
n.a. depicts information that could not be collected from the archived samples. Dates are depicted in day/month/year.
aIn all, 96 individuals were genotyped and only 78 were successfully screened for more than three-fourths of the loci investigated.
Sampling location of A. lupus, in Icelandic waters. Numbers refer to the samples depicted in Table 1 and fishing areas are indicated by coloured areas. The scale indicates the density of catches in 2009 (all gears combined, t nm−2). The archived samples collected during autumn (A1 and A3) were not presented in the figure as they were collected during feeding time and not at a single geographical location.
Muscle samples were collected from each individual and conserved in 99% ethanol. Samples were genotyped at 16 microsatellite loci, namely Alu7, Alu9, Alu10, Alu11, Alu14, Alu21, Alu22, Alu23, Alu24, Alu25, Alu26, Alu27, Alu28, Alu29, Alu30, and Alu31 (McCusker et al., 2008). DNA was isolated using Agowa mag Midi DNA Isolation Kit (Agowa GmbH). Polymerase chain reactions (PCRs) were performed in a 10-µl volume containing 2 µl of DNA, 200 µM of each dNTP, 1× Teg buffer (100 mM Tris–HCl, pH 8.8; 500 mM KCl; 15 mM MgCl2; 1% Triton X-100), 0.9 U Teg polymerase (Matis, Taq comparable; see Ólafsson et al., 2010), 0.0075–0.10 µl (100 µM) of the labelled forward primers, and the same amount of reverse primers fitted with a GTTTCTT PIG tail (Brownstein et al., 1996). The 16 microsatellite loci were run in four multiplex systems: multiplex 1 (ML1): Alu21, Alu24, Alu25, Alu26, and Alu29; ML2: Alu7, Alu10, and Alu11 (Alu7 PCRs were performed separately); ML3: Alu22, Alu27, Alu28, Alu30, and Alu31; ML4: Alu9, Alu14, and Alu23 (Alu9 PCRs were performed separately).
PCRs were performed on a Tetrad2 Peltier (Bio-Rad) thermal cycler as follows: an initial denaturation step of 3 min at 94°C followed by 30 cycles of 30 s at 94°C, 50 s at 58°C, 50 s at 72°C, and a final elongation step of 7 min at 72°C. PCR products were analysed on an ABI PRISM 3730 sequencer using the GeneScan-500 LIZ size standard and genotyped with GeneMapper 4.0 (Applied Biosystems).
Genetic analyses
As the conclusion drawn from microsatellite loci strongly depends on their neutrality, the coalescent-based simulation methods of Beaumont and Nichols (1996) implemented in the software LOSITAN were applied (Antao et al., 2008). The software calculated FST values and heterozygosity for each locus according to Weir and Cockerham (1984) and expected FST values for each locus weighted by their heterozygosity. Coalescent simulations were then performed using samples of the same size as the observed samples and assuming an island model of 100 islands. One hundred thousand independent loci were generated using the infinite allele and stepwise mutation models, respectively. Simulated distribution of FST values conditional on heterozygosity under a neutral model are thus obtained and compared with the observed FST values to identify potential outlier loci.
Genetic diversity of each sample (archived and contemporary) was evaluated using allele frequencies, observed (Ho) and unbiased expected heterozygosity (He) calculated in the GENEPOP'007 (Rousset, 2008). Deviations from Hardy–Weinberg expectation (HWE) were tested using the inbreeding coefficient FIS (Weir and Cockerham, 1984) implemented in GENEPOP and significance assessed with exact tests. Genetic differentiation was estimated using theta estimates (θ; Weir and Cockerham, 1984) implemented in GENEPOP, and significance was assessed using allelic and genotypic frequency homogeneity tests (5000 permutations). The significance levels were adjusted by a simple Bonferroni correction (Rice, 1989) when multiple tests were applied.
STRUCTURE 2.3.2 (Pritchard et al., 2000) was used to assess the potential number of populations within our contemporary samples. Due to the very low genetic differentiation level detected (see the “Results” section), the admixture model with the LOCPRIOR setting was used, which considers location information. This recently developed method (Hubisz et al., 2009) has been suggested to perform better than the traditional STRUCTURE methods when the genetic structure is weak or when the number of loci is low (<20). The model was run with a “burn-in” period of 300 000 iterations and 600 000 Markov chain Monte Carlo iterations. The potential number of populations (K) varied from 1 to 8 and was tested with five independent analyses for each K. Then, the archived samples were incorporated in the analysis.
As previously published biological information suggested differences between the eastern and western populations in Iceland (Gunnarsson et al., 2006), additional analyses based on the possible existence of these two genetic groups were performed, by only using the contemporary samples. First, the program FSTAT (Goudet, 1995) was used to assess potential differences of genetic diversity indices such as allele richness and observed and expected heterozygosity. Then, a locus-by-locus hierarchical analysis of molecular (AMOVA) variance using the program Arlequin was performed (Excoffier et al., 2005). Because previous biological investigation suggested a drastic reduction in population size (Anon., 2010; Figure 1), the estimation of the effective population size (Ne) of Atlantic wolffish population was done by using the linkage disequilibrium (Hill, 1981) and the temporal methods of Waples (1989) implemented in NEESTIMATOR (Peel et al., 2004). For the temporal approach, all the archived samples from all regions were combined and used as a reference point, both for the samples collected at the western and eastern regions. Ne estimates were also calculated for one single panmictic population of Atlantic wolffish. The archived samples were then only used for the temporal methods (Waples, 1989).
Results
Biological and geographical information of the collected samples are presented in Table 1.
All studied microsatellite loci were highly polymorphic. Genetic diversity assessed as the number of alleles per locus was high, ranging from 6 (Alu7, Alu24, and Alu30) to 27 (Alu11; data not shown). The expected heterozygosity (He) per sample ranged from 0.682 (sample 11) to 0.696 (sample 8; Table 2). Genotypic proportions were out of HWE in 14 of 176 exact tests before correction for multiple tests (none of them were significant after correction) and were not attributable to any specific loci or samples. None of the samples collected deviated from HWE (Table 2) significantly. Simulations for selection suggested that none of the observed variation detected at the studied microsatellite loci departed significantly from neutral expectations (data not shown).
Expected (He) and observed (Ho) heterozygosities, number of alleles (Na) for 16 microsatellite loci in 11 samples of Atlantic A. lupus.
| . | 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus . | He . | Ho . | Na . | He . | Ho . | Na . | He . | Ho . | Na . | He . | Ho . | Na . | He . | Ho . | Na . | He . | Ho . | Na . |
| Alu7 | 0.686 | 0.710 | 6 | 0.655 | 0.667 | 6 | 0.655 | 0.686 | 5 | 0.702 | 0.750 | 6 | 0.679 | 0.650 | 6 | 0.631 | 0.620 | 6 |
| Alu9 | 0.703 | 0.729 | 7 | 0.721 | 0.748 | 7 | 0.754 | 0.750 | 7 | 0.700 | 0.7368 | 7 | 0.770 | 0.802 | 8 | 0.722 | 0.745 | 7 |
| Alu10 | 0.711 | 0.640 | 7 | 0.673 | 0.670 | 7 | 0.711 | 0.714 | 8 | 0.720 | 0.750 | 7 | 0.718 | 0.730 | 8 | 0.684 | 0.640 | 7 |
| Alu11 | 0.829 | 0.810 | 21 | 0.853 | 0.880 | 20 | 0.838 | 0.843 | 17 | 0.838 | 0.810 | 14 | 0.871 | 0.850 | 20 | 0.850 | 0.891 | 19 |
| Alu14 | 0.765 | 0.707 | 14 | 0.823 | 0.850 | 14 | 0.779 | 0.800 | 13 | 0.769 | 0.820 | 13 | 0.807 | 0.800 | 14 | 0.765 | 0.790 | 15 |
| Alu21 | 0.700 | 0.780 | 9 | 0.733 | 0.760 | 20 | 0.710 | 0.771 | 16 | 0.711 | 0.640 | 10 | 0.730 | 0.660 | 10 | 0.727 | 0.810 | 9 |
| Alu22 | 0.429 | 0.420 | 6 | 0.428 | 0.412 | 9 | 0.489 | 0.471 | 7 | 0.414 | 0.380 | 7 | 0.427 | 0.410 | 6 | 0.424 | 0.465 | 5 |
| Alu23 | 0.582 | 0.630 | 7 | 0.583 | 0.560 | 8 | 0.594 | 0.529 | 6 | 0.586 | 0.580 | 6 | 0.615 | 0.650 | 6 | 0.586 | 0.680 | 6 |
| Alu24 | 0.661 | 0.750 | 5 | 0.628 | 0.620 | 4 | 0.648 | 0.620 | 4 | 0.663 | 0.604 | 4 | 0.670 | 0.680 | 5 | 0.669 | 0.744 | 5 |
| Alu25 | 0.784 | 0.790 | 12 | 0.780 | 0.790 | 10 | 0.760 | 0.757 | 10 | 0.762 | 0.740 | 8 | 0.769 | 0.760 | 9 | 0.791 | 0.810 | 11 |
| Alu26 | 0.841 | 0.810 | 15 | 0.859 | 0.887 | 16 | 0.846 | 0.814 | 17 | 0.834 | 0.761 | 14 | 0.854 | 0.930 | 16 | 0.863 | 0.895 | 20 |
| Alu27 | 0.822 | 0.818 | 20 | 0.844 | 0.810 | 18 | 0.795 | 0.771 | 17 | 0.868 | 0.887 | 20 | 0.814 | 0.810 | 18 | 0.854 | 0.850 | 20 |
| Alu28 | 0.900 | 0.890 | 16 | 0.904 | 0.920 | 16 | 0.898 | 0.871 | 15 | 0.913 | 0.920 | 15 | 0.912 | 0.910 | 17 | 0.880 | 0.850 | 17 |
| Alu29 | 0.600 | 0.556 | 6 | 0.506 | 0.536 | 7 | 0.540 | 0.544 | 5 | 0.554 | 0.568 | 6 | 0.524 | 0.494 | 8 | 0.532 | 0.604 | 7 |
| Alu30 | 0.174 | 0.170 | 3 | 0.124 | 0.130 | 5 | 0.134 | 0.143 | 4 | 0.141 | 0.140 | 4 | 0.114 | 0.120 | 4 | 0.164 | 0.180 | 2 |
| Alu31 | 0.830 | 0.889 | 12 | 0.841 | 0.860 | 13 | 0.842 | 0.886 | 11 | 0.841 | 0.840 | 13 | 0.839 | 0.760 | 11 | 0.831 | 0.849 | 13 |
| Overall | 0.689 | 0.694 | 9.78 | 0.685 | 0.694 | 9.83 | 0.687 | 0.685 | 9.46 | 0.689 | 0.683 | 9.19 | 0.695 | 0.689 | 9.73 | 0.687 | 0.714 | 9.77 |
| 7 . | 8 . | A1 . | A2 . | A3 . | . | . | . | |||||||||||
| He | Ho | Na | He | Ho | Na | He | Ho | Na | He | Ho | Na | He | Ho | Na | ||||
| Alu7 | 0.670 | 0.590 | 6 | 0.714 | 0.700 | 6 | 0.661 | 0.667 | 6 | 0.643 | 0.653 | 6 | 0.680 | 0.597 | 6 | |||
| Alu9 | 0.743 | 0.806 | 7 | 0.733 | 0.750 | 7 | 0.722 | 0.723 | 7 | 0.762 | 0.731 | 8 | 0.751 | 0.776 | 8 | |||
| Alu10 | 0.692 | 0.760 | 8 | 0.711 | 0.720 | 8 | 0.739 | 0.671 | 8 | 0.716 | 0.842 | 8 | 0.718 | 0.667 | 7 | |||
| Alu11 | 0.857 | 0.790 | 17 | 0.857 | 0.904 | 18 | 0.839 | 0.879 | 19 | 0.817 | 0.800 | 18 | 0.809 | 0.805 | 16 | |||
| Alu14 | 0.793 | 0.780 | 13 | 0.816 | 0.840 | 16 | 0.802 | 0.821 | 13 | 0.774 | 0.800 | 12 | 0.765 | 0.769 | 14 | |||
| Alu21 | 0.697 | 0.690 | 7 | 0.719 | 0.770 | 8 | 0.751 | 0.758 | 11 | 0.687 | 0.670 | 7 | 0.721 | 0.718 | 8 | |||
| Alu22 | 0.365 | 0.340 | 7 | 0.413 | 0.360 | 8 | 0.407 | 0.418 | 7 | 0.440 | 0.451 | 7 | 0.361 | 0.338 | 7 | |||
| Alu23 | 0.616 | 0.590 | 8 | 0.606 | 0.570 | 7 | 0.599 | 0.537 | 6 | 0.603 | 0.632 | 5 | 0.579 | 0.705 | 4 | |||
| Alu24 | 0.651 | 0.700 | 5 | 0.674 | 0.670 | 5 | 0.646 | 0.692 | 5 | 0.655 | 0.733 | 4 | 0.649 | 0.653 | 4 | |||
| Alu25 | 0.762 | 0.750 | 11 | 0.776 | 0.800 | 11 | 0.771 | 0.716 | 10 | 0.764 | 0.716 | 10 | 0.749 | 0.692 | 10 | |||
| Alu26 | 0.843 | 0.880 | 17 | 0.843 | 0.897 | 15 | 0.842 | 0.862 | 16 | 0.851 | 0.843 | 17 | 0.836 | 0.878 | 15 | |||
| Alu27 | 0.831 | 0.860 | 19 | 0.836 | 0.810 | 20 | 0.831 | 0.773 | 19 | 0.826 | 0.898 | 20 | 0.819 | 0.816 | 19 | |||
| Alu28 | 0.900 | 0.890 | 16 | 0.909 | 0.880 | 16 | 0.895 | 0.925 | 14 | 0.904 | 0.913 | 15 | 0.912 | 0.936 | 16 | |||
| Alu29 | 0.550 | 0.540 | 6 | 0.537 | 0.573 | 8 | 0.567 | 0.578 | 6 | 0.536 | 0.578 | 5 | 0.583 | 0.600 | 6 | |||
| Alu30 | 0.165 | 0.180 | 4 | 0.141 | 0.130 | 4 | 0.180 | 0.197 | 3 | 0.102 | 0.106 | 4 | 0.147 | 0.158 | 3 | |||
| Alu31 | 0.852 | 0.870 | 11 | 0.853 | 0.810 | 12 | 0.844 | 0.806 | 12 | 0.848 | 0.842 | 14 | 0.827 | 0.829 | 11 | |||
| Overall | 0.687 | 0.689 | 9.39 | 0.696 | 0.699 | 9.83 | 0.694 | 0.689 | 10.13 | 0.683 | 0.696 | 10.00 | 0.682 | 0.684 | 9.63 | |||
| . | 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus . | He . | Ho . | Na . | He . | Ho . | Na . | He . | Ho . | Na . | He . | Ho . | Na . | He . | Ho . | Na . | He . | Ho . | Na . |
| Alu7 | 0.686 | 0.710 | 6 | 0.655 | 0.667 | 6 | 0.655 | 0.686 | 5 | 0.702 | 0.750 | 6 | 0.679 | 0.650 | 6 | 0.631 | 0.620 | 6 |
| Alu9 | 0.703 | 0.729 | 7 | 0.721 | 0.748 | 7 | 0.754 | 0.750 | 7 | 0.700 | 0.7368 | 7 | 0.770 | 0.802 | 8 | 0.722 | 0.745 | 7 |
| Alu10 | 0.711 | 0.640 | 7 | 0.673 | 0.670 | 7 | 0.711 | 0.714 | 8 | 0.720 | 0.750 | 7 | 0.718 | 0.730 | 8 | 0.684 | 0.640 | 7 |
| Alu11 | 0.829 | 0.810 | 21 | 0.853 | 0.880 | 20 | 0.838 | 0.843 | 17 | 0.838 | 0.810 | 14 | 0.871 | 0.850 | 20 | 0.850 | 0.891 | 19 |
| Alu14 | 0.765 | 0.707 | 14 | 0.823 | 0.850 | 14 | 0.779 | 0.800 | 13 | 0.769 | 0.820 | 13 | 0.807 | 0.800 | 14 | 0.765 | 0.790 | 15 |
| Alu21 | 0.700 | 0.780 | 9 | 0.733 | 0.760 | 20 | 0.710 | 0.771 | 16 | 0.711 | 0.640 | 10 | 0.730 | 0.660 | 10 | 0.727 | 0.810 | 9 |
| Alu22 | 0.429 | 0.420 | 6 | 0.428 | 0.412 | 9 | 0.489 | 0.471 | 7 | 0.414 | 0.380 | 7 | 0.427 | 0.410 | 6 | 0.424 | 0.465 | 5 |
| Alu23 | 0.582 | 0.630 | 7 | 0.583 | 0.560 | 8 | 0.594 | 0.529 | 6 | 0.586 | 0.580 | 6 | 0.615 | 0.650 | 6 | 0.586 | 0.680 | 6 |
| Alu24 | 0.661 | 0.750 | 5 | 0.628 | 0.620 | 4 | 0.648 | 0.620 | 4 | 0.663 | 0.604 | 4 | 0.670 | 0.680 | 5 | 0.669 | 0.744 | 5 |
| Alu25 | 0.784 | 0.790 | 12 | 0.780 | 0.790 | 10 | 0.760 | 0.757 | 10 | 0.762 | 0.740 | 8 | 0.769 | 0.760 | 9 | 0.791 | 0.810 | 11 |
| Alu26 | 0.841 | 0.810 | 15 | 0.859 | 0.887 | 16 | 0.846 | 0.814 | 17 | 0.834 | 0.761 | 14 | 0.854 | 0.930 | 16 | 0.863 | 0.895 | 20 |
| Alu27 | 0.822 | 0.818 | 20 | 0.844 | 0.810 | 18 | 0.795 | 0.771 | 17 | 0.868 | 0.887 | 20 | 0.814 | 0.810 | 18 | 0.854 | 0.850 | 20 |
| Alu28 | 0.900 | 0.890 | 16 | 0.904 | 0.920 | 16 | 0.898 | 0.871 | 15 | 0.913 | 0.920 | 15 | 0.912 | 0.910 | 17 | 0.880 | 0.850 | 17 |
| Alu29 | 0.600 | 0.556 | 6 | 0.506 | 0.536 | 7 | 0.540 | 0.544 | 5 | 0.554 | 0.568 | 6 | 0.524 | 0.494 | 8 | 0.532 | 0.604 | 7 |
| Alu30 | 0.174 | 0.170 | 3 | 0.124 | 0.130 | 5 | 0.134 | 0.143 | 4 | 0.141 | 0.140 | 4 | 0.114 | 0.120 | 4 | 0.164 | 0.180 | 2 |
| Alu31 | 0.830 | 0.889 | 12 | 0.841 | 0.860 | 13 | 0.842 | 0.886 | 11 | 0.841 | 0.840 | 13 | 0.839 | 0.760 | 11 | 0.831 | 0.849 | 13 |
| Overall | 0.689 | 0.694 | 9.78 | 0.685 | 0.694 | 9.83 | 0.687 | 0.685 | 9.46 | 0.689 | 0.683 | 9.19 | 0.695 | 0.689 | 9.73 | 0.687 | 0.714 | 9.77 |
| 7 . | 8 . | A1 . | A2 . | A3 . | . | . | . | |||||||||||
| He | Ho | Na | He | Ho | Na | He | Ho | Na | He | Ho | Na | He | Ho | Na | ||||
| Alu7 | 0.670 | 0.590 | 6 | 0.714 | 0.700 | 6 | 0.661 | 0.667 | 6 | 0.643 | 0.653 | 6 | 0.680 | 0.597 | 6 | |||
| Alu9 | 0.743 | 0.806 | 7 | 0.733 | 0.750 | 7 | 0.722 | 0.723 | 7 | 0.762 | 0.731 | 8 | 0.751 | 0.776 | 8 | |||
| Alu10 | 0.692 | 0.760 | 8 | 0.711 | 0.720 | 8 | 0.739 | 0.671 | 8 | 0.716 | 0.842 | 8 | 0.718 | 0.667 | 7 | |||
| Alu11 | 0.857 | 0.790 | 17 | 0.857 | 0.904 | 18 | 0.839 | 0.879 | 19 | 0.817 | 0.800 | 18 | 0.809 | 0.805 | 16 | |||
| Alu14 | 0.793 | 0.780 | 13 | 0.816 | 0.840 | 16 | 0.802 | 0.821 | 13 | 0.774 | 0.800 | 12 | 0.765 | 0.769 | 14 | |||
| Alu21 | 0.697 | 0.690 | 7 | 0.719 | 0.770 | 8 | 0.751 | 0.758 | 11 | 0.687 | 0.670 | 7 | 0.721 | 0.718 | 8 | |||
| Alu22 | 0.365 | 0.340 | 7 | 0.413 | 0.360 | 8 | 0.407 | 0.418 | 7 | 0.440 | 0.451 | 7 | 0.361 | 0.338 | 7 | |||
| Alu23 | 0.616 | 0.590 | 8 | 0.606 | 0.570 | 7 | 0.599 | 0.537 | 6 | 0.603 | 0.632 | 5 | 0.579 | 0.705 | 4 | |||
| Alu24 | 0.651 | 0.700 | 5 | 0.674 | 0.670 | 5 | 0.646 | 0.692 | 5 | 0.655 | 0.733 | 4 | 0.649 | 0.653 | 4 | |||
| Alu25 | 0.762 | 0.750 | 11 | 0.776 | 0.800 | 11 | 0.771 | 0.716 | 10 | 0.764 | 0.716 | 10 | 0.749 | 0.692 | 10 | |||
| Alu26 | 0.843 | 0.880 | 17 | 0.843 | 0.897 | 15 | 0.842 | 0.862 | 16 | 0.851 | 0.843 | 17 | 0.836 | 0.878 | 15 | |||
| Alu27 | 0.831 | 0.860 | 19 | 0.836 | 0.810 | 20 | 0.831 | 0.773 | 19 | 0.826 | 0.898 | 20 | 0.819 | 0.816 | 19 | |||
| Alu28 | 0.900 | 0.890 | 16 | 0.909 | 0.880 | 16 | 0.895 | 0.925 | 14 | 0.904 | 0.913 | 15 | 0.912 | 0.936 | 16 | |||
| Alu29 | 0.550 | 0.540 | 6 | 0.537 | 0.573 | 8 | 0.567 | 0.578 | 6 | 0.536 | 0.578 | 5 | 0.583 | 0.600 | 6 | |||
| Alu30 | 0.165 | 0.180 | 4 | 0.141 | 0.130 | 4 | 0.180 | 0.197 | 3 | 0.102 | 0.106 | 4 | 0.147 | 0.158 | 3 | |||
| Alu31 | 0.852 | 0.870 | 11 | 0.853 | 0.810 | 12 | 0.844 | 0.806 | 12 | 0.848 | 0.842 | 14 | 0.827 | 0.829 | 11 | |||
| Overall | 0.687 | 0.689 | 9.39 | 0.696 | 0.699 | 9.83 | 0.694 | 0.689 | 10.13 | 0.683 | 0.696 | 10.00 | 0.682 | 0.684 | 9.63 | |||
Emboldened values indicate samples that were not in HWE (exact test, p < 0.05), none remained significant after correction for multiple tests (α = 0.05/176 = 0.00025).
Expected (He) and observed (Ho) heterozygosities, number of alleles (Na) for 16 microsatellite loci in 11 samples of Atlantic A. lupus.
| . | 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus . | He . | Ho . | Na . | He . | Ho . | Na . | He . | Ho . | Na . | He . | Ho . | Na . | He . | Ho . | Na . | He . | Ho . | Na . |
| Alu7 | 0.686 | 0.710 | 6 | 0.655 | 0.667 | 6 | 0.655 | 0.686 | 5 | 0.702 | 0.750 | 6 | 0.679 | 0.650 | 6 | 0.631 | 0.620 | 6 |
| Alu9 | 0.703 | 0.729 | 7 | 0.721 | 0.748 | 7 | 0.754 | 0.750 | 7 | 0.700 | 0.7368 | 7 | 0.770 | 0.802 | 8 | 0.722 | 0.745 | 7 |
| Alu10 | 0.711 | 0.640 | 7 | 0.673 | 0.670 | 7 | 0.711 | 0.714 | 8 | 0.720 | 0.750 | 7 | 0.718 | 0.730 | 8 | 0.684 | 0.640 | 7 |
| Alu11 | 0.829 | 0.810 | 21 | 0.853 | 0.880 | 20 | 0.838 | 0.843 | 17 | 0.838 | 0.810 | 14 | 0.871 | 0.850 | 20 | 0.850 | 0.891 | 19 |
| Alu14 | 0.765 | 0.707 | 14 | 0.823 | 0.850 | 14 | 0.779 | 0.800 | 13 | 0.769 | 0.820 | 13 | 0.807 | 0.800 | 14 | 0.765 | 0.790 | 15 |
| Alu21 | 0.700 | 0.780 | 9 | 0.733 | 0.760 | 20 | 0.710 | 0.771 | 16 | 0.711 | 0.640 | 10 | 0.730 | 0.660 | 10 | 0.727 | 0.810 | 9 |
| Alu22 | 0.429 | 0.420 | 6 | 0.428 | 0.412 | 9 | 0.489 | 0.471 | 7 | 0.414 | 0.380 | 7 | 0.427 | 0.410 | 6 | 0.424 | 0.465 | 5 |
| Alu23 | 0.582 | 0.630 | 7 | 0.583 | 0.560 | 8 | 0.594 | 0.529 | 6 | 0.586 | 0.580 | 6 | 0.615 | 0.650 | 6 | 0.586 | 0.680 | 6 |
| Alu24 | 0.661 | 0.750 | 5 | 0.628 | 0.620 | 4 | 0.648 | 0.620 | 4 | 0.663 | 0.604 | 4 | 0.670 | 0.680 | 5 | 0.669 | 0.744 | 5 |
| Alu25 | 0.784 | 0.790 | 12 | 0.780 | 0.790 | 10 | 0.760 | 0.757 | 10 | 0.762 | 0.740 | 8 | 0.769 | 0.760 | 9 | 0.791 | 0.810 | 11 |
| Alu26 | 0.841 | 0.810 | 15 | 0.859 | 0.887 | 16 | 0.846 | 0.814 | 17 | 0.834 | 0.761 | 14 | 0.854 | 0.930 | 16 | 0.863 | 0.895 | 20 |
| Alu27 | 0.822 | 0.818 | 20 | 0.844 | 0.810 | 18 | 0.795 | 0.771 | 17 | 0.868 | 0.887 | 20 | 0.814 | 0.810 | 18 | 0.854 | 0.850 | 20 |
| Alu28 | 0.900 | 0.890 | 16 | 0.904 | 0.920 | 16 | 0.898 | 0.871 | 15 | 0.913 | 0.920 | 15 | 0.912 | 0.910 | 17 | 0.880 | 0.850 | 17 |
| Alu29 | 0.600 | 0.556 | 6 | 0.506 | 0.536 | 7 | 0.540 | 0.544 | 5 | 0.554 | 0.568 | 6 | 0.524 | 0.494 | 8 | 0.532 | 0.604 | 7 |
| Alu30 | 0.174 | 0.170 | 3 | 0.124 | 0.130 | 5 | 0.134 | 0.143 | 4 | 0.141 | 0.140 | 4 | 0.114 | 0.120 | 4 | 0.164 | 0.180 | 2 |
| Alu31 | 0.830 | 0.889 | 12 | 0.841 | 0.860 | 13 | 0.842 | 0.886 | 11 | 0.841 | 0.840 | 13 | 0.839 | 0.760 | 11 | 0.831 | 0.849 | 13 |
| Overall | 0.689 | 0.694 | 9.78 | 0.685 | 0.694 | 9.83 | 0.687 | 0.685 | 9.46 | 0.689 | 0.683 | 9.19 | 0.695 | 0.689 | 9.73 | 0.687 | 0.714 | 9.77 |
| 7 . | 8 . | A1 . | A2 . | A3 . | . | . | . | |||||||||||
| He | Ho | Na | He | Ho | Na | He | Ho | Na | He | Ho | Na | He | Ho | Na | ||||
| Alu7 | 0.670 | 0.590 | 6 | 0.714 | 0.700 | 6 | 0.661 | 0.667 | 6 | 0.643 | 0.653 | 6 | 0.680 | 0.597 | 6 | |||
| Alu9 | 0.743 | 0.806 | 7 | 0.733 | 0.750 | 7 | 0.722 | 0.723 | 7 | 0.762 | 0.731 | 8 | 0.751 | 0.776 | 8 | |||
| Alu10 | 0.692 | 0.760 | 8 | 0.711 | 0.720 | 8 | 0.739 | 0.671 | 8 | 0.716 | 0.842 | 8 | 0.718 | 0.667 | 7 | |||
| Alu11 | 0.857 | 0.790 | 17 | 0.857 | 0.904 | 18 | 0.839 | 0.879 | 19 | 0.817 | 0.800 | 18 | 0.809 | 0.805 | 16 | |||
| Alu14 | 0.793 | 0.780 | 13 | 0.816 | 0.840 | 16 | 0.802 | 0.821 | 13 | 0.774 | 0.800 | 12 | 0.765 | 0.769 | 14 | |||
| Alu21 | 0.697 | 0.690 | 7 | 0.719 | 0.770 | 8 | 0.751 | 0.758 | 11 | 0.687 | 0.670 | 7 | 0.721 | 0.718 | 8 | |||
| Alu22 | 0.365 | 0.340 | 7 | 0.413 | 0.360 | 8 | 0.407 | 0.418 | 7 | 0.440 | 0.451 | 7 | 0.361 | 0.338 | 7 | |||
| Alu23 | 0.616 | 0.590 | 8 | 0.606 | 0.570 | 7 | 0.599 | 0.537 | 6 | 0.603 | 0.632 | 5 | 0.579 | 0.705 | 4 | |||
| Alu24 | 0.651 | 0.700 | 5 | 0.674 | 0.670 | 5 | 0.646 | 0.692 | 5 | 0.655 | 0.733 | 4 | 0.649 | 0.653 | 4 | |||
| Alu25 | 0.762 | 0.750 | 11 | 0.776 | 0.800 | 11 | 0.771 | 0.716 | 10 | 0.764 | 0.716 | 10 | 0.749 | 0.692 | 10 | |||
| Alu26 | 0.843 | 0.880 | 17 | 0.843 | 0.897 | 15 | 0.842 | 0.862 | 16 | 0.851 | 0.843 | 17 | 0.836 | 0.878 | 15 | |||
| Alu27 | 0.831 | 0.860 | 19 | 0.836 | 0.810 | 20 | 0.831 | 0.773 | 19 | 0.826 | 0.898 | 20 | 0.819 | 0.816 | 19 | |||
| Alu28 | 0.900 | 0.890 | 16 | 0.909 | 0.880 | 16 | 0.895 | 0.925 | 14 | 0.904 | 0.913 | 15 | 0.912 | 0.936 | 16 | |||
| Alu29 | 0.550 | 0.540 | 6 | 0.537 | 0.573 | 8 | 0.567 | 0.578 | 6 | 0.536 | 0.578 | 5 | 0.583 | 0.600 | 6 | |||
| Alu30 | 0.165 | 0.180 | 4 | 0.141 | 0.130 | 4 | 0.180 | 0.197 | 3 | 0.102 | 0.106 | 4 | 0.147 | 0.158 | 3 | |||
| Alu31 | 0.852 | 0.870 | 11 | 0.853 | 0.810 | 12 | 0.844 | 0.806 | 12 | 0.848 | 0.842 | 14 | 0.827 | 0.829 | 11 | |||
| Overall | 0.687 | 0.689 | 9.39 | 0.696 | 0.699 | 9.83 | 0.694 | 0.689 | 10.13 | 0.683 | 0.696 | 10.00 | 0.682 | 0.684 | 9.63 | |||
| . | 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus . | He . | Ho . | Na . | He . | Ho . | Na . | He . | Ho . | Na . | He . | Ho . | Na . | He . | Ho . | Na . | He . | Ho . | Na . |
| Alu7 | 0.686 | 0.710 | 6 | 0.655 | 0.667 | 6 | 0.655 | 0.686 | 5 | 0.702 | 0.750 | 6 | 0.679 | 0.650 | 6 | 0.631 | 0.620 | 6 |
| Alu9 | 0.703 | 0.729 | 7 | 0.721 | 0.748 | 7 | 0.754 | 0.750 | 7 | 0.700 | 0.7368 | 7 | 0.770 | 0.802 | 8 | 0.722 | 0.745 | 7 |
| Alu10 | 0.711 | 0.640 | 7 | 0.673 | 0.670 | 7 | 0.711 | 0.714 | 8 | 0.720 | 0.750 | 7 | 0.718 | 0.730 | 8 | 0.684 | 0.640 | 7 |
| Alu11 | 0.829 | 0.810 | 21 | 0.853 | 0.880 | 20 | 0.838 | 0.843 | 17 | 0.838 | 0.810 | 14 | 0.871 | 0.850 | 20 | 0.850 | 0.891 | 19 |
| Alu14 | 0.765 | 0.707 | 14 | 0.823 | 0.850 | 14 | 0.779 | 0.800 | 13 | 0.769 | 0.820 | 13 | 0.807 | 0.800 | 14 | 0.765 | 0.790 | 15 |
| Alu21 | 0.700 | 0.780 | 9 | 0.733 | 0.760 | 20 | 0.710 | 0.771 | 16 | 0.711 | 0.640 | 10 | 0.730 | 0.660 | 10 | 0.727 | 0.810 | 9 |
| Alu22 | 0.429 | 0.420 | 6 | 0.428 | 0.412 | 9 | 0.489 | 0.471 | 7 | 0.414 | 0.380 | 7 | 0.427 | 0.410 | 6 | 0.424 | 0.465 | 5 |
| Alu23 | 0.582 | 0.630 | 7 | 0.583 | 0.560 | 8 | 0.594 | 0.529 | 6 | 0.586 | 0.580 | 6 | 0.615 | 0.650 | 6 | 0.586 | 0.680 | 6 |
| Alu24 | 0.661 | 0.750 | 5 | 0.628 | 0.620 | 4 | 0.648 | 0.620 | 4 | 0.663 | 0.604 | 4 | 0.670 | 0.680 | 5 | 0.669 | 0.744 | 5 |
| Alu25 | 0.784 | 0.790 | 12 | 0.780 | 0.790 | 10 | 0.760 | 0.757 | 10 | 0.762 | 0.740 | 8 | 0.769 | 0.760 | 9 | 0.791 | 0.810 | 11 |
| Alu26 | 0.841 | 0.810 | 15 | 0.859 | 0.887 | 16 | 0.846 | 0.814 | 17 | 0.834 | 0.761 | 14 | 0.854 | 0.930 | 16 | 0.863 | 0.895 | 20 |
| Alu27 | 0.822 | 0.818 | 20 | 0.844 | 0.810 | 18 | 0.795 | 0.771 | 17 | 0.868 | 0.887 | 20 | 0.814 | 0.810 | 18 | 0.854 | 0.850 | 20 |
| Alu28 | 0.900 | 0.890 | 16 | 0.904 | 0.920 | 16 | 0.898 | 0.871 | 15 | 0.913 | 0.920 | 15 | 0.912 | 0.910 | 17 | 0.880 | 0.850 | 17 |
| Alu29 | 0.600 | 0.556 | 6 | 0.506 | 0.536 | 7 | 0.540 | 0.544 | 5 | 0.554 | 0.568 | 6 | 0.524 | 0.494 | 8 | 0.532 | 0.604 | 7 |
| Alu30 | 0.174 | 0.170 | 3 | 0.124 | 0.130 | 5 | 0.134 | 0.143 | 4 | 0.141 | 0.140 | 4 | 0.114 | 0.120 | 4 | 0.164 | 0.180 | 2 |
| Alu31 | 0.830 | 0.889 | 12 | 0.841 | 0.860 | 13 | 0.842 | 0.886 | 11 | 0.841 | 0.840 | 13 | 0.839 | 0.760 | 11 | 0.831 | 0.849 | 13 |
| Overall | 0.689 | 0.694 | 9.78 | 0.685 | 0.694 | 9.83 | 0.687 | 0.685 | 9.46 | 0.689 | 0.683 | 9.19 | 0.695 | 0.689 | 9.73 | 0.687 | 0.714 | 9.77 |
| 7 . | 8 . | A1 . | A2 . | A3 . | . | . | . | |||||||||||
| He | Ho | Na | He | Ho | Na | He | Ho | Na | He | Ho | Na | He | Ho | Na | ||||
| Alu7 | 0.670 | 0.590 | 6 | 0.714 | 0.700 | 6 | 0.661 | 0.667 | 6 | 0.643 | 0.653 | 6 | 0.680 | 0.597 | 6 | |||
| Alu9 | 0.743 | 0.806 | 7 | 0.733 | 0.750 | 7 | 0.722 | 0.723 | 7 | 0.762 | 0.731 | 8 | 0.751 | 0.776 | 8 | |||
| Alu10 | 0.692 | 0.760 | 8 | 0.711 | 0.720 | 8 | 0.739 | 0.671 | 8 | 0.716 | 0.842 | 8 | 0.718 | 0.667 | 7 | |||
| Alu11 | 0.857 | 0.790 | 17 | 0.857 | 0.904 | 18 | 0.839 | 0.879 | 19 | 0.817 | 0.800 | 18 | 0.809 | 0.805 | 16 | |||
| Alu14 | 0.793 | 0.780 | 13 | 0.816 | 0.840 | 16 | 0.802 | 0.821 | 13 | 0.774 | 0.800 | 12 | 0.765 | 0.769 | 14 | |||
| Alu21 | 0.697 | 0.690 | 7 | 0.719 | 0.770 | 8 | 0.751 | 0.758 | 11 | 0.687 | 0.670 | 7 | 0.721 | 0.718 | 8 | |||
| Alu22 | 0.365 | 0.340 | 7 | 0.413 | 0.360 | 8 | 0.407 | 0.418 | 7 | 0.440 | 0.451 | 7 | 0.361 | 0.338 | 7 | |||
| Alu23 | 0.616 | 0.590 | 8 | 0.606 | 0.570 | 7 | 0.599 | 0.537 | 6 | 0.603 | 0.632 | 5 | 0.579 | 0.705 | 4 | |||
| Alu24 | 0.651 | 0.700 | 5 | 0.674 | 0.670 | 5 | 0.646 | 0.692 | 5 | 0.655 | 0.733 | 4 | 0.649 | 0.653 | 4 | |||
| Alu25 | 0.762 | 0.750 | 11 | 0.776 | 0.800 | 11 | 0.771 | 0.716 | 10 | 0.764 | 0.716 | 10 | 0.749 | 0.692 | 10 | |||
| Alu26 | 0.843 | 0.880 | 17 | 0.843 | 0.897 | 15 | 0.842 | 0.862 | 16 | 0.851 | 0.843 | 17 | 0.836 | 0.878 | 15 | |||
| Alu27 | 0.831 | 0.860 | 19 | 0.836 | 0.810 | 20 | 0.831 | 0.773 | 19 | 0.826 | 0.898 | 20 | 0.819 | 0.816 | 19 | |||
| Alu28 | 0.900 | 0.890 | 16 | 0.909 | 0.880 | 16 | 0.895 | 0.925 | 14 | 0.904 | 0.913 | 15 | 0.912 | 0.936 | 16 | |||
| Alu29 | 0.550 | 0.540 | 6 | 0.537 | 0.573 | 8 | 0.567 | 0.578 | 6 | 0.536 | 0.578 | 5 | 0.583 | 0.600 | 6 | |||
| Alu30 | 0.165 | 0.180 | 4 | 0.141 | 0.130 | 4 | 0.180 | 0.197 | 3 | 0.102 | 0.106 | 4 | 0.147 | 0.158 | 3 | |||
| Alu31 | 0.852 | 0.870 | 11 | 0.853 | 0.810 | 12 | 0.844 | 0.806 | 12 | 0.848 | 0.842 | 14 | 0.827 | 0.829 | 11 | |||
| Overall | 0.687 | 0.689 | 9.39 | 0.696 | 0.699 | 9.83 | 0.694 | 0.689 | 10.13 | 0.683 | 0.696 | 10.00 | 0.682 | 0.684 | 9.63 | |||
Emboldened values indicate samples that were not in HWE (exact test, p < 0.05), none remained significant after correction for multiple tests (α = 0.05/176 = 0.00025).
The overall genetic estimates based on combined archived and contemporary samples did not reveal significant FST (FST = −0.00006, p > 0.05, 95% CI: −0.0005 to 0.0004) and FIS values (FIS = −0.00006, p > 0.05, 95% CI: −0.0086 to 0.0066). This genetic pattern was reflected in the pairwise FST comparisons of samples as none of the comparisons remained significant after Bonferroni correction (Table 3). The pairwise FST comparisons among samples collected in August and September within a spawning ground were therefore not different which suggests the absence of temporal variation for the contemporary time-frame investigated. In addition, none of the archived samples were genetically different from the contemporary ones (Table 3).
Pairwise FST (above diagonal) and values of p (below diagonal) among the 11 samples of A. lupus based on allele frequencies.
| . | 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | 7 . | 8 . | A1 . | A2 . | A3 . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0 | 0.0024 | 0.0001 | −0.0002 | 0.0012 | 0.0004 | 0.0004 | −0.0006 | −0.0016 | 0.0010 | −0.0006 |
| 2 | 0.367 | 0 | 0.001 | 0.0035 | 0.0026 | 0.0022 | 0.0023 | 0.0005 | 0.0025 | 0.0015 | 0.0012 |
| 3 | 0.630 | 0.1189 | 0 | 0.0011 | −0.0006 | 0.0003 | 0.0004 | −0.0008 | −0.0015 | −0.0016 | −0.0004 |
| 4 | 0.494 | 0.001 | 0.031 | 0 | −0.0016 | −0.0001 | −0.0005 | −0.0013 | −0.0009 | −0.0002 | −0.0008 |
| 5 | 0.113 | 0.069 | 0.380 | 0.286 | 0 | −0.0005 | −0.0011 | −0.0018 | −0.0005 | −0.0008 | −0.0011 |
| 6 | 0.338 | 0.029 | 0.536 | 0.228 | 0.725 | 0 | −0.0009 | −0.0001 | −0.0004 | −0.0004 | 0.0003 |
| 7 | 0.250 | 0.024 | 0.368 | 0.358 | 0.609 | 0.736 | 0 | −0.0017 | −0.0014 | −0.0014 | −0.0005 |
| 8 | 0.836 | 0.289 | 0.317 | 0.206 | 0.713 | 0.247 | 0.891 | 0 | −0.0017 | −0.0011 | −0.0012 |
| 9 | 0.838 | 0.104 | 0.752 | 0.537 | 0.363 | 0.544 | 0.926 | 0.738 | 0 | −0.0005 | −0.0001 |
| 10 | 0.453 | 0.326 | 0.656 | 0.666 | 0.304 | 0.304 | 0.983 | 0.749 | 0.737 | 0 | −0.0010 |
| 11 | 0.772 | 0.657 | 0.225 | 0.789 | 0.280 | 0.393 | 0.295 | 0.463 | 0.402 | 0.762 | 0 |
| . | 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | 7 . | 8 . | A1 . | A2 . | A3 . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0 | 0.0024 | 0.0001 | −0.0002 | 0.0012 | 0.0004 | 0.0004 | −0.0006 | −0.0016 | 0.0010 | −0.0006 |
| 2 | 0.367 | 0 | 0.001 | 0.0035 | 0.0026 | 0.0022 | 0.0023 | 0.0005 | 0.0025 | 0.0015 | 0.0012 |
| 3 | 0.630 | 0.1189 | 0 | 0.0011 | −0.0006 | 0.0003 | 0.0004 | −0.0008 | −0.0015 | −0.0016 | −0.0004 |
| 4 | 0.494 | 0.001 | 0.031 | 0 | −0.0016 | −0.0001 | −0.0005 | −0.0013 | −0.0009 | −0.0002 | −0.0008 |
| 5 | 0.113 | 0.069 | 0.380 | 0.286 | 0 | −0.0005 | −0.0011 | −0.0018 | −0.0005 | −0.0008 | −0.0011 |
| 6 | 0.338 | 0.029 | 0.536 | 0.228 | 0.725 | 0 | −0.0009 | −0.0001 | −0.0004 | −0.0004 | 0.0003 |
| 7 | 0.250 | 0.024 | 0.368 | 0.358 | 0.609 | 0.736 | 0 | −0.0017 | −0.0014 | −0.0014 | −0.0005 |
| 8 | 0.836 | 0.289 | 0.317 | 0.206 | 0.713 | 0.247 | 0.891 | 0 | −0.0017 | −0.0011 | −0.0012 |
| 9 | 0.838 | 0.104 | 0.752 | 0.537 | 0.363 | 0.544 | 0.926 | 0.738 | 0 | −0.0005 | −0.0001 |
| 10 | 0.453 | 0.326 | 0.656 | 0.666 | 0.304 | 0.304 | 0.983 | 0.749 | 0.737 | 0 | −0.0010 |
| 11 | 0.772 | 0.657 | 0.225 | 0.789 | 0.280 | 0.393 | 0.295 | 0.463 | 0.402 | 0.762 | 0 |
Emboldened values differ significantly from zero (Fisher's exact test, p < 0.05). None of the comparisons remained significant after Bonferroni correction (α = 0.05/55 = 0.0009).
Pairwise FST (above diagonal) and values of p (below diagonal) among the 11 samples of A. lupus based on allele frequencies.
| . | 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | 7 . | 8 . | A1 . | A2 . | A3 . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0 | 0.0024 | 0.0001 | −0.0002 | 0.0012 | 0.0004 | 0.0004 | −0.0006 | −0.0016 | 0.0010 | −0.0006 |
| 2 | 0.367 | 0 | 0.001 | 0.0035 | 0.0026 | 0.0022 | 0.0023 | 0.0005 | 0.0025 | 0.0015 | 0.0012 |
| 3 | 0.630 | 0.1189 | 0 | 0.0011 | −0.0006 | 0.0003 | 0.0004 | −0.0008 | −0.0015 | −0.0016 | −0.0004 |
| 4 | 0.494 | 0.001 | 0.031 | 0 | −0.0016 | −0.0001 | −0.0005 | −0.0013 | −0.0009 | −0.0002 | −0.0008 |
| 5 | 0.113 | 0.069 | 0.380 | 0.286 | 0 | −0.0005 | −0.0011 | −0.0018 | −0.0005 | −0.0008 | −0.0011 |
| 6 | 0.338 | 0.029 | 0.536 | 0.228 | 0.725 | 0 | −0.0009 | −0.0001 | −0.0004 | −0.0004 | 0.0003 |
| 7 | 0.250 | 0.024 | 0.368 | 0.358 | 0.609 | 0.736 | 0 | −0.0017 | −0.0014 | −0.0014 | −0.0005 |
| 8 | 0.836 | 0.289 | 0.317 | 0.206 | 0.713 | 0.247 | 0.891 | 0 | −0.0017 | −0.0011 | −0.0012 |
| 9 | 0.838 | 0.104 | 0.752 | 0.537 | 0.363 | 0.544 | 0.926 | 0.738 | 0 | −0.0005 | −0.0001 |
| 10 | 0.453 | 0.326 | 0.656 | 0.666 | 0.304 | 0.304 | 0.983 | 0.749 | 0.737 | 0 | −0.0010 |
| 11 | 0.772 | 0.657 | 0.225 | 0.789 | 0.280 | 0.393 | 0.295 | 0.463 | 0.402 | 0.762 | 0 |
| . | 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | 7 . | 8 . | A1 . | A2 . | A3 . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0 | 0.0024 | 0.0001 | −0.0002 | 0.0012 | 0.0004 | 0.0004 | −0.0006 | −0.0016 | 0.0010 | −0.0006 |
| 2 | 0.367 | 0 | 0.001 | 0.0035 | 0.0026 | 0.0022 | 0.0023 | 0.0005 | 0.0025 | 0.0015 | 0.0012 |
| 3 | 0.630 | 0.1189 | 0 | 0.0011 | −0.0006 | 0.0003 | 0.0004 | −0.0008 | −0.0015 | −0.0016 | −0.0004 |
| 4 | 0.494 | 0.001 | 0.031 | 0 | −0.0016 | −0.0001 | −0.0005 | −0.0013 | −0.0009 | −0.0002 | −0.0008 |
| 5 | 0.113 | 0.069 | 0.380 | 0.286 | 0 | −0.0005 | −0.0011 | −0.0018 | −0.0005 | −0.0008 | −0.0011 |
| 6 | 0.338 | 0.029 | 0.536 | 0.228 | 0.725 | 0 | −0.0009 | −0.0001 | −0.0004 | −0.0004 | 0.0003 |
| 7 | 0.250 | 0.024 | 0.368 | 0.358 | 0.609 | 0.736 | 0 | −0.0017 | −0.0014 | −0.0014 | −0.0005 |
| 8 | 0.836 | 0.289 | 0.317 | 0.206 | 0.713 | 0.247 | 0.891 | 0 | −0.0017 | −0.0011 | −0.0012 |
| 9 | 0.838 | 0.104 | 0.752 | 0.537 | 0.363 | 0.544 | 0.926 | 0.738 | 0 | −0.0005 | −0.0001 |
| 10 | 0.453 | 0.326 | 0.656 | 0.666 | 0.304 | 0.304 | 0.983 | 0.749 | 0.737 | 0 | −0.0010 |
| 11 | 0.772 | 0.657 | 0.225 | 0.789 | 0.280 | 0.393 | 0.295 | 0.463 | 0.402 | 0.762 | 0 |
Emboldened values differ significantly from zero (Fisher's exact test, p < 0.05). None of the comparisons remained significant after Bonferroni correction (α = 0.05/55 = 0.0009).
The Bayesian cluster analysis performed on contemporary samples (using location information) confirmed the observed pattern with the pairwise FST comparisons and showed that the most likely number of K was 1 (mean Ln P(D) ± s.d.: K=1, −32 692 ± 85; K = 2, −39 459 ± 709; K = 3, −40 107 ± 2213 K = 4, −42 522 ± 7106; K = 5, −46 172 ± 14 426; K = 6, −50 813 ± 23 676; K = 7, −42 075 ± 5874; K = 8, −40390 ± 2483). The inclusion of archived samples did not affect the results (data not shown).
The locus-by-locus AMOVA also confirmed this pattern as the overall among-groups differentiation was not significant between contemporary samples collected at the east and the west of Icelandic waters (Table 4), even if two microsatellite loci revealed weak significant differences between groups.
Locus-by-locus AMOVA variance among the east and the west group of A. lupus (AEW, FCT), among samples within group (ASWG, FSC), and within samples (WS, FST).
| Locus . | %AEW . | %ASWG . | %WS . | FCT . | FSC . | FST . |
|---|---|---|---|---|---|---|
| Alu7 | −0.0795 | 0.3392 | 100.0964 | −0.0002 | −0.0010 | −0.0008 |
| Alu9 | 0.3841 | 0.3675 | 99.7652 | −0.0015 | 0.0024 | 0.00384* |
| Alu10 | 0.1169 | 0.3529 | 99.8853 | 0.0000 | 0.0012 | 0.0012 |
| Alu11 | 0.0623 | 0.4269 | 100.0156 | −0.0008 | −0.0002 | 0.0006 |
| Alu14 | 0.0269 | 0.3971 | 99.6924 | 0.00281* | 0.0031* | 0.0003 |
| Alu21 | −0.0728 | 0.3600 | 100.1300 | −0.0006 | −0.0013 | −0.0007 |
| Alu22 | 0.0005 | 0.2116 | 100.1475 | −0.0015 | −0.0015 | 0.0000 |
| Alu23 | 0.0149 | 0.2996 | 100.2020 | −0.0022 | −0.0020 | 0.0002 |
| Alu24 | −0.0060 | 0.3309 | 100.0192 | −0.0001 | −0.0002 | −0.0001 |
| Alu25 | −0.0074 | 0.3888 | 99.9614 | 0.0005 | 0.0004 | −0.0001 |
| Alu26 | 0.0010 | 0.4263 | 100.0084 | −0.0001 | −0.0001 | 0.0000 |
| Alu27 | −0.0429 | 0.4193 | 99.9489 | 0.0009 | 0.0005 | −0.0004 |
| Alu28 | 0.0145 | 0.4545 | 99.9026 | 0.0008 | 0.0010 | 0.0001 |
| Alu29 | −0.0214 | 0.2729 | 99.9771 | 0.0004 | 0.0002 | −0.0002 |
| Alu30 | −0.0262 | 0.0730 | 100.1485 | −0.0012 | −0.0015 | −0.0003 |
| Alu31 | −0.1529 | 0.4228 | 99.9284 | 0.0022* | 0.0007 | −0.0015 |
| Total | 0.0133 | 0.3465 | 99.9893 | 0.0002 | 0.0002 | 0.0003 |
| Locus . | %AEW . | %ASWG . | %WS . | FCT . | FSC . | FST . |
|---|---|---|---|---|---|---|
| Alu7 | −0.0795 | 0.3392 | 100.0964 | −0.0002 | −0.0010 | −0.0008 |
| Alu9 | 0.3841 | 0.3675 | 99.7652 | −0.0015 | 0.0024 | 0.00384* |
| Alu10 | 0.1169 | 0.3529 | 99.8853 | 0.0000 | 0.0012 | 0.0012 |
| Alu11 | 0.0623 | 0.4269 | 100.0156 | −0.0008 | −0.0002 | 0.0006 |
| Alu14 | 0.0269 | 0.3971 | 99.6924 | 0.00281* | 0.0031* | 0.0003 |
| Alu21 | −0.0728 | 0.3600 | 100.1300 | −0.0006 | −0.0013 | −0.0007 |
| Alu22 | 0.0005 | 0.2116 | 100.1475 | −0.0015 | −0.0015 | 0.0000 |
| Alu23 | 0.0149 | 0.2996 | 100.2020 | −0.0022 | −0.0020 | 0.0002 |
| Alu24 | −0.0060 | 0.3309 | 100.0192 | −0.0001 | −0.0002 | −0.0001 |
| Alu25 | −0.0074 | 0.3888 | 99.9614 | 0.0005 | 0.0004 | −0.0001 |
| Alu26 | 0.0010 | 0.4263 | 100.0084 | −0.0001 | −0.0001 | 0.0000 |
| Alu27 | −0.0429 | 0.4193 | 99.9489 | 0.0009 | 0.0005 | −0.0004 |
| Alu28 | 0.0145 | 0.4545 | 99.9026 | 0.0008 | 0.0010 | 0.0001 |
| Alu29 | −0.0214 | 0.2729 | 99.9771 | 0.0004 | 0.0002 | −0.0002 |
| Alu30 | −0.0262 | 0.0730 | 100.1485 | −0.0012 | −0.0015 | −0.0003 |
| Alu31 | −0.1529 | 0.4228 | 99.9284 | 0.0022* | 0.0007 | −0.0015 |
| Total | 0.0133 | 0.3465 | 99.9893 | 0.0002 | 0.0002 | 0.0003 |
This analysis was performed on contemporary samples only. The loci that showed significant group comparison are indicated emboldened.
*p < 0.05.
Locus-by-locus AMOVA variance among the east and the west group of A. lupus (AEW, FCT), among samples within group (ASWG, FSC), and within samples (WS, FST).
| Locus . | %AEW . | %ASWG . | %WS . | FCT . | FSC . | FST . |
|---|---|---|---|---|---|---|
| Alu7 | −0.0795 | 0.3392 | 100.0964 | −0.0002 | −0.0010 | −0.0008 |
| Alu9 | 0.3841 | 0.3675 | 99.7652 | −0.0015 | 0.0024 | 0.00384* |
| Alu10 | 0.1169 | 0.3529 | 99.8853 | 0.0000 | 0.0012 | 0.0012 |
| Alu11 | 0.0623 | 0.4269 | 100.0156 | −0.0008 | −0.0002 | 0.0006 |
| Alu14 | 0.0269 | 0.3971 | 99.6924 | 0.00281* | 0.0031* | 0.0003 |
| Alu21 | −0.0728 | 0.3600 | 100.1300 | −0.0006 | −0.0013 | −0.0007 |
| Alu22 | 0.0005 | 0.2116 | 100.1475 | −0.0015 | −0.0015 | 0.0000 |
| Alu23 | 0.0149 | 0.2996 | 100.2020 | −0.0022 | −0.0020 | 0.0002 |
| Alu24 | −0.0060 | 0.3309 | 100.0192 | −0.0001 | −0.0002 | −0.0001 |
| Alu25 | −0.0074 | 0.3888 | 99.9614 | 0.0005 | 0.0004 | −0.0001 |
| Alu26 | 0.0010 | 0.4263 | 100.0084 | −0.0001 | −0.0001 | 0.0000 |
| Alu27 | −0.0429 | 0.4193 | 99.9489 | 0.0009 | 0.0005 | −0.0004 |
| Alu28 | 0.0145 | 0.4545 | 99.9026 | 0.0008 | 0.0010 | 0.0001 |
| Alu29 | −0.0214 | 0.2729 | 99.9771 | 0.0004 | 0.0002 | −0.0002 |
| Alu30 | −0.0262 | 0.0730 | 100.1485 | −0.0012 | −0.0015 | −0.0003 |
| Alu31 | −0.1529 | 0.4228 | 99.9284 | 0.0022* | 0.0007 | −0.0015 |
| Total | 0.0133 | 0.3465 | 99.9893 | 0.0002 | 0.0002 | 0.0003 |
| Locus . | %AEW . | %ASWG . | %WS . | FCT . | FSC . | FST . |
|---|---|---|---|---|---|---|
| Alu7 | −0.0795 | 0.3392 | 100.0964 | −0.0002 | −0.0010 | −0.0008 |
| Alu9 | 0.3841 | 0.3675 | 99.7652 | −0.0015 | 0.0024 | 0.00384* |
| Alu10 | 0.1169 | 0.3529 | 99.8853 | 0.0000 | 0.0012 | 0.0012 |
| Alu11 | 0.0623 | 0.4269 | 100.0156 | −0.0008 | −0.0002 | 0.0006 |
| Alu14 | 0.0269 | 0.3971 | 99.6924 | 0.00281* | 0.0031* | 0.0003 |
| Alu21 | −0.0728 | 0.3600 | 100.1300 | −0.0006 | −0.0013 | −0.0007 |
| Alu22 | 0.0005 | 0.2116 | 100.1475 | −0.0015 | −0.0015 | 0.0000 |
| Alu23 | 0.0149 | 0.2996 | 100.2020 | −0.0022 | −0.0020 | 0.0002 |
| Alu24 | −0.0060 | 0.3309 | 100.0192 | −0.0001 | −0.0002 | −0.0001 |
| Alu25 | −0.0074 | 0.3888 | 99.9614 | 0.0005 | 0.0004 | −0.0001 |
| Alu26 | 0.0010 | 0.4263 | 100.0084 | −0.0001 | −0.0001 | 0.0000 |
| Alu27 | −0.0429 | 0.4193 | 99.9489 | 0.0009 | 0.0005 | −0.0004 |
| Alu28 | 0.0145 | 0.4545 | 99.9026 | 0.0008 | 0.0010 | 0.0001 |
| Alu29 | −0.0214 | 0.2729 | 99.9771 | 0.0004 | 0.0002 | −0.0002 |
| Alu30 | −0.0262 | 0.0730 | 100.1485 | −0.0012 | −0.0015 | −0.0003 |
| Alu31 | −0.1529 | 0.4228 | 99.9284 | 0.0022* | 0.0007 | −0.0015 |
| Total | 0.0133 | 0.3465 | 99.9893 | 0.0002 | 0.0002 | 0.0003 |
This analysis was performed on contemporary samples only. The loci that showed significant group comparison are indicated emboldened.
*p < 0.05.
Finally, the genetic diversity indices comparison among regions (eastern vs. western) using permutation tests implemented in FSTAT did not reveal any significant pattern (Eastern: HO = 0.689, HE = 0.691, AR = 9.563; Western: HO = 0.697, HE = 0.695, AR = 9.679, p > 0.05). Genetic differentiation among eastern and western regions was also not significant (FST = 0.0002, p > 0.05, 95%). In addition, comparison between each region and archived samples using the same test did not reveal any loss of genetic diversity within the period investigated (Archived: HO = 0.690, HE = 0.690, AR = 9.608; P > 0.05).
Estimates of effective population size for each region using several methods are listed in Table 5. The linkage disequilibrium methods (Hill, 1981) led to large 95% CIs for both regions and suggested that Ne was usually higher in the western region than in the eastern one (Table 5). The temporal approach (Waples, 1989) also led to very large estimates of Ne (Table 5).
Estimates of effective population sizes (Ne) using linkage disequilibrium (Hill, 1981) and temporal methods (Waples, 1989).
| Population . | Method applied . | Median . | 95% confidence interval . |
|---|---|---|---|
| East | Hill (1981) | 1 938 | 1 421–2 933 |
| Waples (1989) | ∞ | 469–∞ | |
| West | Hill (1981) | 3 158 | 2 109–6 110 |
| Waples (1989) | ∞ | 1 363–∞ | |
| Archived East | Hill (1981) | 642 | 479–955 |
| Archived West | Hill (1981) | 1 253 | 589–∞ |
| One population | Hill (1981) | 1 817 | 1 574–2 132 |
| Waples (1989) | ∞ | 1 420–∞ |
| Population . | Method applied . | Median . | 95% confidence interval . |
|---|---|---|---|
| East | Hill (1981) | 1 938 | 1 421–2 933 |
| Waples (1989) | ∞ | 469–∞ | |
| West | Hill (1981) | 3 158 | 2 109–6 110 |
| Waples (1989) | ∞ | 1 363–∞ | |
| Archived East | Hill (1981) | 642 | 479–955 |
| Archived West | Hill (1981) | 1 253 | 589–∞ |
| One population | Hill (1981) | 1 817 | 1 574–2 132 |
| Waples (1989) | ∞ | 1 420–∞ |
Estimates of effective population sizes (Ne) using linkage disequilibrium (Hill, 1981) and temporal methods (Waples, 1989).
| Population . | Method applied . | Median . | 95% confidence interval . |
|---|---|---|---|
| East | Hill (1981) | 1 938 | 1 421–2 933 |
| Waples (1989) | ∞ | 469–∞ | |
| West | Hill (1981) | 3 158 | 2 109–6 110 |
| Waples (1989) | ∞ | 1 363–∞ | |
| Archived East | Hill (1981) | 642 | 479–955 |
| Archived West | Hill (1981) | 1 253 | 589–∞ |
| One population | Hill (1981) | 1 817 | 1 574–2 132 |
| Waples (1989) | ∞ | 1 420–∞ |
| Population . | Method applied . | Median . | 95% confidence interval . |
|---|---|---|---|
| East | Hill (1981) | 1 938 | 1 421–2 933 |
| Waples (1989) | ∞ | 469–∞ | |
| West | Hill (1981) | 3 158 | 2 109–6 110 |
| Waples (1989) | ∞ | 1 363–∞ | |
| Archived East | Hill (1981) | 642 | 479–955 |
| Archived West | Hill (1981) | 1 253 | 589–∞ |
| One population | Hill (1981) | 1 817 | 1 574–2 132 |
| Waples (1989) | ∞ | 1 420–∞ |
Discussion
The present study aimed to assess the genetic structure of the Atlantic wolffish, A. lupus, in Icelandic waters using microsatellite loci. The obtained results revealed that this species is not genetically structured around Iceland at neutral markers. The results are supported by the lack of differences in the genetic diversity indices of the collected samples, a non-significant overall FST, the temporal stability of the genetic pattern detected (see FST results), and the absence of distinguishable genetic groups during the Bayesian cluster analysis. The lack of genetic structure for this species is discussed in terms of gene flow, recent isolation of populations, and effective population size.
Indeed, despite the apparent lack of gene flow due to the biology of the species, the observed genetic pattern might be explained by at least two alternative hypotheses detailed below.
(i) One of the most likely explanations for the absence of genetic structure despite the apparent lack of gene flow would be the recent isolation of the west and east Atlantic wolffish populations in Iceland. Given enough time, drift would typically have led to genetic differentiation of subpopulations in such a species, but marine populations around Iceland tend to be young and to originate from ice-free refugia during the last glacial maximum (LGM) some 20–25 Kyr BP (Coyer et al., 2003; Maggs et al., 2008; Pampoulie et al., 2008a, 2011). During LGM, most of the Icelandic waters were covered with an ice-cap, which reached the break of the shelf and was ca. 1500–2000 m thick (Ingólfsson et al., 2009; see Figure 4.1). The ice-cap rapidly collapsed from the seawater and retreated onto present-day dry land between 14.9 and 13.9 Kyr BP (Ingólfsson et al., 2009; see Figure 4.2). The recolonization of ice-free environment by marine organisms could therefore start at around 13 Kyr BP in Iceland, a time, which might not have been sufficient to promote genetic differentiation at neutral markers (Slatkin, 1987; Cavalli-Sforza, 1998). In fact, for marine species such as the Atlantic wolffish with large effective population size and long generation time, drift might not have promoted genetic differentiation among isolated populations since the last glacial event (see Slatkin, 1987 and Pampoulie et al., 2008a, b, 2011, for Icelandic cases). The post-glacial history of recolonization of Icelandic waters has already been suggested to be responsible for the weak genetic differentiation among populations of several commercial fish species in Iceland (Pampoulie et al., 2008a, b, 2011) and for the weak genetic structure of Atlantic wolffish across the North Atlantic ocean (McCusker and Bentzen, 2010a). The recent recolonization of Icelandic waters by the Atlantic wolffish would have resulted in a typical lack of mutation-drift equilibrium due to recent population expansion, hence the lack of genetic differentiation. Therefore, the present study is fully consistent with genetic studies performed on the genus Anarhichas in the North Atlantic, suggesting a very limited genetic structure due to recent isolation of populations (McCusker and Bentzen, 2010a, b, 2011).
(ii) A second possible explanation for the genetic pattern detected might be the observed large effective population size (Ne) of the studied species. Indeed, the apparent lack of the genetic structure and the long-term stability were also reflected in the estimates of population sizes (see Ne in Table 5). Both the linkage disequilibrium and the temporal methods revealed that the lower estimated Ne was around or higher than 2000 for the Eastern and Western populations, even when one panmictic population was considered. Only the archived samples exhibited lower estimates (642 < Ne < 1253). As the magnitude of genetic drift crucially depends on Ne, such large effective population sizes for the Atlantic wolffish might not have favoured genetic differentiation in Icelandic waters (Slatkin, 1987).
Such high estimates of Ne are also commonly interpreted as evidence of conserved evolutionary potential of exploited populations (Franklin, 1980; Poulsen et al., 2006). In addition, the comparison of allele frequencies and the comparison of genetic diversity indices among contemporary and archived samples revealed a lack of genetic differences and did not bring evidence for any loss of genetic diversity despite the previously mentioned decline in population size in Icelandic populations of Atlantic wolffish (Anon., 2010). The present study is therefore in line with several studies performed on neutral genetic diversity and showing a stable temporal pattern despite drastic stocks' overexploitation (Ruzzante et al., 2001; Hauser et al., 2002; Cuveliers et al., 2011; Pujolar et al., 2011). However, detecting a loss of genetic diversity at neutral genetic markers might not be appropriate to assess the potential loss of adaptive genetic variation due to fisheries as mentioned for the Icelandic cod (Jakobsdóttir et al., 2011).
The Atlantic wolffish is currently managed as a single fishing unit in Icelandic waters, and although this study does not suggest the presence of reproductively isolated populations, we recommend that biological parameters, such as variability in mean size, age at maturity, and growth pattern, which vary between Western and Eastern populations, should be taken into account for future management advice as already stated (Gunnarsson et al., 2006), as well as the decrease in recruitment.
Acknowledgements
This research was supported by the Icelandic Fisheries Research Fund in Iceland (Verkefnasjóðs sjávarútvegsins um sjávarrannsóknir á samkeppnissviði). We thank Sif Guðmunsdóttir for her help during sampling, Steinunn Magnúsdóttir and Kristinn Ólafsson for their help during the developmental work performed on microsatellite loci, and Megan R. McCusker for sending us the archived samples. Special thanks are addressed to two anonymous referees for their valuable comments on the manuscript.
References
Author notes
Handling editor: Sarah Kraak