-
PDF
- Split View
-
Views
-
Cite
Cite
J. Spitz, T. Chouvelon, M. Cardinaud, C. Kostecki, P. Lorance, Prey preferences of adult sea bass Dicentrarchus labrax in the northeastern Atlantic: implications for bycatch of common dolphin Delphinus delphis, ICES Journal of Marine Science, Volume 70, Issue 2, March 2013, Pages 452–461, https://doi.org/10.1093/icesjms/fss200
Close - Share Icon Share
Abstract
In the northeastern Atlantic, adult sea bass (Dicentrarchus labrax) is one of largest fish living on the shelf, and this species has important commercial value. However, pelagic trawl fisheries that target sea bass have negative operational interactions with common dolphins (Delphinus delphis). Our goal was to determine the diet of adult sea bass in the Bay of Biscay from stomach-content and stable-isotope analyses, and explore the dietary overlap between sea bass and common dolphins. We found that sea bass primarily target small pelagic fish, most notably mackerel (Scomber scombrus), scads (Trachurus spp.), anchovy (Engraulis encrasicolus), and sardine (Sardina pilchardus). These four species also dominated the diets of common dolphins. This overlap in feeding preferences could increase the risk of dolphins being caught by trawl fisheries while feeding among sea bass, and may be an underlying mechanism to explain the high rate of common dolphin bycatch observed in the pelagic trawl fishery for sea bass in the Bay of Biscay. Understanding the foraging ecology and trophic interactions of predator species is an essential step for identifying and resolving management issues in the northeastern Atlantic and other marine ecosystems.Spitz, J., Chouvelon, T., Cardinaud, M., Kostecki, C., and Lorance, P. 2013. Prey preferences of adult sea bass Dicentrarchus labrax in the northeastern Atlantic: implications for bycatch of common dolphin Delphinus delphis – ICES Journal of Marine Science, 70: 452–461.
Introduction
Marine top predators display various foraging strategies, such as interspecific relationships (e.g. competition or cooperation), as a result of different evolutionary pressures. At the extremes, some predators are opportunistic and consume their prey without selection (i.e. proportionately to prey availability in the environment), whereas other predators are specialized and consume a very narrow range of prey types (Begon et al., 2006). Specialized predators may be more dependent on the availability of their prey and more constrained by their foraging strategies than opportunistic predators. Hence, understanding the trophic interactions in the marine food web and, as a consequence, the identification of pertinent management measures, appears to be strongly dependent on knowledge of predators' foraging ecology.
The Bay of Biscay in the northeastern Atlantic supports a diverse marine fauna (Quéro et al., 2003; Kiszka et al., 2007; Certain et al., 2008) and has been extensively exploited by numerous fisheries over a long period of time (Lorance et al., 2009). The high trophic level predator community in the Bay of Biscay is mainly composed of several species of small cetaceans and seabirds, with only a few species of large fish (Lassalle et al., 2011); this is in contrast to oceanic (tropical) ecosystems where large fish such as tuna and sharks play a more prominent role (Kitchell et al., 1999). In this context, adult European sea bass (Dicentrarchus labrax) appear to be one of the major large fish predators on the continental shelf of the Bay of Biscay. This species has a high landed value, and consequently sea bass are exploited by several fisheries in European waters (e.g. professional liners, trawlers or gillnetters). Unfortunately, operational interactions between common dolphin (Delphinus delphis) and pelagic trawl fishery for sea bass are known to occur seasonally. Since the late 1980s, these interactions have been revealed by extensive strandings of common dolphin along the French coast. Although several fish species are targeted by pelagic trawl fisheries in the Bay of Biscay, cetacean bycatch occurs almost exclusively in the sea bass fishery (Morizur et al., 1999; Northridge et al., 2006) and these mortalities appear to be unevenly distributed over time, suggesting that bycatch events may depend on specific ecological mechanisms.
European sea bass inhabits estuaries and open waters up to 100 m in depth. The species is mainly found in coastal waters, but is known to migrate offshore and to deeper waters during the winter (Pickett and Pawson, 1994). The biology and ecology of sea bass have been extensively studied in estuarine and coastal areas, especially at the juvenile stage, with a particular interest in nursery areas (Aprahamian and Barr, 1985; Cabral and Costa, 2001; Martinho et al., 2008). Juvenile sea bass is generally described as an opportunistic predator (Pickett & Pawson, 1994); however, the ecology of the adult stage has received little attention, particularly in open waters where the diet of adult sea bass has not yet been the subject of a quantitative study.
Here, we postulate that the feeding interactions between sea bass and common dolphin may be an underlying mechanism which increases the bycatch vulnerability of common dolphin in pelagic trawl fisheries for sea bass. To test this hypothesis, we describe for the first time the diet of adult sea bass on the continental shelf of the Bay of Biscay by combining two techniques: analyses of stomach contents and isotopic signatures. Prey selection was explored using two independent approaches: an index of selectivity of feeding based on prey abundance in both the diet and environment, and a Bayesian isotopic mixing model. Additionally, we compared these results with the published diet of common dolphin, with the aim of highlighting the potential dietary overlap between the two species in the context of dolphin bycatch.
Material and methods
Collection and preparation of samples
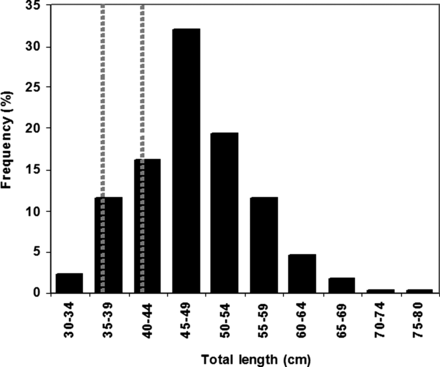
The stomach contents were obtained from 404 sea bass collected on the continental shelf of the Bay of Biscay in the autumn and winter of 2005, 2006 and 2008. The fish were caught during the annual fisheries science EVHOE surveys conducted by Ifremer and from commercial fishing trawlers. During the EVHOE surveys, hauls were performed with a large vertical-opening bottom trawl. The fishing gear used was a GOV 36/47; the gear has a horizontal opening of 20 m, a vertical opening of 4 m and a codend mesh size of 20 mm. The haul duration was 30 min, at a trawl speed of 3.5 knots during daylight. The stomachs of all adult sea bass caught from 21 different hauls were taken, ligatured and individually stored deep-frozen (–20°C) in polythene bags until further analyses. The size of the sea bass sampled ranged from 31–79 cm, with a mean size of 48 ± 7.5 cm (Figure 1); these values correspond well with adult sea bass in which the first maturity occurs at 36 and 42 cm for males and females, respectively (Dorel, 1986; Pawson and Pickett, 1996). A proportion of females are observed to start gonad maturation at 32 cm (Pawson and Pickett, 1996).
Length (cm) distribution of adult sea bass (Dicentrarchus labrax) sampled in autumn/winter of 2005, 2006 and 2008 for stomach content analysis. Vertical dot bars depict the onset of maturity in males (36 cm) and females (42 cm).
For stable isotope analysis, a standard piece of dorsal muscle was sampled from the adult sea bass and other different forage species (Table 1). The sea bass and forage species were caught and sampled from the same hauls during the 2008 survey; the muscle of the adult sea bass and different forage species were sampled at the same time in order to limit temporal variability. After collection, the samples were immediately placed in individual polythene bags, deep-frozen at –20°C and then subsequently freeze-dried. The freeze-dried tissues were ground into a fine powder and stored in individual polythene vials until further analysis.
Carbon (δ13C) and nitrogen (δ15N) stable isotope raw values in the muscle of adult sea bass and forage species on the continental shelf of the Bay of Biscay.
| Species . | N . | Size (mm) . | d13C (‰) . | d15N (‰) . | ||
|---|---|---|---|---|---|---|
| x ± sd . | x ± sd . | p-value . | x ± sd . | p-value . | ||
| PREDATOR | ||||||
| Dicentrarchus labrax | 15 | 585 ± 103 | −16.7 ± 0.6 | – | 14.0 ± 0.6 | – |
| PELAGIC FISH | −18.2 ± 0.6 | − | 11.6 ± 0.3 | – | ||
| Engraulis encrasicolus | 7 | 126 ± 4 | −17.6 ± 0.3 | 0.002 | 12.0 ± 0.4 | <0.001 |
| Sprattus sprattus | 9 | 103 ± 10 | −17.8 ± 0.2 | <0.001 | 11.9 ± 0.3 | <0.001 |
| Sardina pilchardus | 10 | 192 ± 48 | −17.7 ± 0.4 | <0.001 | 11.3 ± 0.8 | <0.001 |
| Trachurus trachurus | 10 | 170 ± 85 | −19.0 ± 0.9 | <0.001 | 11.4 ± 0.9 | <0.001 |
| Scomber scombrus | 10 | 257 ± 63 | −18.7 ± 0.4 | <0.001 | 11.3 ± 0.7 | <0.001 |
| Micromesistius poutassou | 9 | 220 ± 14 | −18.1 ± 0.3 | <0.001 | 11.4 ± 0.3 | <0.001 |
| DEMERSAL AND BENTHIC FISH | −17.0 ± 0.7 | − | 12.7 ± 0.7 | − | ||
| Argentina sphyraena | 6 | 187 ± 10 | −17.4 ± 0.2 | 0.078 | 12.5 ± 0.3 | <0.001 |
| Callionymus lyra | 5 | 222 ± 16 | −16.6 ± 0.3 | 1.000 | 12.5 ± 0.3 | <0.001 |
| Trisopterus minutus | 8 | 201 ± 20 | −17.2 ± 0.4 | 1.000 | 13.0 ± 0.5 | 0.032 |
| Trisopterus luscus | 5 | 184 ± 23 | −16.4 ± 0.1 | 1.000 | 14.1 ± 0.2 | 1.000 |
| Merluccius merluccius | 11 | 186 ± 54 | −18.3 ± 0.2 | <0.001 | 12.3 ± 0.3 | <0.001 |
| Pomatoschistus minutus | 5 | 56 ± 5 | −17.5 ± 0.1 | 0.030 | 12.7 ± 0.3 | 0.011 |
| Solea solea | 5 | 178 ± 13 | −16.3 ± 0.3 | 1.000 | 11.7 ± 0.4 | <0.001 |
| Microchirus variegatus | 5 | 162 ± 8 | −17.3 ± 0.0 | 1.000 | 12.2 ± 0.1 | <0.001 |
| Dicologlossa cuneata | 4 | 190 ± 18 | −16.6 ± 0.4 | 1.000 | 13.4 ± 0.7 | 1.000 |
| COASTAL FISH | −16.6 ± 0.1 | − | 13.6 ± 1.0 | − | ||
| Spondyliosoma cantharus | 5 | 142 ± 37 | −16.6 ± 0.8 | 1.000 | 12.3 ± 0.3 | <0.001 |
| Trachinus draco | 10 | 237 ± 20 | −16.7 ± 0.8 | 1.000 | 13.1 ± 1.3 | 0.039 |
| Merlangius merlangus | 10 | 116 ± 27 | −16.7 ± 0.3 | 1.000 | 13.8 ± 0.3 | 1.000 |
| Hyperoplus lanceolatus | 5 | 340 ± 14 | −16.4 ± 0.3 | 1.000 | 14.3 ± 0.3 | 1.000 |
| Atherina presbyter | 5 | 110 ± 10 | −16.5 ± 0.2 | 1.000 | 14.8 ± 0.4 | 1.000 |
| CEPHALOPODS | −17.2 ± 0.7 | − | 11.9 ± 1.1 | − | ||
| Sepia orbignyana | 5 | 73 ± 18 | −17.7 ± 0.2 | 0.001 | 10.6 ± 0.3 | <0.001 |
| Sepia elegans | 9 | 39 ± 16 | −17.3 ± 0.2 | 0.046 | 11.4 ± 0.7 | <0.001 |
| Sepia officinalis | 5 | 78 ± 11 | −16.2 ± 0.1 | 1.000 | 13.0 ± 0.5 | 0.392 |
| Alloteuthis spp. | 7 | 39 ± 13 | −17.7 ± 0.2 | <0.001 | 12.4 ± 0.4 | <0.001 |
| CRUSTACEANS | −16.1 ± 0.4 | − | 11.8 ± 0.4 | − | ||
| Liocarcinus depurator | 5 | 48 ± 2 | −16.2 ± 0.3 | 1.000 | 11.7 ± 0.7 | <0.001 |
| Polybius henslowii | 5 | 42 ± 3 | −16.5 ± 0.4 | 0.178 | 11.3 ± 0.7 | <0.001 |
| Crangon crangon | 5 | 54 ± 4 | −15.6 ± 0.4 | <0.001 | 12.1 ± 0.3 | <0.001 |
| Crangon allmanni | 5 | 54 ± 5 | −15.9 ± 0.2 | 0.159 | 12.2 ± 0.3 | <0.001 |
| Species . | N . | Size (mm) . | d13C (‰) . | d15N (‰) . | ||
|---|---|---|---|---|---|---|
| x ± sd . | x ± sd . | p-value . | x ± sd . | p-value . | ||
| PREDATOR | ||||||
| Dicentrarchus labrax | 15 | 585 ± 103 | −16.7 ± 0.6 | – | 14.0 ± 0.6 | – |
| PELAGIC FISH | −18.2 ± 0.6 | − | 11.6 ± 0.3 | – | ||
| Engraulis encrasicolus | 7 | 126 ± 4 | −17.6 ± 0.3 | 0.002 | 12.0 ± 0.4 | <0.001 |
| Sprattus sprattus | 9 | 103 ± 10 | −17.8 ± 0.2 | <0.001 | 11.9 ± 0.3 | <0.001 |
| Sardina pilchardus | 10 | 192 ± 48 | −17.7 ± 0.4 | <0.001 | 11.3 ± 0.8 | <0.001 |
| Trachurus trachurus | 10 | 170 ± 85 | −19.0 ± 0.9 | <0.001 | 11.4 ± 0.9 | <0.001 |
| Scomber scombrus | 10 | 257 ± 63 | −18.7 ± 0.4 | <0.001 | 11.3 ± 0.7 | <0.001 |
| Micromesistius poutassou | 9 | 220 ± 14 | −18.1 ± 0.3 | <0.001 | 11.4 ± 0.3 | <0.001 |
| DEMERSAL AND BENTHIC FISH | −17.0 ± 0.7 | − | 12.7 ± 0.7 | − | ||
| Argentina sphyraena | 6 | 187 ± 10 | −17.4 ± 0.2 | 0.078 | 12.5 ± 0.3 | <0.001 |
| Callionymus lyra | 5 | 222 ± 16 | −16.6 ± 0.3 | 1.000 | 12.5 ± 0.3 | <0.001 |
| Trisopterus minutus | 8 | 201 ± 20 | −17.2 ± 0.4 | 1.000 | 13.0 ± 0.5 | 0.032 |
| Trisopterus luscus | 5 | 184 ± 23 | −16.4 ± 0.1 | 1.000 | 14.1 ± 0.2 | 1.000 |
| Merluccius merluccius | 11 | 186 ± 54 | −18.3 ± 0.2 | <0.001 | 12.3 ± 0.3 | <0.001 |
| Pomatoschistus minutus | 5 | 56 ± 5 | −17.5 ± 0.1 | 0.030 | 12.7 ± 0.3 | 0.011 |
| Solea solea | 5 | 178 ± 13 | −16.3 ± 0.3 | 1.000 | 11.7 ± 0.4 | <0.001 |
| Microchirus variegatus | 5 | 162 ± 8 | −17.3 ± 0.0 | 1.000 | 12.2 ± 0.1 | <0.001 |
| Dicologlossa cuneata | 4 | 190 ± 18 | −16.6 ± 0.4 | 1.000 | 13.4 ± 0.7 | 1.000 |
| COASTAL FISH | −16.6 ± 0.1 | − | 13.6 ± 1.0 | − | ||
| Spondyliosoma cantharus | 5 | 142 ± 37 | −16.6 ± 0.8 | 1.000 | 12.3 ± 0.3 | <0.001 |
| Trachinus draco | 10 | 237 ± 20 | −16.7 ± 0.8 | 1.000 | 13.1 ± 1.3 | 0.039 |
| Merlangius merlangus | 10 | 116 ± 27 | −16.7 ± 0.3 | 1.000 | 13.8 ± 0.3 | 1.000 |
| Hyperoplus lanceolatus | 5 | 340 ± 14 | −16.4 ± 0.3 | 1.000 | 14.3 ± 0.3 | 1.000 |
| Atherina presbyter | 5 | 110 ± 10 | −16.5 ± 0.2 | 1.000 | 14.8 ± 0.4 | 1.000 |
| CEPHALOPODS | −17.2 ± 0.7 | − | 11.9 ± 1.1 | − | ||
| Sepia orbignyana | 5 | 73 ± 18 | −17.7 ± 0.2 | 0.001 | 10.6 ± 0.3 | <0.001 |
| Sepia elegans | 9 | 39 ± 16 | −17.3 ± 0.2 | 0.046 | 11.4 ± 0.7 | <0.001 |
| Sepia officinalis | 5 | 78 ± 11 | −16.2 ± 0.1 | 1.000 | 13.0 ± 0.5 | 0.392 |
| Alloteuthis spp. | 7 | 39 ± 13 | −17.7 ± 0.2 | <0.001 | 12.4 ± 0.4 | <0.001 |
| CRUSTACEANS | −16.1 ± 0.4 | − | 11.8 ± 0.4 | − | ||
| Liocarcinus depurator | 5 | 48 ± 2 | −16.2 ± 0.3 | 1.000 | 11.7 ± 0.7 | <0.001 |
| Polybius henslowii | 5 | 42 ± 3 | −16.5 ± 0.4 | 0.178 | 11.3 ± 0.7 | <0.001 |
| Crangon crangon | 5 | 54 ± 4 | −15.6 ± 0.4 | <0.001 | 12.1 ± 0.3 | <0.001 |
| Crangon allmanni | 5 | 54 ± 5 | −15.9 ± 0.2 | 0.159 | 12.2 ± 0.3 | <0.001 |
N = number of individual for each species, x = mean value, sd = standard deviation; p-value, significance of the statistical difference between signature of sea bass and signatures of each forage species. The mean values of forage species type correspond to the data point with standard deviation in Figure 5.
Carbon (δ13C) and nitrogen (δ15N) stable isotope raw values in the muscle of adult sea bass and forage species on the continental shelf of the Bay of Biscay.
| Species . | N . | Size (mm) . | d13C (‰) . | d15N (‰) . | ||
|---|---|---|---|---|---|---|
| x ± sd . | x ± sd . | p-value . | x ± sd . | p-value . | ||
| PREDATOR | ||||||
| Dicentrarchus labrax | 15 | 585 ± 103 | −16.7 ± 0.6 | – | 14.0 ± 0.6 | – |
| PELAGIC FISH | −18.2 ± 0.6 | − | 11.6 ± 0.3 | – | ||
| Engraulis encrasicolus | 7 | 126 ± 4 | −17.6 ± 0.3 | 0.002 | 12.0 ± 0.4 | <0.001 |
| Sprattus sprattus | 9 | 103 ± 10 | −17.8 ± 0.2 | <0.001 | 11.9 ± 0.3 | <0.001 |
| Sardina pilchardus | 10 | 192 ± 48 | −17.7 ± 0.4 | <0.001 | 11.3 ± 0.8 | <0.001 |
| Trachurus trachurus | 10 | 170 ± 85 | −19.0 ± 0.9 | <0.001 | 11.4 ± 0.9 | <0.001 |
| Scomber scombrus | 10 | 257 ± 63 | −18.7 ± 0.4 | <0.001 | 11.3 ± 0.7 | <0.001 |
| Micromesistius poutassou | 9 | 220 ± 14 | −18.1 ± 0.3 | <0.001 | 11.4 ± 0.3 | <0.001 |
| DEMERSAL AND BENTHIC FISH | −17.0 ± 0.7 | − | 12.7 ± 0.7 | − | ||
| Argentina sphyraena | 6 | 187 ± 10 | −17.4 ± 0.2 | 0.078 | 12.5 ± 0.3 | <0.001 |
| Callionymus lyra | 5 | 222 ± 16 | −16.6 ± 0.3 | 1.000 | 12.5 ± 0.3 | <0.001 |
| Trisopterus minutus | 8 | 201 ± 20 | −17.2 ± 0.4 | 1.000 | 13.0 ± 0.5 | 0.032 |
| Trisopterus luscus | 5 | 184 ± 23 | −16.4 ± 0.1 | 1.000 | 14.1 ± 0.2 | 1.000 |
| Merluccius merluccius | 11 | 186 ± 54 | −18.3 ± 0.2 | <0.001 | 12.3 ± 0.3 | <0.001 |
| Pomatoschistus minutus | 5 | 56 ± 5 | −17.5 ± 0.1 | 0.030 | 12.7 ± 0.3 | 0.011 |
| Solea solea | 5 | 178 ± 13 | −16.3 ± 0.3 | 1.000 | 11.7 ± 0.4 | <0.001 |
| Microchirus variegatus | 5 | 162 ± 8 | −17.3 ± 0.0 | 1.000 | 12.2 ± 0.1 | <0.001 |
| Dicologlossa cuneata | 4 | 190 ± 18 | −16.6 ± 0.4 | 1.000 | 13.4 ± 0.7 | 1.000 |
| COASTAL FISH | −16.6 ± 0.1 | − | 13.6 ± 1.0 | − | ||
| Spondyliosoma cantharus | 5 | 142 ± 37 | −16.6 ± 0.8 | 1.000 | 12.3 ± 0.3 | <0.001 |
| Trachinus draco | 10 | 237 ± 20 | −16.7 ± 0.8 | 1.000 | 13.1 ± 1.3 | 0.039 |
| Merlangius merlangus | 10 | 116 ± 27 | −16.7 ± 0.3 | 1.000 | 13.8 ± 0.3 | 1.000 |
| Hyperoplus lanceolatus | 5 | 340 ± 14 | −16.4 ± 0.3 | 1.000 | 14.3 ± 0.3 | 1.000 |
| Atherina presbyter | 5 | 110 ± 10 | −16.5 ± 0.2 | 1.000 | 14.8 ± 0.4 | 1.000 |
| CEPHALOPODS | −17.2 ± 0.7 | − | 11.9 ± 1.1 | − | ||
| Sepia orbignyana | 5 | 73 ± 18 | −17.7 ± 0.2 | 0.001 | 10.6 ± 0.3 | <0.001 |
| Sepia elegans | 9 | 39 ± 16 | −17.3 ± 0.2 | 0.046 | 11.4 ± 0.7 | <0.001 |
| Sepia officinalis | 5 | 78 ± 11 | −16.2 ± 0.1 | 1.000 | 13.0 ± 0.5 | 0.392 |
| Alloteuthis spp. | 7 | 39 ± 13 | −17.7 ± 0.2 | <0.001 | 12.4 ± 0.4 | <0.001 |
| CRUSTACEANS | −16.1 ± 0.4 | − | 11.8 ± 0.4 | − | ||
| Liocarcinus depurator | 5 | 48 ± 2 | −16.2 ± 0.3 | 1.000 | 11.7 ± 0.7 | <0.001 |
| Polybius henslowii | 5 | 42 ± 3 | −16.5 ± 0.4 | 0.178 | 11.3 ± 0.7 | <0.001 |
| Crangon crangon | 5 | 54 ± 4 | −15.6 ± 0.4 | <0.001 | 12.1 ± 0.3 | <0.001 |
| Crangon allmanni | 5 | 54 ± 5 | −15.9 ± 0.2 | 0.159 | 12.2 ± 0.3 | <0.001 |
| Species . | N . | Size (mm) . | d13C (‰) . | d15N (‰) . | ||
|---|---|---|---|---|---|---|
| x ± sd . | x ± sd . | p-value . | x ± sd . | p-value . | ||
| PREDATOR | ||||||
| Dicentrarchus labrax | 15 | 585 ± 103 | −16.7 ± 0.6 | – | 14.0 ± 0.6 | – |
| PELAGIC FISH | −18.2 ± 0.6 | − | 11.6 ± 0.3 | – | ||
| Engraulis encrasicolus | 7 | 126 ± 4 | −17.6 ± 0.3 | 0.002 | 12.0 ± 0.4 | <0.001 |
| Sprattus sprattus | 9 | 103 ± 10 | −17.8 ± 0.2 | <0.001 | 11.9 ± 0.3 | <0.001 |
| Sardina pilchardus | 10 | 192 ± 48 | −17.7 ± 0.4 | <0.001 | 11.3 ± 0.8 | <0.001 |
| Trachurus trachurus | 10 | 170 ± 85 | −19.0 ± 0.9 | <0.001 | 11.4 ± 0.9 | <0.001 |
| Scomber scombrus | 10 | 257 ± 63 | −18.7 ± 0.4 | <0.001 | 11.3 ± 0.7 | <0.001 |
| Micromesistius poutassou | 9 | 220 ± 14 | −18.1 ± 0.3 | <0.001 | 11.4 ± 0.3 | <0.001 |
| DEMERSAL AND BENTHIC FISH | −17.0 ± 0.7 | − | 12.7 ± 0.7 | − | ||
| Argentina sphyraena | 6 | 187 ± 10 | −17.4 ± 0.2 | 0.078 | 12.5 ± 0.3 | <0.001 |
| Callionymus lyra | 5 | 222 ± 16 | −16.6 ± 0.3 | 1.000 | 12.5 ± 0.3 | <0.001 |
| Trisopterus minutus | 8 | 201 ± 20 | −17.2 ± 0.4 | 1.000 | 13.0 ± 0.5 | 0.032 |
| Trisopterus luscus | 5 | 184 ± 23 | −16.4 ± 0.1 | 1.000 | 14.1 ± 0.2 | 1.000 |
| Merluccius merluccius | 11 | 186 ± 54 | −18.3 ± 0.2 | <0.001 | 12.3 ± 0.3 | <0.001 |
| Pomatoschistus minutus | 5 | 56 ± 5 | −17.5 ± 0.1 | 0.030 | 12.7 ± 0.3 | 0.011 |
| Solea solea | 5 | 178 ± 13 | −16.3 ± 0.3 | 1.000 | 11.7 ± 0.4 | <0.001 |
| Microchirus variegatus | 5 | 162 ± 8 | −17.3 ± 0.0 | 1.000 | 12.2 ± 0.1 | <0.001 |
| Dicologlossa cuneata | 4 | 190 ± 18 | −16.6 ± 0.4 | 1.000 | 13.4 ± 0.7 | 1.000 |
| COASTAL FISH | −16.6 ± 0.1 | − | 13.6 ± 1.0 | − | ||
| Spondyliosoma cantharus | 5 | 142 ± 37 | −16.6 ± 0.8 | 1.000 | 12.3 ± 0.3 | <0.001 |
| Trachinus draco | 10 | 237 ± 20 | −16.7 ± 0.8 | 1.000 | 13.1 ± 1.3 | 0.039 |
| Merlangius merlangus | 10 | 116 ± 27 | −16.7 ± 0.3 | 1.000 | 13.8 ± 0.3 | 1.000 |
| Hyperoplus lanceolatus | 5 | 340 ± 14 | −16.4 ± 0.3 | 1.000 | 14.3 ± 0.3 | 1.000 |
| Atherina presbyter | 5 | 110 ± 10 | −16.5 ± 0.2 | 1.000 | 14.8 ± 0.4 | 1.000 |
| CEPHALOPODS | −17.2 ± 0.7 | − | 11.9 ± 1.1 | − | ||
| Sepia orbignyana | 5 | 73 ± 18 | −17.7 ± 0.2 | 0.001 | 10.6 ± 0.3 | <0.001 |
| Sepia elegans | 9 | 39 ± 16 | −17.3 ± 0.2 | 0.046 | 11.4 ± 0.7 | <0.001 |
| Sepia officinalis | 5 | 78 ± 11 | −16.2 ± 0.1 | 1.000 | 13.0 ± 0.5 | 0.392 |
| Alloteuthis spp. | 7 | 39 ± 13 | −17.7 ± 0.2 | <0.001 | 12.4 ± 0.4 | <0.001 |
| CRUSTACEANS | −16.1 ± 0.4 | − | 11.8 ± 0.4 | − | ||
| Liocarcinus depurator | 5 | 48 ± 2 | −16.2 ± 0.3 | 1.000 | 11.7 ± 0.7 | <0.001 |
| Polybius henslowii | 5 | 42 ± 3 | −16.5 ± 0.4 | 0.178 | 11.3 ± 0.7 | <0.001 |
| Crangon crangon | 5 | 54 ± 4 | −15.6 ± 0.4 | <0.001 | 12.1 ± 0.3 | <0.001 |
| Crangon allmanni | 5 | 54 ± 5 | −15.9 ± 0.2 | 0.159 | 12.2 ± 0.3 | <0.001 |
N = number of individual for each species, x = mean value, sd = standard deviation; p-value, significance of the statistical difference between signature of sea bass and signatures of each forage species. The mean values of forage species type correspond to the data point with standard deviation in Figure 5.
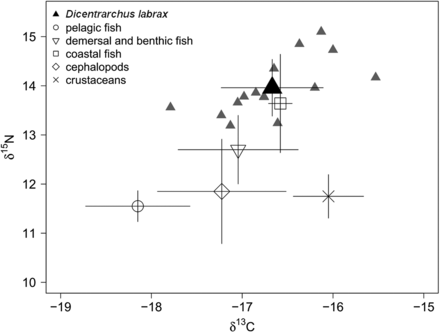
Carbon (δ13C) and nitrogen (δ15N) isotope signatures in muscle of adult sea bass (black triangles) and forage species on the shelf of the Bay of Biscay; data are mean (‰) ± standard deviation, all individual data points for sea bass are shown (grey triangles).
Stomach content analysis
Stomach content analysis describes the diet in terms of prey occurrence, relative abundance, calculated mass and size distribution, following a standard procedure for marine top predators (Pierce and Boyle, 1991; Pusineri et al., 2005; Spitz et al., 2006). The stomach contents were washed through a 0.2-mm mesh sieve. The diagnostic parts were recovered and stored dry (fish bones and otoliths) or in 70% ethanol (cephalopod beaks, crustacean remains and any remains with flesh attached). The items were identified to the lowest taxonomic level using published guides (Lagardère, 1971; Clarke, 1986; Härkönen, 1986) and our reference collection of specimens caught in the Bay of Biscay and adjacent Atlantic areas. The total number of food items was estimated as the highest number, given by either the number of paired structures (e.g. otoliths, opercula, and hyomandibular, dentary and premaxillary bones for fishes, upper and lower beaks for cephalopods, and eyes for crustaceans) or unpaired structures (e.g. parasphenoid for fishes, gladius for cephalopods, and carapace and telson for crustaceans). Diagnostic hard parts such as beaks, otoliths and carapaces were measured using digital vernier callipers (± 0.02 mm) following standards (Clarke, 1986; Härkönen, 1986). Individual prey body length and body mass were back-calculated using relationships from the literature (Clarke, 1986; Härkönen, 1986) or by fitting to measurements performed on the specimens in our reference collection. Body size distribution per prey species was defined as the body size for all individuals from each prey species, irrespective of the predator size. The prey size distributions were constructed in both number and biomass per size class, since these two variables convey different information about the importance of prey species in the diet.
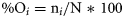
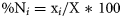
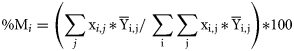
Selectivity index
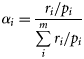
Stable isotope analysis
Muscle is the reference tissue used in food web studies based on stable isotope analyses (Hobson and Welch, 1992; Chouvelon et al., 2011). Stable isotope analysis of muscle allows a comparison of the isotopic signatures between different individuals and taxa, minimizing inter-tissue differences in terms of biochemical and physiological properties such as protein turnover rate and metabolic routing (Cherel et al., 2009). As lipids are highly depleted in 13C relative to other tissue components (DeNiro and Epstein, 1977), lipids were extracted from the muscle samples using cyclohexane (Chouvelon et al., 2011). Subsamples (0.40 ± 0.05 mg) of lipid-free powder were finally weighed in tin cups for stable isotope analyses. Isotopic analyses were performed using an elemental analyser coupled to an Isoprime (Micromass) continuous-flow isotope-ratio mass spectrometer (CF IR-MS). The results are presented as the usual δ notation relative to the deviation from standards (Pee Dee Belemnite for δ13C and atmospheric nitrogen for δ15N) in parts per thousand (‰). The experimental precision based on replicate measurements of internal laboratory standards was ±0.15‰ and ±0.20‰ for δ13C and δ15N, respectively. The significance of the differences between sea bass and each forage species was investigated using a Kruskal-Wallis test followed by a multiple comparison test with the Holm adjustment method.
Isotopic mixing model
A Bayesian isotopic mixing model was applied using the SIAR package (Stable Isotope Analysis in R) (Parnell et al., 2010) to estimate the proportional contribution of prey (sources) to the isotopic signature of adult sea bass (mixture). SIAR takes the isotopic signatures of a predator and its potential prey and fits a Bayesian model to generate the probability of each source proportion in the diet of the predator, based upon a Gaussian likelihood with a mixture Dirichlet-distributed prior on the mean. A strong advantage of the use of SIAR in isotopic modelling is its ability to account for variation in trophic enrichment factors (TEFs), as well as variation in prey and predator isotopic signatures. Hence, SIAR has become the most popular tool for interpreting prey-predator relationships from stable isotope signatures (Jaeger et al., 2009; Eguchi et al., 2011; Mèndez-Fernandez et al., 2012). Here, in order to create accurate mixing models, the prey were first grouped into five forage species types according to their habitat and ecology. The five groups were denoted as: pelagic fish from the shelf, demersal and benthic fish from the shelf, coastal fish, cephalopods, and crustaceans (Table 1). Secondly, as SIAR models are sensitive to assumptions regarding TEFs, we performed three mixing models using three different TEFs for fish muscle tissue from the literature (Pinnegar and Polunin, 1999; Trueman et al., 2005; Sweeting et al., 2007a, 2007b) (see values in Table 3). All of the tested models were in good agreement with mixing polygon assumptions.
Results of SIAR mixing models applied with different TEFs, showing the mean proportion (%) and standard deviation of each probable source in diet of adult sea bass on the shelf of the Bay of Biscay.
| . | Model 1 . | Model 2 . | Model 3 . |
|---|---|---|---|
| TEFs | Sweeting et al. 2007ab | Pinnegar and Polunin 1999 | Trueman et al. 2005 |
| Δδ13C | 1.7 ± 1.1 | 2.5 ± 0.1 | 2.1 ± 0.1 |
| Δδ15N | 3.2 ± 1.3 | 3.3 ± 0.2 | 2.3 ± 0.3 |
| POTENTIAL CONTRIBUTIONS | |||
| Pelagic fish | 69.1 ± 10.4 | 72.4 ± 15.8 | 74.7 ± 13.7 |
| Demersal and benthic fish | 6.5 ± 5.8 | 5.9 ± 6.0 | 5.8 ± 5.3 |
| Coastal fish | 3.6 ± 3.3 | 3.5 ± 3.5 | 3.6 ± 3.2 |
| Cephalopods | 14.2 ± 10.4 | 12.0 ± 11.1 | 11.1 ± 10.6 |
| Crustaceans | 6.6 ± 5.5 | 6.2 ± 6.5 | 4.8 ± 5.1 |
| . | Model 1 . | Model 2 . | Model 3 . |
|---|---|---|---|
| TEFs | Sweeting et al. 2007ab | Pinnegar and Polunin 1999 | Trueman et al. 2005 |
| Δδ13C | 1.7 ± 1.1 | 2.5 ± 0.1 | 2.1 ± 0.1 |
| Δδ15N | 3.2 ± 1.3 | 3.3 ± 0.2 | 2.3 ± 0.3 |
| POTENTIAL CONTRIBUTIONS | |||
| Pelagic fish | 69.1 ± 10.4 | 72.4 ± 15.8 | 74.7 ± 13.7 |
| Demersal and benthic fish | 6.5 ± 5.8 | 5.9 ± 6.0 | 5.8 ± 5.3 |
| Coastal fish | 3.6 ± 3.3 | 3.5 ± 3.5 | 3.6 ± 3.2 |
| Cephalopods | 14.2 ± 10.4 | 12.0 ± 11.1 | 11.1 ± 10.6 |
| Crustaceans | 6.6 ± 5.5 | 6.2 ± 6.5 | 4.8 ± 5.1 |
Results of SIAR mixing models applied with different TEFs, showing the mean proportion (%) and standard deviation of each probable source in diet of adult sea bass on the shelf of the Bay of Biscay.
| . | Model 1 . | Model 2 . | Model 3 . |
|---|---|---|---|
| TEFs | Sweeting et al. 2007ab | Pinnegar and Polunin 1999 | Trueman et al. 2005 |
| Δδ13C | 1.7 ± 1.1 | 2.5 ± 0.1 | 2.1 ± 0.1 |
| Δδ15N | 3.2 ± 1.3 | 3.3 ± 0.2 | 2.3 ± 0.3 |
| POTENTIAL CONTRIBUTIONS | |||
| Pelagic fish | 69.1 ± 10.4 | 72.4 ± 15.8 | 74.7 ± 13.7 |
| Demersal and benthic fish | 6.5 ± 5.8 | 5.9 ± 6.0 | 5.8 ± 5.3 |
| Coastal fish | 3.6 ± 3.3 | 3.5 ± 3.5 | 3.6 ± 3.2 |
| Cephalopods | 14.2 ± 10.4 | 12.0 ± 11.1 | 11.1 ± 10.6 |
| Crustaceans | 6.6 ± 5.5 | 6.2 ± 6.5 | 4.8 ± 5.1 |
| . | Model 1 . | Model 2 . | Model 3 . |
|---|---|---|---|
| TEFs | Sweeting et al. 2007ab | Pinnegar and Polunin 1999 | Trueman et al. 2005 |
| Δδ13C | 1.7 ± 1.1 | 2.5 ± 0.1 | 2.1 ± 0.1 |
| Δδ15N | 3.2 ± 1.3 | 3.3 ± 0.2 | 2.3 ± 0.3 |
| POTENTIAL CONTRIBUTIONS | |||
| Pelagic fish | 69.1 ± 10.4 | 72.4 ± 15.8 | 74.7 ± 13.7 |
| Demersal and benthic fish | 6.5 ± 5.8 | 5.9 ± 6.0 | 5.8 ± 5.3 |
| Coastal fish | 3.6 ± 3.3 | 3.5 ± 3.5 | 3.6 ± 3.2 |
| Cephalopods | 14.2 ± 10.4 | 12.0 ± 11.1 | 11.1 ± 10.6 |
| Crustaceans | 6.6 ± 5.5 | 6.2 ± 6.5 | 4.8 ± 5.1 |
Dietary overlap between sea bass and common dolphin
The dietary composition of common dolphin used in the present work comes from a previous analysis of stomach contents performed on dolphins stranded along the Atlantic coasts of the Bay of Biscay between 1999 and 2002 (Meynier et al., 2008) (summarized in Table 4). Briefly, the stomach contents from 71 common dolphins were analysed by prey occurrence, number and mass, following similar methods to the present work. The diet was dominated by small pelagic fish, mainly sardine, anchovy, sprat and horse mackerel.
Percent biomass of the main prey species found in stomach contents of adult sea bass and common dolphin on the continental shelf of the Bay of Biscay, northeastern Atlantic (prey species below 2% biomass excluded).
| Species . | Biomass M% . | |
|---|---|---|
| Sea Bass . | Common dolphin . | |
| (This study) . | (Meynier et al. 2008) . | |
| PELAGIC FISH | 78.8 | 78.6 |
| Clupeidae | ||
| Sardina pilchardus | 7.4 | 36.2 |
| Sprattus sprattus | 1.0 | 4.2 |
| Engraulidae | ||
| Engraulis encrasicolus | 10.2 | 12.4 |
| Carangidae | ||
| Trachurus trachurus | 20.1 | 19.2 |
| Scombridae | ||
| Scomber scombrus | 40.1 | 6.6 |
| DEMERSAL AND BENTHIC FISH | 8.4 | 14.3 |
| Merluccidae | ||
| Merluccius merluccius | 1.3 | 2.2 |
| Gadidae | ||
| Trisopterus spp. | 2.3 | 3.9 |
| Merlangius merlangus | 1.5 | 2.2 |
| Micromesistius poutassou | 3.3 | 6 |
| CEPHALOPODS | 2.5 | 2.6 |
| Loliginidae | ||
| Loligo spp. | 0.1 | 2.5 |
| Sepiidae | ||
| Sepia spp. | 2.4 | <0.1 |
| Species . | Biomass M% . | |
|---|---|---|
| Sea Bass . | Common dolphin . | |
| (This study) . | (Meynier et al. 2008) . | |
| PELAGIC FISH | 78.8 | 78.6 |
| Clupeidae | ||
| Sardina pilchardus | 7.4 | 36.2 |
| Sprattus sprattus | 1.0 | 4.2 |
| Engraulidae | ||
| Engraulis encrasicolus | 10.2 | 12.4 |
| Carangidae | ||
| Trachurus trachurus | 20.1 | 19.2 |
| Scombridae | ||
| Scomber scombrus | 40.1 | 6.6 |
| DEMERSAL AND BENTHIC FISH | 8.4 | 14.3 |
| Merluccidae | ||
| Merluccius merluccius | 1.3 | 2.2 |
| Gadidae | ||
| Trisopterus spp. | 2.3 | 3.9 |
| Merlangius merlangus | 1.5 | 2.2 |
| Micromesistius poutassou | 3.3 | 6 |
| CEPHALOPODS | 2.5 | 2.6 |
| Loliginidae | ||
| Loligo spp. | 0.1 | 2.5 |
| Sepiidae | ||
| Sepia spp. | 2.4 | <0.1 |
Percent biomass of the main prey species found in stomach contents of adult sea bass and common dolphin on the continental shelf of the Bay of Biscay, northeastern Atlantic (prey species below 2% biomass excluded).
| Species . | Biomass M% . | |
|---|---|---|
| Sea Bass . | Common dolphin . | |
| (This study) . | (Meynier et al. 2008) . | |
| PELAGIC FISH | 78.8 | 78.6 |
| Clupeidae | ||
| Sardina pilchardus | 7.4 | 36.2 |
| Sprattus sprattus | 1.0 | 4.2 |
| Engraulidae | ||
| Engraulis encrasicolus | 10.2 | 12.4 |
| Carangidae | ||
| Trachurus trachurus | 20.1 | 19.2 |
| Scombridae | ||
| Scomber scombrus | 40.1 | 6.6 |
| DEMERSAL AND BENTHIC FISH | 8.4 | 14.3 |
| Merluccidae | ||
| Merluccius merluccius | 1.3 | 2.2 |
| Gadidae | ||
| Trisopterus spp. | 2.3 | 3.9 |
| Merlangius merlangus | 1.5 | 2.2 |
| Micromesistius poutassou | 3.3 | 6 |
| CEPHALOPODS | 2.5 | 2.6 |
| Loliginidae | ||
| Loligo spp. | 0.1 | 2.5 |
| Sepiidae | ||
| Sepia spp. | 2.4 | <0.1 |
| Species . | Biomass M% . | |
|---|---|---|
| Sea Bass . | Common dolphin . | |
| (This study) . | (Meynier et al. 2008) . | |
| PELAGIC FISH | 78.8 | 78.6 |
| Clupeidae | ||
| Sardina pilchardus | 7.4 | 36.2 |
| Sprattus sprattus | 1.0 | 4.2 |
| Engraulidae | ||
| Engraulis encrasicolus | 10.2 | 12.4 |
| Carangidae | ||
| Trachurus trachurus | 20.1 | 19.2 |
| Scombridae | ||
| Scomber scombrus | 40.1 | 6.6 |
| DEMERSAL AND BENTHIC FISH | 8.4 | 14.3 |
| Merluccidae | ||
| Merluccius merluccius | 1.3 | 2.2 |
| Gadidae | ||
| Trisopterus spp. | 2.3 | 3.9 |
| Merlangius merlangus | 1.5 | 2.2 |
| Micromesistius poutassou | 3.3 | 6 |
| CEPHALOPODS | 2.5 | 2.6 |
| Loliginidae | ||
| Loligo spp. | 0.1 | 2.5 |
| Sepiidae | ||
| Sepia spp. | 2.4 | <0.1 |
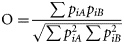
Results
Sea bass diet composition and local prey selectivity
Identifiable material was retrieved from 280 of the 404 stomachs. A total of 770 prey individuals were found, accounting for a total estimated biomass of approximately 16 kg. Fish, cephalopods, crustaceans, tubeworms and bivalves were identified and represented a species richness of at least 40 (24, 3, 11, 1 and 1 species, respectively). Fish dominated the diet (Table 2) both by number (87%) and mass (95%). Crustaceans reached a relative abundance of 9%, but accounted for a low fraction of the diet by reconstructed biomass (3%). Cephalopods accounted for a low fraction of the diet by both number and biomass. Tubeworms and bivalves were negligible in the diet.
Prey found in stomach contents of adult sea bass on the continental shelf of the Bay of Biscay, northeastern Atlantic.
| Species . | Occurrence . | Abundance . | Body length (mm) . | Body mass (g) . | Biomass . | ||
|---|---|---|---|---|---|---|---|
| O% . | N% . | x ± sd . | range . | x ± sd . | range . | M% . | |
| PELAGIC FISH | |||||||
| Clupeidae | |||||||
| Sardina pilchardus | 11.2 | 7.6 | 115 ± 33 | [69,231] | 20.9 ± 28.7 | [3.5,130.8] | 7.4 |
| Sprattus sprattus | 1.7 | 2.5 | 101 ± 10 | [84,126] | 8.2 ± 2.6 | [4.3,16.7] | 1.0 |
| Unid. Clupeidae | 0.8 | 0.3 | 8.5 | 0.1 | |||
| Engraulidae | |||||||
| Engraulis encrasicolus | 14.0 | 32.0 | 94 ± 14 | [62,266] | 6.8 ± 3.0 | [2.3,109.0] | 10.2 |
| Carangidae | |||||||
| Trachurus trachurus | 26.0 | 18.9 | 109 ± 44 | [52,295] | 22.7 ± 34.1 | [2.8,228.8] | 20.1 |
| Scombridae | |||||||
| Scomber scombrus | 11.6 | 5.6 | 237 ± 37 | [172,336] | 152.9 ± 71.6 | [35.4,300.7] | 40.1 |
| DEMERSAL AND BENTHIC FISH | |||||||
| Argentinidae | |||||||
| Argentina spp. | 1.2 | 0.6 | 177 ± 23 | [137,197] | 39.2 ± 14.4 | [15.3,53.4] | 1.0 |
| Callionymidae | |||||||
| Callionymus spp. | 8.7 | 8.4 | 76 ± 23 | [32,170] | 3.6 ± 5.1 | [0.2,33.6] | 1.4 |
| Gobiidae | |||||||
| Unid. Gobiidae | 4.5 | 1.8 | 55 ± 10 | [35,77] | 2.5 ± 1.7 | [0.4,5.3] | 0.2 |
| Merluccidae | |||||||
| Merluccius merluccius | 2.1 | 1.3 | 143 ± 24 | [102,190] | 21.8 ± 10.6 | [9.0,46.0] | 1.3 |
| Gadidae | |||||||
| Trisopterus spp. | 2.9 | 1.8 | 136 ± 32 | [85,192] | 27.3 ± 15.2 | [5.4,60.5] | 2.3 |
| Merlangius merlangus | 0.4 | 0.1 | 292.0 | 234.8 | 1.5 | ||
| Micromesistius poutassou | 3.7 | 1.3 | 200 ± 50 | [119,268] | 56.1 ± 37.4 | [11.1,131.4] | 3.3 |
| Unid. Gadidae | 0.4 | 0.1 | 51.2 | 0.3 | |||
| Soleidae | |||||||
| Unid. Soleidae | 0.4 | 0.1 | 173.0 | 45.0 | 0.3 | ||
| COASTAL FISH | |||||||
| Ammodytidae | |||||||
| Unid. Ammodytidae | 2.1 | 1.7 | 160 ±19 | [125,189] | 11.7 ± 3.4 | [4.8,17.2] | 0.9 |
| Atherinidae | |||||||
| Atherina presbyter | 0.8 | 0.3 | 53 ± 11 | [42,64] | 1.0 ± 0.6 | [0.4,1.6] | 0.0 |
| Sparidae | |||||||
| Pagellus sp. | 0.4 | 0.1 | 101.0 | 16.3 | 0.1 | ||
| Spondyliosoma cantharus | 0.6 | 0.4 | 138 ± 10 | [128,156] | 37.6 ± 9.3 | [29.1,54.6] | 0.7 |
| Unid. Sparidae | 0.4 | 0.1 | 122 ± 7 | [115,130] | 25.6 ± 4.8 | [20.7,30.4] | 0.2 |
| Syngnatidae | |||||||
| Unid. Syngnatidae | 0.8 | 0.3 | 189 ± 9 | [183,195] | 0.6 ± 0.1 | [0.6,0.6] | 0.0 |
| Trachinidae | |||||||
| Trachinus draco | 0.4 | 0.1 | 200 | 50 | 0.3 | ||
| Sciaenidae | |||||||
| Argyrosomus regius | 0.4 | 0.1 | 150.0 | 250.0 | 1.6 | ||
| OTHER FISH | |||||||
| Unid. Fish | 1.7 | 0.6 | |||||
| Larva | 0.4 | 1.0 | 27 ± 8 | [11,36] | 0.0 | ||
| CEPHALOPODS | |||||||
| Loliginidae | |||||||
| Alloteuthis spp. | 3.3 | 1.3 | 49 ± 23 | [26,104] | 3 ± 3.2 | [1.1,11.6] | 0.2 |
| Sepiidae | |||||||
| Sepia spp. | 5.4 | 1.8 | 22 ± 52 | [42,110] | 28.5 ± 37.7 | [2.1,93.0] | 2.4 |
| Sepiolidae | |||||||
| Unid. Sepiolidae | 0.8 | 0.3 | 20 | 2 | 0.0 | ||
| CRUSTACEANS | |||||||
| Brachyura | |||||||
| Atelecyclus undecimdentatus | 0.4 | 0.1 | |||||
| Corystes cassivelaunus | 0.8 | 0.4 | 23 ± 3 | [21,25] | 3.7 ± 0.4 | [3.3,4.1] | 0.1 |
| Macropodia spp. | 1.2 | 1.7 | 6 ± 1 | [4,8] | |||
| Pisidia longicornis | 0.8 | 0.3 | |||||
| Liocarcinus spp. | 3.7 | 1.7 | 29 ± 14 | [11,50] | 9.6 ± 9.1 | [0.2,23.2] | 0.8 |
| Necora puber | 0.8 | 0.3 | 31 ± 6 | [27,36] | 12.6 ± 0.5 | [12.2,12.9] | 0.2 |
| Polybius henslowi | 2.1 | 1.1 | 45 ± 4 | [37,50] | 18 ± 5.5 | [8.6,23.1] | 0.9 |
| Unid. Brachyura | 2.5 | 1.3 | 24 ± 11 | [8,33] | 4 ± 0.8 | [3.0,5.0] | 0.2 |
| Others crustaceans | |||||||
| Unid. Gammaridae | 2.5 | 0.8 | 10 | ||||
| Unid. Shrimps | 23.6 | - | 6 | 0.7 | |||
| Unid. Crustaceans | 3.3 | 1.1 | |||||
| OTHERS PREY | |||||||
| Tubeworm Annelida | 0.8 | 0.4 | 111 ± 47 | [62,155] | 2.7 ± 1.5 | [1.6,4.3] | 0.1 |
| Unid. Bivalves | 0.8 | 0.3 | |||||
| Species . | Occurrence . | Abundance . | Body length (mm) . | Body mass (g) . | Biomass . | ||
|---|---|---|---|---|---|---|---|
| O% . | N% . | x ± sd . | range . | x ± sd . | range . | M% . | |
| PELAGIC FISH | |||||||
| Clupeidae | |||||||
| Sardina pilchardus | 11.2 | 7.6 | 115 ± 33 | [69,231] | 20.9 ± 28.7 | [3.5,130.8] | 7.4 |
| Sprattus sprattus | 1.7 | 2.5 | 101 ± 10 | [84,126] | 8.2 ± 2.6 | [4.3,16.7] | 1.0 |
| Unid. Clupeidae | 0.8 | 0.3 | 8.5 | 0.1 | |||
| Engraulidae | |||||||
| Engraulis encrasicolus | 14.0 | 32.0 | 94 ± 14 | [62,266] | 6.8 ± 3.0 | [2.3,109.0] | 10.2 |
| Carangidae | |||||||
| Trachurus trachurus | 26.0 | 18.9 | 109 ± 44 | [52,295] | 22.7 ± 34.1 | [2.8,228.8] | 20.1 |
| Scombridae | |||||||
| Scomber scombrus | 11.6 | 5.6 | 237 ± 37 | [172,336] | 152.9 ± 71.6 | [35.4,300.7] | 40.1 |
| DEMERSAL AND BENTHIC FISH | |||||||
| Argentinidae | |||||||
| Argentina spp. | 1.2 | 0.6 | 177 ± 23 | [137,197] | 39.2 ± 14.4 | [15.3,53.4] | 1.0 |
| Callionymidae | |||||||
| Callionymus spp. | 8.7 | 8.4 | 76 ± 23 | [32,170] | 3.6 ± 5.1 | [0.2,33.6] | 1.4 |
| Gobiidae | |||||||
| Unid. Gobiidae | 4.5 | 1.8 | 55 ± 10 | [35,77] | 2.5 ± 1.7 | [0.4,5.3] | 0.2 |
| Merluccidae | |||||||
| Merluccius merluccius | 2.1 | 1.3 | 143 ± 24 | [102,190] | 21.8 ± 10.6 | [9.0,46.0] | 1.3 |
| Gadidae | |||||||
| Trisopterus spp. | 2.9 | 1.8 | 136 ± 32 | [85,192] | 27.3 ± 15.2 | [5.4,60.5] | 2.3 |
| Merlangius merlangus | 0.4 | 0.1 | 292.0 | 234.8 | 1.5 | ||
| Micromesistius poutassou | 3.7 | 1.3 | 200 ± 50 | [119,268] | 56.1 ± 37.4 | [11.1,131.4] | 3.3 |
| Unid. Gadidae | 0.4 | 0.1 | 51.2 | 0.3 | |||
| Soleidae | |||||||
| Unid. Soleidae | 0.4 | 0.1 | 173.0 | 45.0 | 0.3 | ||
| COASTAL FISH | |||||||
| Ammodytidae | |||||||
| Unid. Ammodytidae | 2.1 | 1.7 | 160 ±19 | [125,189] | 11.7 ± 3.4 | [4.8,17.2] | 0.9 |
| Atherinidae | |||||||
| Atherina presbyter | 0.8 | 0.3 | 53 ± 11 | [42,64] | 1.0 ± 0.6 | [0.4,1.6] | 0.0 |
| Sparidae | |||||||
| Pagellus sp. | 0.4 | 0.1 | 101.0 | 16.3 | 0.1 | ||
| Spondyliosoma cantharus | 0.6 | 0.4 | 138 ± 10 | [128,156] | 37.6 ± 9.3 | [29.1,54.6] | 0.7 |
| Unid. Sparidae | 0.4 | 0.1 | 122 ± 7 | [115,130] | 25.6 ± 4.8 | [20.7,30.4] | 0.2 |
| Syngnatidae | |||||||
| Unid. Syngnatidae | 0.8 | 0.3 | 189 ± 9 | [183,195] | 0.6 ± 0.1 | [0.6,0.6] | 0.0 |
| Trachinidae | |||||||
| Trachinus draco | 0.4 | 0.1 | 200 | 50 | 0.3 | ||
| Sciaenidae | |||||||
| Argyrosomus regius | 0.4 | 0.1 | 150.0 | 250.0 | 1.6 | ||
| OTHER FISH | |||||||
| Unid. Fish | 1.7 | 0.6 | |||||
| Larva | 0.4 | 1.0 | 27 ± 8 | [11,36] | 0.0 | ||
| CEPHALOPODS | |||||||
| Loliginidae | |||||||
| Alloteuthis spp. | 3.3 | 1.3 | 49 ± 23 | [26,104] | 3 ± 3.2 | [1.1,11.6] | 0.2 |
| Sepiidae | |||||||
| Sepia spp. | 5.4 | 1.8 | 22 ± 52 | [42,110] | 28.5 ± 37.7 | [2.1,93.0] | 2.4 |
| Sepiolidae | |||||||
| Unid. Sepiolidae | 0.8 | 0.3 | 20 | 2 | 0.0 | ||
| CRUSTACEANS | |||||||
| Brachyura | |||||||
| Atelecyclus undecimdentatus | 0.4 | 0.1 | |||||
| Corystes cassivelaunus | 0.8 | 0.4 | 23 ± 3 | [21,25] | 3.7 ± 0.4 | [3.3,4.1] | 0.1 |
| Macropodia spp. | 1.2 | 1.7 | 6 ± 1 | [4,8] | |||
| Pisidia longicornis | 0.8 | 0.3 | |||||
| Liocarcinus spp. | 3.7 | 1.7 | 29 ± 14 | [11,50] | 9.6 ± 9.1 | [0.2,23.2] | 0.8 |
| Necora puber | 0.8 | 0.3 | 31 ± 6 | [27,36] | 12.6 ± 0.5 | [12.2,12.9] | 0.2 |
| Polybius henslowi | 2.1 | 1.1 | 45 ± 4 | [37,50] | 18 ± 5.5 | [8.6,23.1] | 0.9 |
| Unid. Brachyura | 2.5 | 1.3 | 24 ± 11 | [8,33] | 4 ± 0.8 | [3.0,5.0] | 0.2 |
| Others crustaceans | |||||||
| Unid. Gammaridae | 2.5 | 0.8 | 10 | ||||
| Unid. Shrimps | 23.6 | - | 6 | 0.7 | |||
| Unid. Crustaceans | 3.3 | 1.1 | |||||
| OTHERS PREY | |||||||
| Tubeworm Annelida | 0.8 | 0.4 | 111 ± 47 | [62,155] | 2.7 ± 1.5 | [1.6,4.3] | 0.1 |
| Unid. Bivalves | 0.8 | 0.3 | |||||
Prey found in stomach contents of adult sea bass on the continental shelf of the Bay of Biscay, northeastern Atlantic.
| Species . | Occurrence . | Abundance . | Body length (mm) . | Body mass (g) . | Biomass . | ||
|---|---|---|---|---|---|---|---|
| O% . | N% . | x ± sd . | range . | x ± sd . | range . | M% . | |
| PELAGIC FISH | |||||||
| Clupeidae | |||||||
| Sardina pilchardus | 11.2 | 7.6 | 115 ± 33 | [69,231] | 20.9 ± 28.7 | [3.5,130.8] | 7.4 |
| Sprattus sprattus | 1.7 | 2.5 | 101 ± 10 | [84,126] | 8.2 ± 2.6 | [4.3,16.7] | 1.0 |
| Unid. Clupeidae | 0.8 | 0.3 | 8.5 | 0.1 | |||
| Engraulidae | |||||||
| Engraulis encrasicolus | 14.0 | 32.0 | 94 ± 14 | [62,266] | 6.8 ± 3.0 | [2.3,109.0] | 10.2 |
| Carangidae | |||||||
| Trachurus trachurus | 26.0 | 18.9 | 109 ± 44 | [52,295] | 22.7 ± 34.1 | [2.8,228.8] | 20.1 |
| Scombridae | |||||||
| Scomber scombrus | 11.6 | 5.6 | 237 ± 37 | [172,336] | 152.9 ± 71.6 | [35.4,300.7] | 40.1 |
| DEMERSAL AND BENTHIC FISH | |||||||
| Argentinidae | |||||||
| Argentina spp. | 1.2 | 0.6 | 177 ± 23 | [137,197] | 39.2 ± 14.4 | [15.3,53.4] | 1.0 |
| Callionymidae | |||||||
| Callionymus spp. | 8.7 | 8.4 | 76 ± 23 | [32,170] | 3.6 ± 5.1 | [0.2,33.6] | 1.4 |
| Gobiidae | |||||||
| Unid. Gobiidae | 4.5 | 1.8 | 55 ± 10 | [35,77] | 2.5 ± 1.7 | [0.4,5.3] | 0.2 |
| Merluccidae | |||||||
| Merluccius merluccius | 2.1 | 1.3 | 143 ± 24 | [102,190] | 21.8 ± 10.6 | [9.0,46.0] | 1.3 |
| Gadidae | |||||||
| Trisopterus spp. | 2.9 | 1.8 | 136 ± 32 | [85,192] | 27.3 ± 15.2 | [5.4,60.5] | 2.3 |
| Merlangius merlangus | 0.4 | 0.1 | 292.0 | 234.8 | 1.5 | ||
| Micromesistius poutassou | 3.7 | 1.3 | 200 ± 50 | [119,268] | 56.1 ± 37.4 | [11.1,131.4] | 3.3 |
| Unid. Gadidae | 0.4 | 0.1 | 51.2 | 0.3 | |||
| Soleidae | |||||||
| Unid. Soleidae | 0.4 | 0.1 | 173.0 | 45.0 | 0.3 | ||
| COASTAL FISH | |||||||
| Ammodytidae | |||||||
| Unid. Ammodytidae | 2.1 | 1.7 | 160 ±19 | [125,189] | 11.7 ± 3.4 | [4.8,17.2] | 0.9 |
| Atherinidae | |||||||
| Atherina presbyter | 0.8 | 0.3 | 53 ± 11 | [42,64] | 1.0 ± 0.6 | [0.4,1.6] | 0.0 |
| Sparidae | |||||||
| Pagellus sp. | 0.4 | 0.1 | 101.0 | 16.3 | 0.1 | ||
| Spondyliosoma cantharus | 0.6 | 0.4 | 138 ± 10 | [128,156] | 37.6 ± 9.3 | [29.1,54.6] | 0.7 |
| Unid. Sparidae | 0.4 | 0.1 | 122 ± 7 | [115,130] | 25.6 ± 4.8 | [20.7,30.4] | 0.2 |
| Syngnatidae | |||||||
| Unid. Syngnatidae | 0.8 | 0.3 | 189 ± 9 | [183,195] | 0.6 ± 0.1 | [0.6,0.6] | 0.0 |
| Trachinidae | |||||||
| Trachinus draco | 0.4 | 0.1 | 200 | 50 | 0.3 | ||
| Sciaenidae | |||||||
| Argyrosomus regius | 0.4 | 0.1 | 150.0 | 250.0 | 1.6 | ||
| OTHER FISH | |||||||
| Unid. Fish | 1.7 | 0.6 | |||||
| Larva | 0.4 | 1.0 | 27 ± 8 | [11,36] | 0.0 | ||
| CEPHALOPODS | |||||||
| Loliginidae | |||||||
| Alloteuthis spp. | 3.3 | 1.3 | 49 ± 23 | [26,104] | 3 ± 3.2 | [1.1,11.6] | 0.2 |
| Sepiidae | |||||||
| Sepia spp. | 5.4 | 1.8 | 22 ± 52 | [42,110] | 28.5 ± 37.7 | [2.1,93.0] | 2.4 |
| Sepiolidae | |||||||
| Unid. Sepiolidae | 0.8 | 0.3 | 20 | 2 | 0.0 | ||
| CRUSTACEANS | |||||||
| Brachyura | |||||||
| Atelecyclus undecimdentatus | 0.4 | 0.1 | |||||
| Corystes cassivelaunus | 0.8 | 0.4 | 23 ± 3 | [21,25] | 3.7 ± 0.4 | [3.3,4.1] | 0.1 |
| Macropodia spp. | 1.2 | 1.7 | 6 ± 1 | [4,8] | |||
| Pisidia longicornis | 0.8 | 0.3 | |||||
| Liocarcinus spp. | 3.7 | 1.7 | 29 ± 14 | [11,50] | 9.6 ± 9.1 | [0.2,23.2] | 0.8 |
| Necora puber | 0.8 | 0.3 | 31 ± 6 | [27,36] | 12.6 ± 0.5 | [12.2,12.9] | 0.2 |
| Polybius henslowi | 2.1 | 1.1 | 45 ± 4 | [37,50] | 18 ± 5.5 | [8.6,23.1] | 0.9 |
| Unid. Brachyura | 2.5 | 1.3 | 24 ± 11 | [8,33] | 4 ± 0.8 | [3.0,5.0] | 0.2 |
| Others crustaceans | |||||||
| Unid. Gammaridae | 2.5 | 0.8 | 10 | ||||
| Unid. Shrimps | 23.6 | - | 6 | 0.7 | |||
| Unid. Crustaceans | 3.3 | 1.1 | |||||
| OTHERS PREY | |||||||
| Tubeworm Annelida | 0.8 | 0.4 | 111 ± 47 | [62,155] | 2.7 ± 1.5 | [1.6,4.3] | 0.1 |
| Unid. Bivalves | 0.8 | 0.3 | |||||
| Species . | Occurrence . | Abundance . | Body length (mm) . | Body mass (g) . | Biomass . | ||
|---|---|---|---|---|---|---|---|
| O% . | N% . | x ± sd . | range . | x ± sd . | range . | M% . | |
| PELAGIC FISH | |||||||
| Clupeidae | |||||||
| Sardina pilchardus | 11.2 | 7.6 | 115 ± 33 | [69,231] | 20.9 ± 28.7 | [3.5,130.8] | 7.4 |
| Sprattus sprattus | 1.7 | 2.5 | 101 ± 10 | [84,126] | 8.2 ± 2.6 | [4.3,16.7] | 1.0 |
| Unid. Clupeidae | 0.8 | 0.3 | 8.5 | 0.1 | |||
| Engraulidae | |||||||
| Engraulis encrasicolus | 14.0 | 32.0 | 94 ± 14 | [62,266] | 6.8 ± 3.0 | [2.3,109.0] | 10.2 |
| Carangidae | |||||||
| Trachurus trachurus | 26.0 | 18.9 | 109 ± 44 | [52,295] | 22.7 ± 34.1 | [2.8,228.8] | 20.1 |
| Scombridae | |||||||
| Scomber scombrus | 11.6 | 5.6 | 237 ± 37 | [172,336] | 152.9 ± 71.6 | [35.4,300.7] | 40.1 |
| DEMERSAL AND BENTHIC FISH | |||||||
| Argentinidae | |||||||
| Argentina spp. | 1.2 | 0.6 | 177 ± 23 | [137,197] | 39.2 ± 14.4 | [15.3,53.4] | 1.0 |
| Callionymidae | |||||||
| Callionymus spp. | 8.7 | 8.4 | 76 ± 23 | [32,170] | 3.6 ± 5.1 | [0.2,33.6] | 1.4 |
| Gobiidae | |||||||
| Unid. Gobiidae | 4.5 | 1.8 | 55 ± 10 | [35,77] | 2.5 ± 1.7 | [0.4,5.3] | 0.2 |
| Merluccidae | |||||||
| Merluccius merluccius | 2.1 | 1.3 | 143 ± 24 | [102,190] | 21.8 ± 10.6 | [9.0,46.0] | 1.3 |
| Gadidae | |||||||
| Trisopterus spp. | 2.9 | 1.8 | 136 ± 32 | [85,192] | 27.3 ± 15.2 | [5.4,60.5] | 2.3 |
| Merlangius merlangus | 0.4 | 0.1 | 292.0 | 234.8 | 1.5 | ||
| Micromesistius poutassou | 3.7 | 1.3 | 200 ± 50 | [119,268] | 56.1 ± 37.4 | [11.1,131.4] | 3.3 |
| Unid. Gadidae | 0.4 | 0.1 | 51.2 | 0.3 | |||
| Soleidae | |||||||
| Unid. Soleidae | 0.4 | 0.1 | 173.0 | 45.0 | 0.3 | ||
| COASTAL FISH | |||||||
| Ammodytidae | |||||||
| Unid. Ammodytidae | 2.1 | 1.7 | 160 ±19 | [125,189] | 11.7 ± 3.4 | [4.8,17.2] | 0.9 |
| Atherinidae | |||||||
| Atherina presbyter | 0.8 | 0.3 | 53 ± 11 | [42,64] | 1.0 ± 0.6 | [0.4,1.6] | 0.0 |
| Sparidae | |||||||
| Pagellus sp. | 0.4 | 0.1 | 101.0 | 16.3 | 0.1 | ||
| Spondyliosoma cantharus | 0.6 | 0.4 | 138 ± 10 | [128,156] | 37.6 ± 9.3 | [29.1,54.6] | 0.7 |
| Unid. Sparidae | 0.4 | 0.1 | 122 ± 7 | [115,130] | 25.6 ± 4.8 | [20.7,30.4] | 0.2 |
| Syngnatidae | |||||||
| Unid. Syngnatidae | 0.8 | 0.3 | 189 ± 9 | [183,195] | 0.6 ± 0.1 | [0.6,0.6] | 0.0 |
| Trachinidae | |||||||
| Trachinus draco | 0.4 | 0.1 | 200 | 50 | 0.3 | ||
| Sciaenidae | |||||||
| Argyrosomus regius | 0.4 | 0.1 | 150.0 | 250.0 | 1.6 | ||
| OTHER FISH | |||||||
| Unid. Fish | 1.7 | 0.6 | |||||
| Larva | 0.4 | 1.0 | 27 ± 8 | [11,36] | 0.0 | ||
| CEPHALOPODS | |||||||
| Loliginidae | |||||||
| Alloteuthis spp. | 3.3 | 1.3 | 49 ± 23 | [26,104] | 3 ± 3.2 | [1.1,11.6] | 0.2 |
| Sepiidae | |||||||
| Sepia spp. | 5.4 | 1.8 | 22 ± 52 | [42,110] | 28.5 ± 37.7 | [2.1,93.0] | 2.4 |
| Sepiolidae | |||||||
| Unid. Sepiolidae | 0.8 | 0.3 | 20 | 2 | 0.0 | ||
| CRUSTACEANS | |||||||
| Brachyura | |||||||
| Atelecyclus undecimdentatus | 0.4 | 0.1 | |||||
| Corystes cassivelaunus | 0.8 | 0.4 | 23 ± 3 | [21,25] | 3.7 ± 0.4 | [3.3,4.1] | 0.1 |
| Macropodia spp. | 1.2 | 1.7 | 6 ± 1 | [4,8] | |||
| Pisidia longicornis | 0.8 | 0.3 | |||||
| Liocarcinus spp. | 3.7 | 1.7 | 29 ± 14 | [11,50] | 9.6 ± 9.1 | [0.2,23.2] | 0.8 |
| Necora puber | 0.8 | 0.3 | 31 ± 6 | [27,36] | 12.6 ± 0.5 | [12.2,12.9] | 0.2 |
| Polybius henslowi | 2.1 | 1.1 | 45 ± 4 | [37,50] | 18 ± 5.5 | [8.6,23.1] | 0.9 |
| Unid. Brachyura | 2.5 | 1.3 | 24 ± 11 | [8,33] | 4 ± 0.8 | [3.0,5.0] | 0.2 |
| Others crustaceans | |||||||
| Unid. Gammaridae | 2.5 | 0.8 | 10 | ||||
| Unid. Shrimps | 23.6 | - | 6 | 0.7 | |||
| Unid. Crustaceans | 3.3 | 1.1 | |||||
| OTHERS PREY | |||||||
| Tubeworm Annelida | 0.8 | 0.4 | 111 ± 47 | [62,155] | 2.7 ± 1.5 | [1.6,4.3] | 0.1 |
| Unid. Bivalves | 0.8 | 0.3 | |||||
The diet of adult sea bass on the shelf of the Bay of Biscay was comprised mainly of a combination of pelagic fish (Table 2). Four fish species made up 77.8% of the biomass. Despite a low relative abundance (5.6%), mackerel (Scomber scombrus) was the most important prey in term of ingested biomass (40.1%), followed by scads (Trachurus trachurus and/or T. mediterraneus, 20.1%), anchovy (Engraulis encrasicolus, 10.2%) and sardine (Sardina pilchardus, 7.4%). The other 36 prey species accounted for less than 2% of the diet by either number or biomass, with the exception of sprat (Sprattus sprattus, 2.5% in number), poor cod and codling, (Trisopterus luscus and/or minutus, 2.3% in weight) and blue whiting (Micromesistius poutassou, 3.3% in weight).
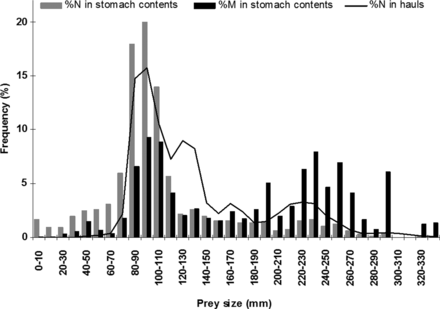
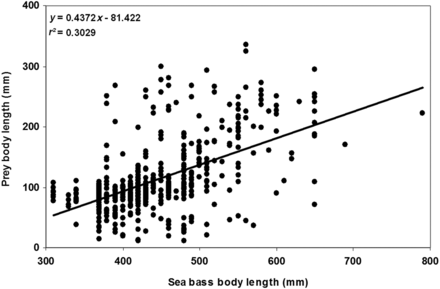
The overall prey size distribution ranged from 43–35 mm (Figure 2). The distribution by number displayed a mode at 70–120 mm; this mode resulted from the high relative abundance of smaller fish; mainly anchovy, scads, sardine and dragonets (Callionymus spp.). In contrast, the distribution by mass showed two modes at 80–120 mm and 200–300 mm; the second mode mostly resulted from the importance of large fish by mass, such as mackerel and larger individuals of scads or sardine. Overall, 77% of all prey individuals were < 120 mm and 7% had a body length > 220 mm. Conversely, prey individuals < 120 mm represented only 34% of the reconstructed biomass, whereas those > 220 mm accounted for 41% of the biomass. Analysis of the relationship between individual sea bass and prey body length revealed a slight increase in prey size during the adult ontogeny of sea bass (r2 = 0.3029, Pearson correlation test p < 0.001; Figure 3). However, the size diversity appeared to be relatively wide (∼ 20 cm) and constant.
Overall prey-size distributions expressed as percent number in stomach contents of sea bass (black bars), percent mass in stomach contents of sea bass (grey bars), and percent number in hauls where sea bass were caught in 2005, 2006 and 2008 (black line).
Relationship between individual sea bass and prey body length.
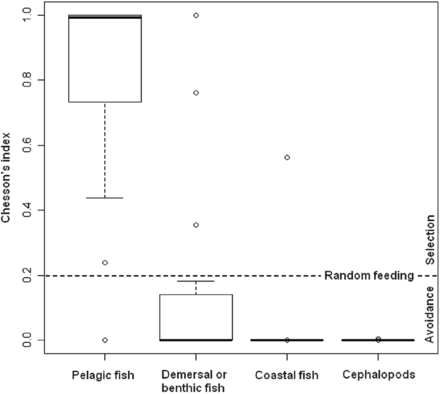
Evaluation of local prey selectivity (trawl by trawl), as given by Chesson's index, revealed that sea bass strongly selected pelagic fish. The median Chesson's index was > 0.9 for pelagic fish; and the value of Chesson's index is under the limit of positive selection for only one trawl (Figure 4). Other prey types were avoided, with the exception of demersal or benthic fish which may be randomly preyed upon or secondarily selected. Furthermore, the length distribution of all catch (in the hauls in which sea bass were caught) was close to the length distribution of the prey found in the sea bass stomach contents (Figure 2), suggesting that the prey field was correctly sampled in the trawls, at least in terms of potential prey size.
Boxplot of Chesson's index from 15 scientific hauls where stomachs of sea bass were sampled and diagnostic of prey selection (Chesson's index close to the horizontal dot line represents a random feeding, value above the line represent a positive prey selection, value below an avoidance). The bold solid line within each box is the median, and the bottom and top of each box represent the 25th and 75th percentiles, respectively. The whiskers represent the 10th and 90th percentiles, respectively, and values outside this range are plotted as individual outliers.
Stable isotope signatures and mixing model
The mean δ15N value for adult sea bass muscle was 13.8 ± 0.5 and the mean δ13C value was –17.0 ± 0.5 (Figure 5). The stable isotope signatures of costal fish were close to those of sea bass. Cephalopods, demersal and benthic fish exhibited lower δ15N values and similar δ13C values to sea bass. Crustaceans showed lower δ15N values and higher δ13C values. Almost all pelagic fish species had significantly lower stable isotope signatures than sea bass for both δ15N and δ13C.
Consequently, the mixing model estimated that pelagic fish were the main source in the sea bass diet. Using three different mixing models to evaluate three different TEFs revealed that the mean pelagic fish contribution ranged from 69.1 ± 10.4% to 74.7 ± 13.7% (Table 3). All mixing models suggested that other forage species were less important in the diet of adult sea bass. The potential contribution of cephalopods ranked second, with a mean contribution of 11.1 ± 10.6% to 14.2 ± 10.4%; the potential contributions of demersal and benthic fish, coastal fish and crustaceans were < 7%.
Dietary overlap
On the continental shelf of the Bay of Biscay, the diet of common dolphin is dominated by small pelagic fish; mainly scads, sardine and anchovy (Table 4). Thus, small pelagic fish constitute the core of both the common dolphin and sea bass diet. The dietary overlap, as estimated by the Pianka index, revealed a high degree of overlap: 0.52 at the prey species level and 0.99 at the prey group level. This first comparison of the diet of common dolphin and sea bass suggested that these marine top predators share similar feeding niches.
Discussion
Feeding ecology of adult sea bass
The present work is the first quantitative study of adult sea bass in the northeastern Atlantic, outside of estuarine and coastal waters. We suggest that sea bass are mainly piscivorous and preferentially feed on small pelagic fish: mainly mackerel, sardine, anchovy and scads. This prey choice is supported by both the Chesson's index of prey selection and the isotopic signatures. Furthermore, the δ13C signature of adult sea bass is more characteristic of the open waters of the continental shelf, rather than the coastal waters in the Bay of Biscay (Chouvelon et al., 2011). Species living in estuarine and coastal habitats exhibit an enriched δ13C signature due to differential carbon fixation by benthic algae in coastal areas and offshore phytoplankton (France, 1995). Thus, based on the rate of muscle turnover (Buchheister and Latour, 2010), the sea bass sampled in this study appear to have reliably exploited the offshore area of the continental shelf, at least during the previous several weeks to a few months.
Nevertheless, several limitations are inherent in the approaches used in this study. Stomach content analysis is based on the recovery and identification of undigested remains. The representativeness of the diet described by stomach content analysis can be undermined by the difficulty of controlling the sampling design and the differential digestion of ingested prey (Pierce and Boyle, 1991; Tollit et al., 1997). However, stomach content analysis is regarded as the best and most widely used method to quantitatively evaluate the prey composition of top marine predators. The prey choice observed in the stomach content analysis in this study was confirmed on a fine scale by the Chesson's index of prey selection. This index is generally used for analysis of a two-prey system, by determining the global dietary composition of the predator, and surveying prey availability within the distribution area of the studied predator (Pinnegar et al., 2003; Spitz et al., 2010). Here, the novelty is the haul was used as the sampling unit; hence, Chesson's index provides a test of instantaneous prey choice, as it compares the prey ingested by the sea bass with the local abundance of forage species that were actually available during foraging. However, the representativeness of the prey field available for sea bass from trawling can be affected by methodological constraints (i.e. trawl dimensions, mesh size, haul duration) and the differential escape capabilities of target species for a given trawl (Wardle, 1993). Thus, both stomach content analysis and trawling are subject to their own selectivity and biases, which could affect our perception of sea bass diets and forage species abundance. However, despite these sources of uncertainty and the low number of trawl hauls, the calculation of prey selectivity provided consistent results; therefore, the values for Chesson's index provided in the present work reveal the general patterns of prey selectivity, rather than represent a precise measure of prey selectivity.
Stable isotope analysis also has a number of limitations, in particular because different prey compositions may lead to the same isotopic signature in the predator's tissues (Bearhop et al., 2004), and some forage species which are absent in the diet of a given predator could have similar isotopic signatures as the prey eaten by the predator. These limitations, and the assumptions associated with TEFs, increase the uncertainty of isotopic mixing modelling to determine potential dietary contributions (Parnell et al., 2010; Bond and Diamond, 2011). In the present study, the results of three different isotopic models were consistent with the stomach contents of the sea bass. The confidence intervals of the potential contributions provided by the three mixing models, which applied three different TEFs, included the relative proportions of each prey type provided by the stomach content analysis. Thus, the mixed models confirmed the selectivity of sea bass for small pelagic fish, as suggested by the stomach content analysis, though the contributions of other prey types were higher than that suggested by the stomach content analysis, especially for cephalopods. However, the mean dietary contribution values proposed by SIAR need to be interpreted with caution, as mixing models can only generate potential contributions. The mean dietary contribution of each prey type should not be directly compared with the relative proportion of prey found in the stomach contents for three reasons: first, the potential sources of uncertainty (e.g. reliability of species grouping, TEF, sample size); second, the isotopic signatures reflect the assimilated food and not the ingested food; and finally, isotopic signatures and stomach contents express dietary preferences over two distinct time scales. Additionally, given the seasonal or annual variability in prey abundance and the potential biases in both the dietary and fish community descriptions, the values obtained in the present work should be interpreted as revealing the general patterns of sea bass prey preferences for small pelagic fish. Despite these limitations, this study reveals the usefulness of stable isotope analysis and mixing models, in combination with stomach content analysis, in assessing the prey preferences in the diet of top predators.
Comparison with previous studies
The diet and dietary resources of juvenile sea bass have been extensively studied in estuaries and coastal areas (Aprahamian and Barr, 1985; Cabral and Costa, 2001; Laffaille et al., 2001; Riley et al., 2011). Sea bass has been described as a demersal predator feeding on planktonic crustaceans during its juvenile stage. At later stages, its diet was thought to include a diverse epibenthic fauna and some fish for the largest individuals. Even so, sea bass is generally described as an opportunistic feeder at each stage, i.e. its diet would reflect prey availability in its foraging area (Pickett and Pawson, 1994), and prey diversity would be larger for adults than for juveniles (Rogdakis et al., 2010). A shift from benthic crustaceans in the juvenile stage to pelagic fish in adult sea bass was suggested; however, this shift had not yet been supported by quantitative data on adult feeding.
Our quantitative analysis of adult sea bass feeding runs counter to the generally accepted view. Here, almost 80% of the ingested biomass comprised only four pelagic fish species, although more than 40 species were found in the stomachs of the sea bass, including fish, cephalopods, crustaceans, tubeworms and bivalves. Hence, studies that form conclusions on the basis of prey occurrence or prey abundance may lead to a false picture of the diet of top marine predators. Therefore, our results complete the previous knowledge and reveal a shift from pelagic and benthic invertebrates in the diet of juvenile sea bass to a piscivorous diet relying on small pelagic fish in adult sea bass.
Interaction between sea bass and common dolphin
Our findings on the foraging ecology of adult sea bass could have implications for the management of a protected top predator, the common dolphin (Delphinus delphis). Indeed, the selective feeding of sea bass on small pelagic fish could also explain the operational fishery interaction with the common dolphin. Like sea bass, the diet of common dolphin in the Bay of Biscay is dominated by small pelagic fish (Meynier et al., 2008); this study suggests a considerable dietary overlap between these predator species both in terms of prey species and also in prey size (see Figure 3 in Meynier et al., 2008 for common dolphin prey size distribution). Moreover, analysis of the stomachs of dolphins taken as bycatch revealed a very high proportion of samples with fresh remains (Spitz, unpublished data), indicating the dolphins were feeding just prior to their death. The similar diets and bycatch of dolphins in the pelagic fishery for sea bass suggests the simultaneous foraging of these species. Therefore, some behavioural interactions could occur, such as the cooperative feeding observed between dolphin species and tuna in oceanic areas (Clua and Grosvalet, 2001). The precise foraging strategies of common dolphin and adult sea bass are yet to be fully described, and this hypothesis requires further studies, possibly using acoustic and video recording, in order to better understand the potential interaction. However, we suggest that the bycatch risk of common dolphin in pelagic sea bass fisheries is closely linked to the similar foraging strategies of these predator species. Improved understanding of the ecological or behavioural processes occurring between sea bass and common dolphin would allow the identification of strategies to minimize dolphin bycatch.
Funding
EVHOE surveys are funded by the European Union in application of the Council Regulation (EC) No.199/2008 of 25 February 2008. This research has been supported by the European projects REPRODUCE (ERAC-CT-2006-025989, FP7) and FACTS (No. 244966, FP7) and by the CPER 2009–2013 (Contrat de Projet Etat-Région).
Acknowledgements
The work was part of a larger research programme on the forage species–top predators interactions in the Bay of Biscay and adjacent Atlantic areas. We are particularly grateful to J-C. Mahé, R. Bellail and J-P. Leauté (Ifremer), and the crew of the R/V “Thalassa” for their support during EVHOE surveys, and also to the crews of professional pelagic trawlers which allowed us to extend our sampling design. Many thanks go to G. Gautier†, W. Dabin and H. Peltier for their help in sampling or stomach content analysis, and to G. Guillou for his help on stable isotope analysis. Finally, the authors also thank the two reviewers for their helpful comments that greatly improved the manuscript.
References
Author notes
Handling editor: Francis Juanes