-
PDF
- Split View
-
Views
-
Cite
Cite
Hojun Song, Ricardo Mariño-Pérez, Derek A Woller, Maria Marta Cigliano, Evolution, Diversification, and Biogeography of Grasshoppers (Orthoptera: Acrididae), Insect Systematics and Diversity, Volume 2, Issue 4, July 2018, 3, https://doi.org/10.1093/isd/ixy008
Close - Share Icon Share
Abstract
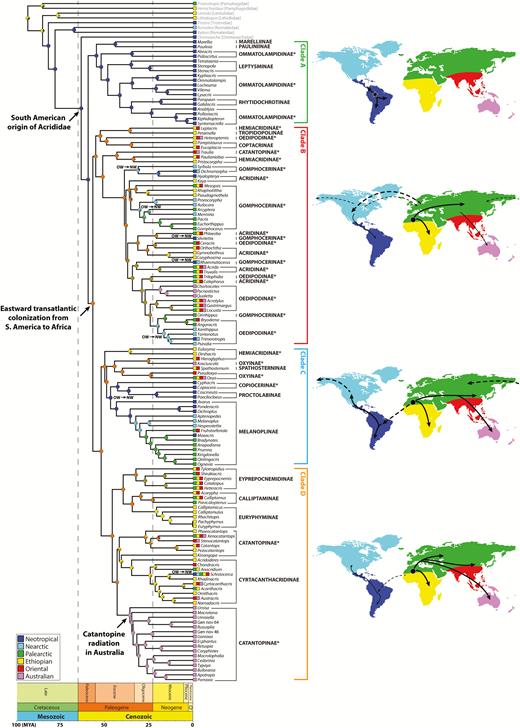
The grasshopper family Acrididae is one of the most diverse lineages within Orthoptera, including more than 6,700 valid species distributed worldwide. Grasshoppers are dominant herbivores, which have diversified into grassland, desert, semi-aquatic, alpine, and tropical forest habitats, and exhibit a wide array of morphological, ecological, and behavioral diversity. Nevertheless, the phylogeny of Acrididae as a whole has never been proposed. In this study, we present the first comprehensive phylogeny of Acrididae based on mitochondrial genomes and nuclear genes to test monophyly of the family and different subfamilies as well as to understand the evolutionary relationships among them. We recovered the monophyletic Acrididae and identified four major clades as well as several well-characterized subfamilies, but we also found that paraphyly is rampant across many subfamilies, highlighting the need for a taxonomic revision of the family. We found that Acrididae originated in the Paleocene of the Cenozoic period (59.3 million years ago) and, because the separation of South America and Africa predates the origin of the family, we hypothesize that the current cosmopolitan distribution of Acrididae was largely achieved by dispersal. We also inferred that the common ancestor of modern grasshoppers originated in South America, contrary to a popular belief that they originated in Africa, based on a biogeographical analysis. We estimate that there have been a number of colonization and recolonization events between the New World and the Old World throughout the diversification of Acrididae, and, thus, the current diversity in any given region is a reflection of this complex history.
Grasshoppers (Orthoptera: Acrididae) are among the most recognizable and familiar insects in terrestrial habitats around the world. They are dominant herbivores and represent a ubiquitous component of grasslands around the world (Uvarov 1966, Mitchell and Pfadt 1974, Gangwere et al. 1997, Cigliano et al. 2000, Guo et al. 2006). In grassland ecosystems, grasshoppers contribute to more than half of the total arthropod biomass in the above ground grass layer (Gillon 1983). They exert a significant ecological impact in grasslands in terms of nutrient cycling (Mitchell and Pfadt 1974, Belovsky and Slade 1993, Gangwere et al. 1997) and provide an important source of nutrition for both invertebrates (Joern et al. 2006) and vertebrates (Gandar 1982), thus supporting other biological components of the ecosystem (Belovsky and Slade 1993). Grasshoppers can also be excellent monitors of landscape use as they are ecologically sensitive and yet sufficiently mobile and abundant to serve as bioindicators (Samways and Sergeev 1997, Gebeyehu and Samways 2002, Bazelet and Samways 2014). Several species of grasshoppers are considered major pests, especially when they periodically develop into local and large-scale outbreaks, causing enormous economic damage (COPR 1982). Some of the most important insect pests around the world are locusts, which are grasshoppers that can form dense migrating swarms and exhibit density-dependent phase polyphenism (Uvarov 1966, Pener 1983, Pener and Simpson 2009, Cullen et al. 2017). While many grasshopper species are pests, some species are beneficial, such as Cornops aquaticum (Bruner, 1906) (Leptysiminae), which has been used as a successful biocontrol agent of water hyacinth in South Africa (Bownes et al. 2011, Coetzee et al. 2011), and Hesperotettix viridis (Thomas, 1872) (Melanoplinae), which prefers to feed on noxious snakeweeds that can harm cattle and other livestock (Thompson and Richman 1993).
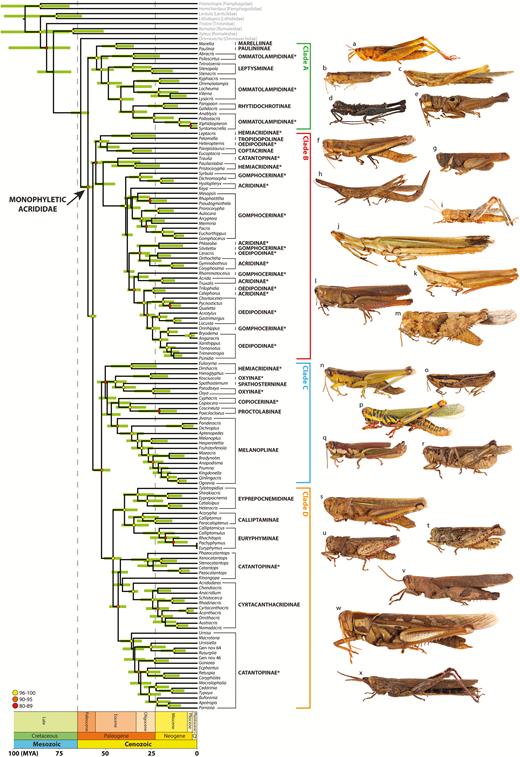
Although grasshoppers are often thought to be associated with grasslands, many species are actually found in tropical forests, shrublands, deserts, wetlands, and alpine regions around the world. For example, Urnisiella rubropunctata Sjöstedt, 1930 (Catantopinae) is highly adapted to the sandy habitat in the Australian outback, where it can withstand high temperatures and uses its long middle legs to sweep sand over its body to bury itself when it is threatened (Rentz 1996). An aquatic grasshopper from South America, Marellia remipes Uvarov, 1929 (Marelliinae), lives on broad, floating leaves of aquatic plants and its hind tibiae are modified and expanded to be oar-like, which help it swim underwater (Carbonell 1957). Many specialized grasshopper species in the subfamilies Proctolabinae and Ommatolampidinae (= Ommatolampinae) live in the canopies of tropical rainforests in the Amazon (Descamps 1976, Amédégnato and Descamps 1978, Descamps 1978). A number of alpine grasshoppers in the subfamily Melanoplinae have diversified in isolated mountain ranges, such as Melanoplus Stål, 1873 in the Rocky Mountains of the United States (Knowles 2001, Knowles and Richards 2005), and Orotettix Ronderos & Carbonell, 1994, Jivarus Giglio-Tos, 1898, and Maeacris Ronderos, 1983 in the Andes in South America (Cigliano and Amédégnato 2010, Cigliano et al. 2011, Pocco et al. 2015), which are typically characterized by short-wings and have limited dispersal abilities. Indeed, grasshoppers are extremely diverse in terms of size, body shape, feeding biology, ecology, and life-history traits (Fig. 1) (Uvarov 1977, Chapman and Joern 1990).
Diversity of Acrididae: (A) Anacridium aegyptium (Linnaeus, 1764) (Cyrtacanthacridinae), France; (B) Dactylotum bicolor Charpentier, 1845 (Melanoplinae), Mexico; (C) Kosciuscola tristis Sjöstedt, 1934 (Oxyinae), Australia; (D) Adimantus ornatissimus (Burmeister, 1838) (Copiocerinae), Argentina; (E) Calliptamus italicus (Linnaeus, 1758) (Calliptaminae), France; (F) Proctolabus mexicanus (Saussure, 1859) (Proctolabinae), Mexico; (G) Marellia remipes Uvarov, 1929 (Marelliinae), Colombia; (H) Paulinia acuminata (De Geer, 1773) (Pauliniinae), Colombia; (I) Acrida sp. (Acridinae), Vietnam; (J) Hylopedetes surdus Descamps & Rowell, 1978 (Rhytidochrotinae), Costa Rica; (K) Trimerotropis pallidipennis (Burmeister, 1838) (Oedipodinae), Mexico; (L) Stenopola puncticeps (Stål, 1861) (Leptysminae), Argentina; (M) Rhammatocerus pictus (Bruner, 1900) (Gomphocerinae), Argentina; (N) Abracris flavolineata (De Geer, 1773) (Ommatolampidinae), Costa Rica; (O) Hemiacris fervens Walker, 1870 (Hemiacridinae), Mozambique. Photo credits. A, E, I, N: Ruben Foquet; B, O: Ricardo Mariño-Pérez; C, J: Hojun Song; D, L, M: Maria Marta Cigliano; F, K: Paolo Fontana; G, H: Juan Manuel Cardona.
The family Acrididae includes more than 6,700 valid species and represents the most diverse lineage within the orthopteran suborder Caelifera (Cigliano et al. 2018). It is hypothesized to have originated in the early Cenozoic Era and diversified through the mid to late Cenozoic (Song et al. 2015). By this time, major continents had already separated, which suggests that dispersal might have played an important role in forming current biogeographical patterns. There are currently 26 recognized subfamilies within Acrididae (Table 1), of which only five subfamilies (Acridinae, Cyrtacanthacridinae, Gomphocerinae, Melanoplinae, and Oedipodinae) have a cosmopolitan distribution, while others have more restricted distributions (Cigliano et al. 2018). Of the remaining subfamilies, 14 are found exclusively in the Old World, while seven are only found in the New World, mostly in Central and South America. To explain this pattern, Carbonell (1977), Amédégnato and Descamps (1979), Jago (1979), Rowell (1987), (Vickery 1987, 1989), and Amedegnato (1993) proposed various biogeographic hypotheses regarding the origin and diversification of different acridid lineages, but these hypotheses have never been formally tested.
The 26 currently recognized subfamilies within Acrididae, number of genera and species, and distribution
| Subfamily . | Number of genera . | Number of species . | Distribution . |
|---|---|---|---|
| Acridinae | 141 | 483 | Cosmopolitan |
| Calliptaminae | 12 | 92 | Africa, Europe, Middle East, Central Asia, India |
| Catantopinae | 341 | 1,077 | Africa, Middle East, Asia, Australia |
| Copiocerinae | 21 | 90 | Central and South America, Caribbean |
| Coptacrinae | 20 | 116 | Sub-Saharan Africa, India, Southeast Asia |
| Cyrtacanthacridinae | 36 | 162 | Cosmopolitan |
| Egnatiinae* | 9 | 36 | North Africa, Middle East, Central Asia |
| Eremogryllinae* | 2 | 5 | Northwestern Africa |
| Euryphyminae | 23 | 87 | Southern Africa |
| Eyprepocnemidinae | 26 | 159 | Africa, Middle East, Southern Asia, Southeast Asia, Eastern Asia |
| Gomphocerinae | 192 | 1,274 | Cosmopolitan |
| Habrocneminae* | 2 | 3 | Southeast Asia |
| Hemiacridinae | 38 | 122 | Sub-Saharan Africa, Southern Asia, Southeast Asia |
| Leptysminae | 21 | 79 | North, Central, and South America, Caribbean |
| Marelliinae | 1 | 1 | South America |
| Melanoplinae | 145 | 1,173 | North, Central, and South America, Asia, Europe |
| Oedipodinae | 137 | 792 | Cosmopolitan |
| Ommatolampidinae | 114 | 292 | Central and South America, Caribbean |
| Oxyinae | 37 | 307 | Sub-Saharan Africa, Asia, Australia |
| Pauliniinae | 1 | 1 | Central and South America |
| Pezotettiginae* | 2 | 10 | Europe, Northwestern Africa |
| Proctolabinae | 29 | 215 | Central and South America |
| Rhytidochrotinae | 20 | 47 | Northern South America |
| Spathosterninae | 3 | 12 | Sub-Saharan Africa, Southern Asia, Southeast Asia, Australia |
| Teratodinae* | 8 | 24 | India, Middle East, Central eastern Africa |
| Tropidopolinae | 11 | 34 | Africa, Middle East, Southeast Asia |
| Subfamily . | Number of genera . | Number of species . | Distribution . |
|---|---|---|---|
| Acridinae | 141 | 483 | Cosmopolitan |
| Calliptaminae | 12 | 92 | Africa, Europe, Middle East, Central Asia, India |
| Catantopinae | 341 | 1,077 | Africa, Middle East, Asia, Australia |
| Copiocerinae | 21 | 90 | Central and South America, Caribbean |
| Coptacrinae | 20 | 116 | Sub-Saharan Africa, India, Southeast Asia |
| Cyrtacanthacridinae | 36 | 162 | Cosmopolitan |
| Egnatiinae* | 9 | 36 | North Africa, Middle East, Central Asia |
| Eremogryllinae* | 2 | 5 | Northwestern Africa |
| Euryphyminae | 23 | 87 | Southern Africa |
| Eyprepocnemidinae | 26 | 159 | Africa, Middle East, Southern Asia, Southeast Asia, Eastern Asia |
| Gomphocerinae | 192 | 1,274 | Cosmopolitan |
| Habrocneminae* | 2 | 3 | Southeast Asia |
| Hemiacridinae | 38 | 122 | Sub-Saharan Africa, Southern Asia, Southeast Asia |
| Leptysminae | 21 | 79 | North, Central, and South America, Caribbean |
| Marelliinae | 1 | 1 | South America |
| Melanoplinae | 145 | 1,173 | North, Central, and South America, Asia, Europe |
| Oedipodinae | 137 | 792 | Cosmopolitan |
| Ommatolampidinae | 114 | 292 | Central and South America, Caribbean |
| Oxyinae | 37 | 307 | Sub-Saharan Africa, Asia, Australia |
| Pauliniinae | 1 | 1 | Central and South America |
| Pezotettiginae* | 2 | 10 | Europe, Northwestern Africa |
| Proctolabinae | 29 | 215 | Central and South America |
| Rhytidochrotinae | 20 | 47 | Northern South America |
| Spathosterninae | 3 | 12 | Sub-Saharan Africa, Southern Asia, Southeast Asia, Australia |
| Teratodinae* | 8 | 24 | India, Middle East, Central eastern Africa |
| Tropidopolinae | 11 | 34 | Africa, Middle East, Southeast Asia |
The numbers of genera and species are from OSF (Cigliano et al. 2018). Asterisks indicate those subfamilies not included in the present study due to unavailability of DNA-grade specimens.
The 26 currently recognized subfamilies within Acrididae, number of genera and species, and distribution
| Subfamily . | Number of genera . | Number of species . | Distribution . |
|---|---|---|---|
| Acridinae | 141 | 483 | Cosmopolitan |
| Calliptaminae | 12 | 92 | Africa, Europe, Middle East, Central Asia, India |
| Catantopinae | 341 | 1,077 | Africa, Middle East, Asia, Australia |
| Copiocerinae | 21 | 90 | Central and South America, Caribbean |
| Coptacrinae | 20 | 116 | Sub-Saharan Africa, India, Southeast Asia |
| Cyrtacanthacridinae | 36 | 162 | Cosmopolitan |
| Egnatiinae* | 9 | 36 | North Africa, Middle East, Central Asia |
| Eremogryllinae* | 2 | 5 | Northwestern Africa |
| Euryphyminae | 23 | 87 | Southern Africa |
| Eyprepocnemidinae | 26 | 159 | Africa, Middle East, Southern Asia, Southeast Asia, Eastern Asia |
| Gomphocerinae | 192 | 1,274 | Cosmopolitan |
| Habrocneminae* | 2 | 3 | Southeast Asia |
| Hemiacridinae | 38 | 122 | Sub-Saharan Africa, Southern Asia, Southeast Asia |
| Leptysminae | 21 | 79 | North, Central, and South America, Caribbean |
| Marelliinae | 1 | 1 | South America |
| Melanoplinae | 145 | 1,173 | North, Central, and South America, Asia, Europe |
| Oedipodinae | 137 | 792 | Cosmopolitan |
| Ommatolampidinae | 114 | 292 | Central and South America, Caribbean |
| Oxyinae | 37 | 307 | Sub-Saharan Africa, Asia, Australia |
| Pauliniinae | 1 | 1 | Central and South America |
| Pezotettiginae* | 2 | 10 | Europe, Northwestern Africa |
| Proctolabinae | 29 | 215 | Central and South America |
| Rhytidochrotinae | 20 | 47 | Northern South America |
| Spathosterninae | 3 | 12 | Sub-Saharan Africa, Southern Asia, Southeast Asia, Australia |
| Teratodinae* | 8 | 24 | India, Middle East, Central eastern Africa |
| Tropidopolinae | 11 | 34 | Africa, Middle East, Southeast Asia |
| Subfamily . | Number of genera . | Number of species . | Distribution . |
|---|---|---|---|
| Acridinae | 141 | 483 | Cosmopolitan |
| Calliptaminae | 12 | 92 | Africa, Europe, Middle East, Central Asia, India |
| Catantopinae | 341 | 1,077 | Africa, Middle East, Asia, Australia |
| Copiocerinae | 21 | 90 | Central and South America, Caribbean |
| Coptacrinae | 20 | 116 | Sub-Saharan Africa, India, Southeast Asia |
| Cyrtacanthacridinae | 36 | 162 | Cosmopolitan |
| Egnatiinae* | 9 | 36 | North Africa, Middle East, Central Asia |
| Eremogryllinae* | 2 | 5 | Northwestern Africa |
| Euryphyminae | 23 | 87 | Southern Africa |
| Eyprepocnemidinae | 26 | 159 | Africa, Middle East, Southern Asia, Southeast Asia, Eastern Asia |
| Gomphocerinae | 192 | 1,274 | Cosmopolitan |
| Habrocneminae* | 2 | 3 | Southeast Asia |
| Hemiacridinae | 38 | 122 | Sub-Saharan Africa, Southern Asia, Southeast Asia |
| Leptysminae | 21 | 79 | North, Central, and South America, Caribbean |
| Marelliinae | 1 | 1 | South America |
| Melanoplinae | 145 | 1,173 | North, Central, and South America, Asia, Europe |
| Oedipodinae | 137 | 792 | Cosmopolitan |
| Ommatolampidinae | 114 | 292 | Central and South America, Caribbean |
| Oxyinae | 37 | 307 | Sub-Saharan Africa, Asia, Australia |
| Pauliniinae | 1 | 1 | Central and South America |
| Pezotettiginae* | 2 | 10 | Europe, Northwestern Africa |
| Proctolabinae | 29 | 215 | Central and South America |
| Rhytidochrotinae | 20 | 47 | Northern South America |
| Spathosterninae | 3 | 12 | Sub-Saharan Africa, Southern Asia, Southeast Asia, Australia |
| Teratodinae* | 8 | 24 | India, Middle East, Central eastern Africa |
| Tropidopolinae | 11 | 34 | Africa, Middle East, Southeast Asia |
The numbers of genera and species are from OSF (Cigliano et al. 2018). Asterisks indicate those subfamilies not included in the present study due to unavailability of DNA-grade specimens.
The taxonomy of Acrididae has had a tumultuous history (Song 2010). Throughout much of the 19th and 20th centuries, there was no clear definition of what should constitute Acrididae, and the family was used as a taxonomic dumping ground for groups when authors did not know where to place them (Eades 2000). For example, Robert’s (1941) comparative study of male genitalia treated the current families Pyrgomorphidae, Pamphagidae, Ommexechidae, and Romaleidae as subfamilies of Acrididae, but they have since been shown to be quite distinct families from Acrididae. Likewise Dirsh’s (1961) preliminary revision of Acrididae included the currently recognized families Dericorythidae, Tristiridae, Romaleidae, and Lithidiidae as subfamilies of Acrididae. Although early taxonomists relied on external morphological characters, such as stridulatory structures, prosternal process (a short spine located ventrally on the prosternum between the two front coxae), sculpting patterns on head and pronotum, hind legs, and wings for classifying grasshoppers (Rehn and Grant 1961, Bei-Bienko and Mishchenko 1963), later authors regarded male phallic structures as the single most important characters for higher-level classification (Dirsh 1973, Amedegnato 1976, Eades 2000). However, too much reliance on these phallic structures led to over-splitting of taxonomic concepts, especially when Dirsh (1975) elevated several subfamilies to family level, resulting in four families and 40 subfamilies.
Initially, taxonomic research on grasshopper diversity focused on faunas in Europe, Africa, Eurasia, and North America, and, thus, earlier classification schemes were established based on the specimens collected from these regions (Rehn and Grant 1961, Bei-Bienko and Mishchenko 1963, Dirsh 1965). During 1960s and 1970s, taxonomists began exploring South America and discovered previously unknown grasshopper lineages, which led to the erection of several new subfamilies (Amedegnato 1974, Amédégnato and Descamps 1978). In fact, the faunas in Southeast Asia and Australia still have not been fully explored (Key 1992, Rentz 1996, Song 2010). While many of the debates on how to classify different grasshopper groups have been made by European and North American taxonomists, Chinese taxonomists have adopted a different classification scheme (Zheng 1993, Xia 1994, Zheng and Xia 1998, Yin and Xia 2003) based on the species found in China, which they continue to use currently. In an effort to produce a unified classification scheme, Otte (1995a,b) published the Orthoptera Species File (OSF), which later became the basis for an electronic version (Cigliano et al. 2018), which most orthopterists accept. Currently, the OSF recognizes 26 subfamilies (Table 1) and some unplaced tribes and genera for Acrididae.
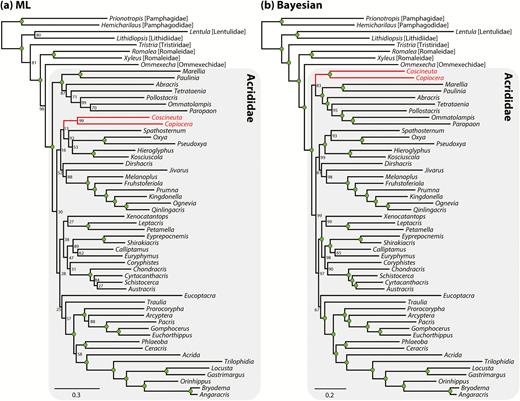
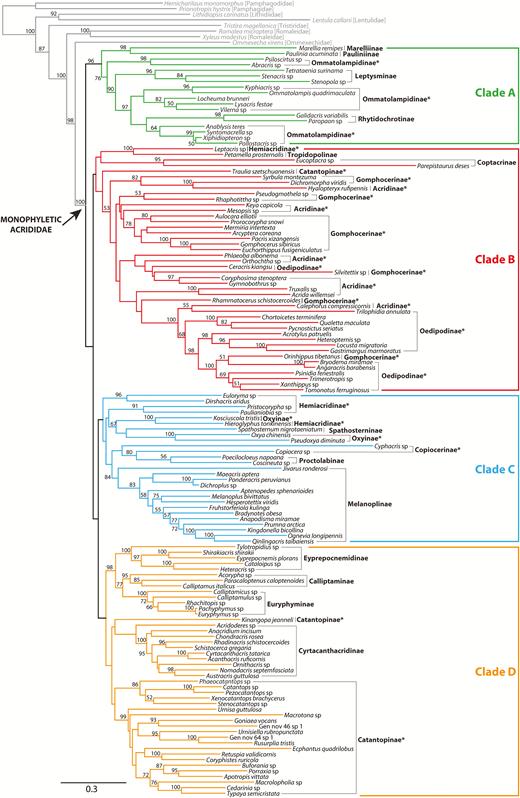
There has never been a comprehensive phylogeny proposed for Acrididae, although several studies have focused on the phylogenetic relationships at subfamily level using either morphology or molecular data (Chapco et al. 2001, Litzenberger and Chapco 2001, Amédégnato et al. 2003, Litzenberger and Chapco 2003, Rowell and Flook 2004, Bugrov et al. 2006, Contreras and Chapco 2006, Fries et al. 2007, Song and Wenzel 2008, Chapco and Contreras 2011, Chintauan-Marquier et al. 2011, Li et al. 2011, Nattier et al. 2011, Chintauan-Marquier et al. 2014). Flook and Rowell (1997) presented the first molecular phylogeny of Caelifera based on fragments of mitochondrial ribosomal RNA genes, which included 12 acridids belonging to four subfamilies, but they did not recover monophyly of Acrididae because Pamphagidae was nested within Acrididae. Based on the investigation of male genitalia across Acridoidea, Eades (2000) proposed that all acridids have a strongly developed arch sclerite in the male phallic complex, which is not found in other families within the Acridoidea except the Pamphagodidae (= Charilaidae), apparently having evolved a similar structure independently. Liu et al. (2008) proposed a phylogeny of Acrididae using 24 Chinese species based on two mitochondrial ribosomal genes and found that Acridinae and Catantopinae were paraphyletic, while Cyrtacanthacridinae, Oxyinae, and Oedipodinae were monophyletic. Li et al. (2011) published a morphological phylogeny of Catantopidae, which Chinese authors recognize as a valid family that includes grasshoppers with the prosternal process, based on an analysis of 87 genera and 88 characters. They recovered monophyletic Catantopidae, but because they did not include any grasshoppers without the prosternal process (such as Acridinae, Gomphocerinae, and Oedipodinae) or the New World endemic groups that possess this structure (such as Ommatolampidinae, Leptysminae, Rhytidochrotinae, Copiocerinae, and Proctolabinae), their inferences need to be viewed with caution. Leavitt et al. (2013) tested the monophyly of Acrididae using complete mitochondrial genome (mtgenome) sequences and 34 caeliferan taxa (including 16 acridid species) and recovered strong monophyly of the family, but only eight subfamilies were included, none of which were from South America. Most recently, Song et al. (2015) published a phylogeny of Orthoptera based on 254 taxa and four nuclear genes (18S and 28S rRNA, histone 3, and wingless) and complete mtgenome sequences for 69 backbone terminals, which included 87 acridid taxa covering the phylogenetic and geographic diversity of the family. While they recovered monophyletic Acrididae with strong support based only on mtgenome data, the family was rendered paraphyletic in a total evidence analysis. They noted that branch lengths were very short within Acrididae, suggesting that the nuclear genes used in the analysis were too conserved and did not have enough phylogenetic signal to accurately resolve the phylogeny of Acrididae.
The two primary objectives of this study are 1) to present the first large-scale molecular phylogeny of Acrididae to test monophyly of the family and different subfamilies, as well as 2) to understand the evolutionary relationships among these groups. Based on the resulting phylogeny and divergence time estimates, we also propose a novel biogeographical hypothesis regarding the origin and diversification of different lineages of Acrididae. This will provide a framework for future phylogeny-based classification of Acrididae and a reference for studying interesting biology and evolutionary patterns within this family.
Materials and Methods
Taxon and Character Sampling
We followed the classification scheme adopted by the OSF (Cigliano et al. 2018) in order to test it with our phylogenetic analysis. We sampled a total of 142 taxa, including 8 outgroup and 134 ingroup taxa covering the phylogenetic diversity within Acrididae (Table 2, Supplementary Table 1). The outgroups included seven representative families within Acridoidea based on our previous findings on the higher-level relationships (Leavitt et al. 2013, Song et al. 2015). Of these outgroup taxa, four are Old World families: Pamphagidae, Pamphagodidae, Lithidiidae, and Lentulidae, and three are endemic to the New World: Tristiridae, Romaleidae (two representatives included), and Ommexechidae. For ingroup sampling, we included 21 of the 26 currently recognized acridid subfamilies. Due to the difficulty in obtaining DNA-grade specimens, we did not include these five subfamilies in our analysis: Egnatiinae, Eremogryllinae, Habrocneminae, Pezotettiginae, and Teratodinae. We included multiple representatives of each subfamily to test monophyly except Spathosterninae, Tropidopolinae, Marelliinae, and Pauliniinae, the latter two of which are monotypic. For 58 terminals, which represented key taxa for understanding higher-level relationships, we included partial or complete mtgenome data, 24 of which were newly sequenced for this study. The remaining mtgenomes were either previously generated by us (Fenn et al. 2008, Sheffield et al. 2010, Leavitt et al. 2013, Song et al. 2015) or obtained from GenBank (Table 2). For all taxa, we generated complete sequences of 18S and 28S ribosomal RNA genes and histone 3 (H3) genes, as well as full-length sequences of mitochondrial cytochrome c oxidase 1 and 2 (COI and COII). For the 19 taxa for which we obtained mtgenome sequences from GenBank, we were unable to generate the three nuclear genes due to an obvious lack of access to specimens.
Taxonomic information and Genbank accession numbers for 142 taxa used in total evidence analysis
| Family . | Subfamily . | Species . | Voucher # (TAMUIC-IGC-#) . | mtgenome . | 18S . | 28S . | H3 . | COI . | COII . |
|---|---|---|---|---|---|---|---|---|---|
| Acrididae | Acridinae | Acrida willemsei | OR059 | NC_011303 | KM853177 | KM853512 | KM853687 | mtgenome | mtgenome |
| Calephorus compressicornis | OR192 | N/A | KM853192 | KM853498 | KM853673 | MG888076 | MG888143 | ||
| Coryphosima stenoptera | OR512 | N/A | MG888284 | MG888333 | MG888241 | MG888120 | MG888187 | ||
| Gymnobothrus sp | OR511 | N/A | MG888283 | MG888332 | MG888240 | MG888119 | MG888186 | ||
| Hyalopteryx rufipennis | OR240 | N/A | KM853210 | KM853480 | KM853655 | MG888088 | MG888155 | ||
| Keya capicola | OR514 | N/A | MG888286 | MG888335 | MG888243 | MG888122 | MG888189 | ||
| Orthochtha sp | OR513 | N/A | MG888285 | MG888334 | MG888242 | MG888121 | MG888188 | ||
| Phlaeoba albonema | N/A | NC_011827 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Truxalis sp | OR510 | N/A | KM853325 | KM853367 | KM853543 | MG888118 | N/A | ||
| Calliptaminae | Acorypha sp | OR195 | N/A | MG888254 | MG888300 | MG888208 | MG888078 | MG888145 | |
| Calliptamus italicus | OR193 | NC_011305 | KM853193 | KM853497 | KM853672 | mtgenome | mtgenome | ||
| Paracaloptenus caloptenoides | OR194 | N/A | KM853194 | KM853496 | KM853671 | MG888077 | MG888144 | ||
| Catantopinae | Apotropis vittata | OR493 | N/A | MG888277 | MG888325 | MG888233 | MG888106 | MG888173 | |
| Buforania sp | OR500 | N/A | MG888278 | MG888327 | MG888235 | MG888113 | MG888180 | ||
| Catantops sp | OR237 | N/A | KM853209 | KM853481 | KM853656 | MG888086 | MG888153 | ||
| Cedarinia sp | OR490 | N/A | MG888274 | MG888322 | MG888230 | MG888103 | MG888170 | ||
| Coryphistes ruricola | OR503 | MG993389, MG993390, MG993403, MG993406 | MG888281 | MG888330 | MG888238 | mtgenome | mtgenome | ||
| Ecphantus quadrilobus | OR495 | N/A | MG888295 | MG888326 | MG888234 | MG888108 | MG888175 | ||
| Gen nov. 46 sp. 1 | OR491 | N/A | MG888275 | MG888323 | MG888231 | MG888104 | MG888171 | ||
| Gen nov. 64 sp. 1 | OR489 | N/A | MG888273 | MG888321 | MG888229 | MG888102 | MG888169 | ||
| Goniaea vocans | OR502 | N/A | MG888280 | MG888329 | MG888237 | MG888115 | MG888182 | ||
| Kinangopa jeanneli | OR574 | N/A | KM853345 | KM853348 | KM853523 | N/A | MG888205 | ||
| Macrolopholia sp | OR235 | N/A | KM853208 | KM853482 | KM853657 | MG888085 | MG888152 | ||
| Macrotona sp | OR488 | N/A | MG888272 | MG888320 | MG888228 | MG888101 | MG888168 | ||
| Pezocatantops sp | OR505 | N/A | KM853321 | KM853372 | KM853548 | MG888117 | MG888184 | ||
| Phaeocatantops sp | OR504 | N/A | MG888282 | MG888331 | MG888239 | MG888116 | MG888183 | ||
| Porraxia sp | OR494 | N/A | KM853316 | KM853377 | KM853553 | MG888107 | MG888174 | ||
| Retuspia validicornis | OR496 | N/A | KM853317 | KM853376 | KM853552 | MG888109 | MG888176 | ||
| Rusurplia tristis | OR497 | N/A | KM853318 | KM853375 | KM853551 | MG888110 | MG888177 | ||
| Stenocatantops vitripennis | OR498 | N/A | KM853319 | KM853374 | KM853550 | MG888111 | MG888178 | ||
| Traulia szetschuanensis | N/A | NC_013826 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Typaya semicristata | OR492 | N/A | MG888276 | MG888324 | MG888232 | MG888105 | MG888172 | ||
| Urnisa guttulosa | OR501 | N/A | MG888279 | MG888328 | MG888236 | MG888114 | MG888181 | ||
| Urnisiella rubropunctata | OR499 | N/A | KM853320 | KM853373 | KM853549 | MG888112 | MG888179 | ||
| Xenocatantops brachycerus | OR236 | NC_021609 | MG888296 | MG888303 | MG888211 | mtgenome | mtgenome | ||
| Copiocerinae | Copiocera sp | OR333 | MG993384 | KM853250 | KM853440 | KM853616 | mtgenome | mtgenome | |
| Cyphacris sp | OR334 | N/A | KM853251 | KM853439 | KM853615 | MG888096 | MG888163 | ||
| Coptacrinae | Eucoptacra sp | OR509 | MG993445 | KM853324 | KM853368 | KM853544 | mtgenome | mtgenome | |
| Parepistaurus deses | OR508 | N/A | KM853323 | KM853369 | KM853545 | N/A | MG888185 | ||
| Cyrtacanthacridinae | Acanthacris ruficornis | OR183 | N/A | MG888253 | MG888299 | MG888207 | MG888071 | MG888138 | |
| Acridoderes sp | OR546 | N/A | MG888293 | MG888342 | MG888250 | MG888137 | MG888203 | ||
| Anacridium incisum | OR184 | N/A | KM853185 | KM853505 | KM853680 | MG888072 | MG888139 | ||
| Acrididae | Cyrtacanthacridinae | Austracris guttulosa | OR182 | MG993415 | MG888252 | MG888298 | MG888206 | mtgenome | mtgenome |
| Chondracris rosea | N/A | NC_019993 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Cyrtacanthacris tatarica | OR181 | MG993444 | KM853184 | KM853506 | KM853681 | mtgenome | mtgenome | ||
| Nomadacris septemfasciata | OR545 | N/A | KM853340 | KM853352 | KM853528 | MG888136 | N/A | ||
| Ornithacris sp | OR544 | N/A | KM853339 | KM853353 | KM853529 | MG888135 | MG888202 | ||
| Rhadinacris schistocercoides | OR547 | N/A | KM853341 | KM853351 | KM853527 | N/A | MG888204 | ||
| Schistocerca gregaria | OR185 | NC_013240 | KM853186 | KM853504 | KM853679 | mtgenome | mtgenome | ||
| Euryphyminae | Calliptamicus sp | OR313 | N/A | MG888261 | MG888308 | MG888216 | N/A | N/A | |
| Calliptamulus sp | OR311 | N/A | KM853241 | KM853449 | KM853625 | MG888094 | MG888161 | ||
| Euryphymus sp | OR314 | MG993388, MG993422, MG993436 | KM853243 | KM853447 | KM853623 | mtgenome | mtgenome | ||
| Pachyphymus sp | OR308 | N/A | MG888260 | MG888307 | MG888215 | MG888092 | MG888159 | ||
| Rhachitopis sp | OR312 | N/A | KM853242 | KM853448 | KM853624 | N/A | N/A | ||
| Eyprepocnemidinae | Cataloipus sp | OR218 | N/A | KM853201 | KM853489 | KM853664 | MG888079 | MG888146 | |
| Eyprepocnemis plorans | OR309 | MG993386, MG993418,MG993424, MG993425,MG993427, MG993433,MG993437, MG993450 | KM853239 | KM853451 | KM853627 | mtgenome | mtgenome | ||
| Heteracris sp | OR310 | N/A | KM853240 | KM853450 | KM853626 | MG888093 | MG888160 | ||
| Shirakiacris shirakii | N/A | NC_021610 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Tylotropidius sp | OR219 | N/A | MG888255 | MG888301 | MG888209 | MG888080 | MG888147 | ||
| Gomphocerinae | Arcyptera coreana | N/A | NC_013805 | N/A | N/A | N/A | mtgenome | mtgenome | |
| Aulocara elliotii | OR521 | N/A | KM853329 | KM853363 | KM853539 | MG888129 | MG888196 | ||
| Dichromorpha viridis | OR226 | N/A | KM853205 | KM853485 | KM853660 | MG888083 | MG888150 | ||
| Euchorthippus fusigeniculatus | N/A | NC_014449 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Gomphocerus sibiricus | N/A | NC_015478 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Mermiria intertexta | OR520 | N/A | KM853328 | KM853364 | KM853540 | MG888128 | MG888195 | ||
| Mesopsis sp | OR239 | N/A | MG888257 | MG888304 | MG888212 | MG888087 | MG888154 | ||
| Orinhippus tibetanus | N/A | NC_023467 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Pacris xizangensis | N/A | NC_023919 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Prorocorypha snowi | OR214 | MG993438, MG993452, MG993453 | KM853199 | KM853491 | KM853666 | mtgenome | mtgenome | ||
| Pseudogmothela sp | OR519 | N/A | MG888289 | MG888338 | MG888246 | MG888127 | MG888194 | ||
| Rhammatocerus schistocercoides | OR346 | N/A | KM853258 | KM853432 | KM853608 | MG888098 | MG888164 | ||
| Rhaphotittha sp | OR518 | N/A | MG888288 | MG888337 | MG888245 | MG888126 | MG888193 | ||
| Silvitettix sp | OR343 | N/A | MG888267 | MG888314 | MG888222 | N/A | N/A | ||
| Syrbula montezuma | OR227 | N/A | KM853206 | KM853484 | KM853659 | MG888084 | MG888151 | ||
| Acrididae | Hemiacridinae | Dirshacris aridus | OR305 | MG993398, MG993399, MG993410, MG993417, MG993420, MG993434, MG993435, MG993451 | MG888259 | MG888306 | MG888214 | mtgenome | mtgenome |
| Euroryma sp | OR302 | N/A | KM853236 | KM853454 | KM853630 | MG888090 | MG888157 | ||
| Hieroglyphus tonkinensis | N/A | NC_030587 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Leptacris sp | OR304 | MG993429 | KM853238 | KM853452 | KM853628 | mtgenome | mtgenome | ||
| Paulianiobia hirsuta | OR301 | N/A | MG888258 | MG888305 | MG888213 | MG888089 | MG888156 | ||
| Pristocorypha sp | OR303 | N/A | KM853237 | KM853453 | KM853629 | MG888091 | MG888158 | ||
| Leptysminae | Stenacris sp | OR342 | N/A | KM853255 | KM853435 | KM853611 | N/A | N/A | |
| Stenopola sp | OR220 | N/A | MG888256 | MG888302 | MG888210 | MG888081 | MG888148 | ||
| Tetrataenia surinama | OR338 | MG993385, MG993395, MG993396, MG993404, MG993407, MG993409, MG993432, MG993448 | KM853254 | KM853436 | KM853612 | mtgenome | mtgenome | ||
| Marelliinae | Marellia remipes | OR344 | MG993387, MG993423, MG993442, MG993447 | KM853256 | KM853434 | KM853610 | mtgenome | mtgenome | |
| Melanoplinae | Anapodisma miramae | OR356 | N/A | KM853265 | KM853425 | KM853601 | MG888100 | MG888166 | |
| Aptenopedes sphenarioides | OR516 | N/A | MG888287 | MG888336 | MG888244 | MG888124 | MG888191 | ||
| Bradynotes obesa | OR515 | N/A | KM853326 | KM853366 | KM853542 | MG888123 | MG888190 | ||
| Dichroplus sp | OR325 | N/A | KM853248 | KM853442 | KM853618 | MG888095 | MG888162 | ||
| Fruhstorferiola kulinga | N/A | NC_026716 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Hesperotettix viridis | OR517 | N/A | KM853327 | KM853365 | KM853541 | MG888125 | MG888192 | ||
| Jivarus ronderosi | OR328 | MG993400, MG993405 | KM853249 | KM853441 | KM853617 | mtgenome | mtgenome | ||
| Kingdonella bicollina | N/A | NC_023920 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Maeacris aptera | OR329 | N/A | MG888266 | MG888313 | MG888221 | N/A | N/A | ||
| Melanoplus bivittatus | OR245 | MG993426 | KM853211 | KM853479 | KM853654 | mtgenome | mtgenome | ||
| Ognevia longipennis | OR394 | NC_013701 | MG888297 | MG888319 | MG888227 | mtgenome | mtgenome | ||
| Ponderacris peruvianus | OR324 | N/A | MG888265 | MG888312 | MG888220 | N/A | N/A | ||
| Prumna arctica | OR395 | NC_013835 | KM853277 | KM853412 | KM853589 | mtgenome | mtgenome | ||
| Qinlingacris taibaiensis | N/A | NC_027187 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Oedipodinae | Acrotylus patruelis | OR190 | N/A | KM853190 | KM853500 | KM853675 | MG888075 | MG888142 | |
| Acrididae | Oedipodinae | Angaracris barabensis | N/A | NC_025946 | N/A | N/A | N/A | mtgenome | mtgenome |
| Bryodema miramae miramae | N/A | KP889242 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Ceracris kiangsu | N/A | NC_019994 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Chortoicetes terminifera | OR524 | N/A | MG888290 | MG888339 | MG888247 | MG888132 | MG888199 | ||
| Gastrimargus marmoratus | N/A | NC_011114 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Heteropternis sp | OR225 | N/A | KM853204 | KM853486 | KM853661 | MG888082 | MG888149 | ||
| Locusta migratoria | OR191 | NC_001712 | KM853191 | KM853499 | KM853674 | mtgenome | mtgenome | ||
| Psinidia fenestralis | OR522 | N/A | KM853330 | KM853362 | KM853538 | MG888130 | MG888197 | ||
| Pycnostictus seriatus | OR525 | N/A | MG888291 | MG888340 | MG888248 | MG888133 | MG888200 | ||
| Qualetta maculata | OR526 | N/A | MG888292 | MG888341 | MG888249 | MG888134 | MG888201 | ||
| Tomonotus ferruginosus | OR523 | N/A | KM853331 | KM853361 | KM853537 | MG888131 | MG888198 | ||
| Trilophidia annulata | N/A | NC_027179 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Trimerotropis sp | OR186 | N/A | KM853187 | KM853503 | KM853678 | MG888073 | MG888140 | ||
| Xanthippus sp | OR187 | N/A | KM853188 | KM853502 | KM853677 | MG888074 | MG888141 | ||
| Ommatolampidinae | Abracris sp | OR222 | MG993440 | KM853202 | KM853488 | KM853663 | mtgenome | mtgenome | |
| Anablysis teres | OR362 | N/A | MG888269 | MG888316 | MG888224 | N/A | N/A | ||
| Kyphiacris sp | OR363 | N/A | MG888270 | MG888317 | MG888225 | N/A | N/A | ||
| Locheuma brunneri | OR366 | N/A | KM853268 | KM853422 | KM853598 | N/A | N/A | ||
| Lysacris festae | OR365 | N/A | MG888271 | MG888318 | MG888226 | N/A | N/A | ||
| Ommatolampis quadrimaculata | OR364 | MG993443 | KM853267 | KM853423 | KM853599 | mtgenome | mtgenome | ||
| Pollostacris sp | OR322 | MG993391, MG993411, MG993413 | MG888263 | MG888310 | MG888218 | mtgenome | mtgenome | ||
| Psiloscirtus sp | OR348 | N/A | MG888268 | MG888315 | MG888223 | MG888099 | MG888165 | ||
| Syntomacrella sp | OR323 | N/A | MG888264 | MG888311 | MG888219 | N/A | N/A | ||
| Vilerna sp | OR336 | N/A | KM853252 | KM853438 | KM853614 | MG888097 | N/A | ||
| Xiphidiopteron sp | OR321 | N/A | MG888262 | MG888309 | MG888217 | N/A | N/A | ||
| Oxyinae | Kosciuscola tristis | OR396 | MG993402, MG993408, MG993414 | KM853278 | KM853411 | KM853588 | mtgenome | mtgenome | |
| Oxya chinensis | OR315 | NC_010219 | KM853244 | KM853446 | KM853622 | mtgenome | mtgenome | ||
| Pseudoxya diminuta | N/A | NC_025765 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Pauliniinae | Paulinia acuminata | OR345 | MG993401, MG993416, MG993419, MG993430, MG993431, MG993446 | KM853257 | KM853433 | KM853609 | mtgenome | mtgenome | |
| Proctolabinae | Coscineuta sp | OR249 | MG993441 | KM853212 | KM853478 | KM853653 | mtgenome | mtgenome | |
| Poecilocloeus napoana | OR368 | N/A | KM853270 | KM853420 | KM853596 | N/A | N/A | ||
| Rhytidochrotinae | Galidacris variabilis | OR371 | N/A | KM853271 | KM853419 | KM853595 | N/A | MG888167 | |
| Acrididae | Rhytidochrotinae | Paropaon sp | OR337 | MG993393, MG993397, MG993421, MG993428, MG993449 | KM853253 | KM853437 | KM853613 | mtgenome | mtgenome |
| Spathosterninae | Spathosternum nigrotaeniatum | OR224 | MG993439 | KM853203 | KM853487 | KM853662 | mtgenome | mtgenome | |
| Tropidopolinae | Petamella prosternalis | OR560 | MG993412 | KM853343 | KM853349 | KM853525 | mtgenome | mtgenome | |
| Lentulidae | Lentulinae | Lentula callani | OR295 | NC_020774 | KM853234 | KM853456 | KM853632 | mtgenome | mtgenome |
| Lithidiidae | Lithidiinae | Lithidiopsis carinatus | OR316 | NC_020775 | KM853245 | KM853445 | KM853621 | mtgenome | mtgenome |
| Ommexechidae | Ommexechinae | Ommexecha virens | OR367 | NC_020778 | KM853269 | KM853421 | KM853597 | mtgenome | mtgenome |
| Pamphagidae | Thrinchinae | Prionotropis hystrix | OR151 | JX913764 | KM853180 | KM853509 | KM853684 | mtgenome | mtgenome |
| Pamphagodidae | Unplaced | Hemicharilaus monomorphus | OR540 | JX913773 | KM853337 | KM853355 | KM853531 | mtgenome | mtgenome |
| Romaleidae | Romaleinae | Romalea microptera | OR1000 | MG993392, MG993394, MG993454, MG993455, MG993456, MG993457 | MG888294 | MG888343 | MG888251 | mtgenome | mtgenome |
| Xyleus modestus | OR265 | NC_014490 | KM853221 | KM853469 | KM853644 | mtgenome | mtgenome | ||
| Tristiridae | Tristirinae | Tristira magellanica | OR204 | NC_020773 | KM853197 | KM853493 | KM853668 | mtgenome | mtgenome |
| Family . | Subfamily . | Species . | Voucher # (TAMUIC-IGC-#) . | mtgenome . | 18S . | 28S . | H3 . | COI . | COII . |
|---|---|---|---|---|---|---|---|---|---|
| Acrididae | Acridinae | Acrida willemsei | OR059 | NC_011303 | KM853177 | KM853512 | KM853687 | mtgenome | mtgenome |
| Calephorus compressicornis | OR192 | N/A | KM853192 | KM853498 | KM853673 | MG888076 | MG888143 | ||
| Coryphosima stenoptera | OR512 | N/A | MG888284 | MG888333 | MG888241 | MG888120 | MG888187 | ||
| Gymnobothrus sp | OR511 | N/A | MG888283 | MG888332 | MG888240 | MG888119 | MG888186 | ||
| Hyalopteryx rufipennis | OR240 | N/A | KM853210 | KM853480 | KM853655 | MG888088 | MG888155 | ||
| Keya capicola | OR514 | N/A | MG888286 | MG888335 | MG888243 | MG888122 | MG888189 | ||
| Orthochtha sp | OR513 | N/A | MG888285 | MG888334 | MG888242 | MG888121 | MG888188 | ||
| Phlaeoba albonema | N/A | NC_011827 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Truxalis sp | OR510 | N/A | KM853325 | KM853367 | KM853543 | MG888118 | N/A | ||
| Calliptaminae | Acorypha sp | OR195 | N/A | MG888254 | MG888300 | MG888208 | MG888078 | MG888145 | |
| Calliptamus italicus | OR193 | NC_011305 | KM853193 | KM853497 | KM853672 | mtgenome | mtgenome | ||
| Paracaloptenus caloptenoides | OR194 | N/A | KM853194 | KM853496 | KM853671 | MG888077 | MG888144 | ||
| Catantopinae | Apotropis vittata | OR493 | N/A | MG888277 | MG888325 | MG888233 | MG888106 | MG888173 | |
| Buforania sp | OR500 | N/A | MG888278 | MG888327 | MG888235 | MG888113 | MG888180 | ||
| Catantops sp | OR237 | N/A | KM853209 | KM853481 | KM853656 | MG888086 | MG888153 | ||
| Cedarinia sp | OR490 | N/A | MG888274 | MG888322 | MG888230 | MG888103 | MG888170 | ||
| Coryphistes ruricola | OR503 | MG993389, MG993390, MG993403, MG993406 | MG888281 | MG888330 | MG888238 | mtgenome | mtgenome | ||
| Ecphantus quadrilobus | OR495 | N/A | MG888295 | MG888326 | MG888234 | MG888108 | MG888175 | ||
| Gen nov. 46 sp. 1 | OR491 | N/A | MG888275 | MG888323 | MG888231 | MG888104 | MG888171 | ||
| Gen nov. 64 sp. 1 | OR489 | N/A | MG888273 | MG888321 | MG888229 | MG888102 | MG888169 | ||
| Goniaea vocans | OR502 | N/A | MG888280 | MG888329 | MG888237 | MG888115 | MG888182 | ||
| Kinangopa jeanneli | OR574 | N/A | KM853345 | KM853348 | KM853523 | N/A | MG888205 | ||
| Macrolopholia sp | OR235 | N/A | KM853208 | KM853482 | KM853657 | MG888085 | MG888152 | ||
| Macrotona sp | OR488 | N/A | MG888272 | MG888320 | MG888228 | MG888101 | MG888168 | ||
| Pezocatantops sp | OR505 | N/A | KM853321 | KM853372 | KM853548 | MG888117 | MG888184 | ||
| Phaeocatantops sp | OR504 | N/A | MG888282 | MG888331 | MG888239 | MG888116 | MG888183 | ||
| Porraxia sp | OR494 | N/A | KM853316 | KM853377 | KM853553 | MG888107 | MG888174 | ||
| Retuspia validicornis | OR496 | N/A | KM853317 | KM853376 | KM853552 | MG888109 | MG888176 | ||
| Rusurplia tristis | OR497 | N/A | KM853318 | KM853375 | KM853551 | MG888110 | MG888177 | ||
| Stenocatantops vitripennis | OR498 | N/A | KM853319 | KM853374 | KM853550 | MG888111 | MG888178 | ||
| Traulia szetschuanensis | N/A | NC_013826 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Typaya semicristata | OR492 | N/A | MG888276 | MG888324 | MG888232 | MG888105 | MG888172 | ||
| Urnisa guttulosa | OR501 | N/A | MG888279 | MG888328 | MG888236 | MG888114 | MG888181 | ||
| Urnisiella rubropunctata | OR499 | N/A | KM853320 | KM853373 | KM853549 | MG888112 | MG888179 | ||
| Xenocatantops brachycerus | OR236 | NC_021609 | MG888296 | MG888303 | MG888211 | mtgenome | mtgenome | ||
| Copiocerinae | Copiocera sp | OR333 | MG993384 | KM853250 | KM853440 | KM853616 | mtgenome | mtgenome | |
| Cyphacris sp | OR334 | N/A | KM853251 | KM853439 | KM853615 | MG888096 | MG888163 | ||
| Coptacrinae | Eucoptacra sp | OR509 | MG993445 | KM853324 | KM853368 | KM853544 | mtgenome | mtgenome | |
| Parepistaurus deses | OR508 | N/A | KM853323 | KM853369 | KM853545 | N/A | MG888185 | ||
| Cyrtacanthacridinae | Acanthacris ruficornis | OR183 | N/A | MG888253 | MG888299 | MG888207 | MG888071 | MG888138 | |
| Acridoderes sp | OR546 | N/A | MG888293 | MG888342 | MG888250 | MG888137 | MG888203 | ||
| Anacridium incisum | OR184 | N/A | KM853185 | KM853505 | KM853680 | MG888072 | MG888139 | ||
| Acrididae | Cyrtacanthacridinae | Austracris guttulosa | OR182 | MG993415 | MG888252 | MG888298 | MG888206 | mtgenome | mtgenome |
| Chondracris rosea | N/A | NC_019993 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Cyrtacanthacris tatarica | OR181 | MG993444 | KM853184 | KM853506 | KM853681 | mtgenome | mtgenome | ||
| Nomadacris septemfasciata | OR545 | N/A | KM853340 | KM853352 | KM853528 | MG888136 | N/A | ||
| Ornithacris sp | OR544 | N/A | KM853339 | KM853353 | KM853529 | MG888135 | MG888202 | ||
| Rhadinacris schistocercoides | OR547 | N/A | KM853341 | KM853351 | KM853527 | N/A | MG888204 | ||
| Schistocerca gregaria | OR185 | NC_013240 | KM853186 | KM853504 | KM853679 | mtgenome | mtgenome | ||
| Euryphyminae | Calliptamicus sp | OR313 | N/A | MG888261 | MG888308 | MG888216 | N/A | N/A | |
| Calliptamulus sp | OR311 | N/A | KM853241 | KM853449 | KM853625 | MG888094 | MG888161 | ||
| Euryphymus sp | OR314 | MG993388, MG993422, MG993436 | KM853243 | KM853447 | KM853623 | mtgenome | mtgenome | ||
| Pachyphymus sp | OR308 | N/A | MG888260 | MG888307 | MG888215 | MG888092 | MG888159 | ||
| Rhachitopis sp | OR312 | N/A | KM853242 | KM853448 | KM853624 | N/A | N/A | ||
| Eyprepocnemidinae | Cataloipus sp | OR218 | N/A | KM853201 | KM853489 | KM853664 | MG888079 | MG888146 | |
| Eyprepocnemis plorans | OR309 | MG993386, MG993418,MG993424, MG993425,MG993427, MG993433,MG993437, MG993450 | KM853239 | KM853451 | KM853627 | mtgenome | mtgenome | ||
| Heteracris sp | OR310 | N/A | KM853240 | KM853450 | KM853626 | MG888093 | MG888160 | ||
| Shirakiacris shirakii | N/A | NC_021610 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Tylotropidius sp | OR219 | N/A | MG888255 | MG888301 | MG888209 | MG888080 | MG888147 | ||
| Gomphocerinae | Arcyptera coreana | N/A | NC_013805 | N/A | N/A | N/A | mtgenome | mtgenome | |
| Aulocara elliotii | OR521 | N/A | KM853329 | KM853363 | KM853539 | MG888129 | MG888196 | ||
| Dichromorpha viridis | OR226 | N/A | KM853205 | KM853485 | KM853660 | MG888083 | MG888150 | ||
| Euchorthippus fusigeniculatus | N/A | NC_014449 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Gomphocerus sibiricus | N/A | NC_015478 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Mermiria intertexta | OR520 | N/A | KM853328 | KM853364 | KM853540 | MG888128 | MG888195 | ||
| Mesopsis sp | OR239 | N/A | MG888257 | MG888304 | MG888212 | MG888087 | MG888154 | ||
| Orinhippus tibetanus | N/A | NC_023467 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Pacris xizangensis | N/A | NC_023919 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Prorocorypha snowi | OR214 | MG993438, MG993452, MG993453 | KM853199 | KM853491 | KM853666 | mtgenome | mtgenome | ||
| Pseudogmothela sp | OR519 | N/A | MG888289 | MG888338 | MG888246 | MG888127 | MG888194 | ||
| Rhammatocerus schistocercoides | OR346 | N/A | KM853258 | KM853432 | KM853608 | MG888098 | MG888164 | ||
| Rhaphotittha sp | OR518 | N/A | MG888288 | MG888337 | MG888245 | MG888126 | MG888193 | ||
| Silvitettix sp | OR343 | N/A | MG888267 | MG888314 | MG888222 | N/A | N/A | ||
| Syrbula montezuma | OR227 | N/A | KM853206 | KM853484 | KM853659 | MG888084 | MG888151 | ||
| Acrididae | Hemiacridinae | Dirshacris aridus | OR305 | MG993398, MG993399, MG993410, MG993417, MG993420, MG993434, MG993435, MG993451 | MG888259 | MG888306 | MG888214 | mtgenome | mtgenome |
| Euroryma sp | OR302 | N/A | KM853236 | KM853454 | KM853630 | MG888090 | MG888157 | ||
| Hieroglyphus tonkinensis | N/A | NC_030587 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Leptacris sp | OR304 | MG993429 | KM853238 | KM853452 | KM853628 | mtgenome | mtgenome | ||
| Paulianiobia hirsuta | OR301 | N/A | MG888258 | MG888305 | MG888213 | MG888089 | MG888156 | ||
| Pristocorypha sp | OR303 | N/A | KM853237 | KM853453 | KM853629 | MG888091 | MG888158 | ||
| Leptysminae | Stenacris sp | OR342 | N/A | KM853255 | KM853435 | KM853611 | N/A | N/A | |
| Stenopola sp | OR220 | N/A | MG888256 | MG888302 | MG888210 | MG888081 | MG888148 | ||
| Tetrataenia surinama | OR338 | MG993385, MG993395, MG993396, MG993404, MG993407, MG993409, MG993432, MG993448 | KM853254 | KM853436 | KM853612 | mtgenome | mtgenome | ||
| Marelliinae | Marellia remipes | OR344 | MG993387, MG993423, MG993442, MG993447 | KM853256 | KM853434 | KM853610 | mtgenome | mtgenome | |
| Melanoplinae | Anapodisma miramae | OR356 | N/A | KM853265 | KM853425 | KM853601 | MG888100 | MG888166 | |
| Aptenopedes sphenarioides | OR516 | N/A | MG888287 | MG888336 | MG888244 | MG888124 | MG888191 | ||
| Bradynotes obesa | OR515 | N/A | KM853326 | KM853366 | KM853542 | MG888123 | MG888190 | ||
| Dichroplus sp | OR325 | N/A | KM853248 | KM853442 | KM853618 | MG888095 | MG888162 | ||
| Fruhstorferiola kulinga | N/A | NC_026716 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Hesperotettix viridis | OR517 | N/A | KM853327 | KM853365 | KM853541 | MG888125 | MG888192 | ||
| Jivarus ronderosi | OR328 | MG993400, MG993405 | KM853249 | KM853441 | KM853617 | mtgenome | mtgenome | ||
| Kingdonella bicollina | N/A | NC_023920 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Maeacris aptera | OR329 | N/A | MG888266 | MG888313 | MG888221 | N/A | N/A | ||
| Melanoplus bivittatus | OR245 | MG993426 | KM853211 | KM853479 | KM853654 | mtgenome | mtgenome | ||
| Ognevia longipennis | OR394 | NC_013701 | MG888297 | MG888319 | MG888227 | mtgenome | mtgenome | ||
| Ponderacris peruvianus | OR324 | N/A | MG888265 | MG888312 | MG888220 | N/A | N/A | ||
| Prumna arctica | OR395 | NC_013835 | KM853277 | KM853412 | KM853589 | mtgenome | mtgenome | ||
| Qinlingacris taibaiensis | N/A | NC_027187 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Oedipodinae | Acrotylus patruelis | OR190 | N/A | KM853190 | KM853500 | KM853675 | MG888075 | MG888142 | |
| Acrididae | Oedipodinae | Angaracris barabensis | N/A | NC_025946 | N/A | N/A | N/A | mtgenome | mtgenome |
| Bryodema miramae miramae | N/A | KP889242 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Ceracris kiangsu | N/A | NC_019994 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Chortoicetes terminifera | OR524 | N/A | MG888290 | MG888339 | MG888247 | MG888132 | MG888199 | ||
| Gastrimargus marmoratus | N/A | NC_011114 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Heteropternis sp | OR225 | N/A | KM853204 | KM853486 | KM853661 | MG888082 | MG888149 | ||
| Locusta migratoria | OR191 | NC_001712 | KM853191 | KM853499 | KM853674 | mtgenome | mtgenome | ||
| Psinidia fenestralis | OR522 | N/A | KM853330 | KM853362 | KM853538 | MG888130 | MG888197 | ||
| Pycnostictus seriatus | OR525 | N/A | MG888291 | MG888340 | MG888248 | MG888133 | MG888200 | ||
| Qualetta maculata | OR526 | N/A | MG888292 | MG888341 | MG888249 | MG888134 | MG888201 | ||
| Tomonotus ferruginosus | OR523 | N/A | KM853331 | KM853361 | KM853537 | MG888131 | MG888198 | ||
| Trilophidia annulata | N/A | NC_027179 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Trimerotropis sp | OR186 | N/A | KM853187 | KM853503 | KM853678 | MG888073 | MG888140 | ||
| Xanthippus sp | OR187 | N/A | KM853188 | KM853502 | KM853677 | MG888074 | MG888141 | ||
| Ommatolampidinae | Abracris sp | OR222 | MG993440 | KM853202 | KM853488 | KM853663 | mtgenome | mtgenome | |
| Anablysis teres | OR362 | N/A | MG888269 | MG888316 | MG888224 | N/A | N/A | ||
| Kyphiacris sp | OR363 | N/A | MG888270 | MG888317 | MG888225 | N/A | N/A | ||
| Locheuma brunneri | OR366 | N/A | KM853268 | KM853422 | KM853598 | N/A | N/A | ||
| Lysacris festae | OR365 | N/A | MG888271 | MG888318 | MG888226 | N/A | N/A | ||
| Ommatolampis quadrimaculata | OR364 | MG993443 | KM853267 | KM853423 | KM853599 | mtgenome | mtgenome | ||
| Pollostacris sp | OR322 | MG993391, MG993411, MG993413 | MG888263 | MG888310 | MG888218 | mtgenome | mtgenome | ||
| Psiloscirtus sp | OR348 | N/A | MG888268 | MG888315 | MG888223 | MG888099 | MG888165 | ||
| Syntomacrella sp | OR323 | N/A | MG888264 | MG888311 | MG888219 | N/A | N/A | ||
| Vilerna sp | OR336 | N/A | KM853252 | KM853438 | KM853614 | MG888097 | N/A | ||
| Xiphidiopteron sp | OR321 | N/A | MG888262 | MG888309 | MG888217 | N/A | N/A | ||
| Oxyinae | Kosciuscola tristis | OR396 | MG993402, MG993408, MG993414 | KM853278 | KM853411 | KM853588 | mtgenome | mtgenome | |
| Oxya chinensis | OR315 | NC_010219 | KM853244 | KM853446 | KM853622 | mtgenome | mtgenome | ||
| Pseudoxya diminuta | N/A | NC_025765 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Pauliniinae | Paulinia acuminata | OR345 | MG993401, MG993416, MG993419, MG993430, MG993431, MG993446 | KM853257 | KM853433 | KM853609 | mtgenome | mtgenome | |
| Proctolabinae | Coscineuta sp | OR249 | MG993441 | KM853212 | KM853478 | KM853653 | mtgenome | mtgenome | |
| Poecilocloeus napoana | OR368 | N/A | KM853270 | KM853420 | KM853596 | N/A | N/A | ||
| Rhytidochrotinae | Galidacris variabilis | OR371 | N/A | KM853271 | KM853419 | KM853595 | N/A | MG888167 | |
| Acrididae | Rhytidochrotinae | Paropaon sp | OR337 | MG993393, MG993397, MG993421, MG993428, MG993449 | KM853253 | KM853437 | KM853613 | mtgenome | mtgenome |
| Spathosterninae | Spathosternum nigrotaeniatum | OR224 | MG993439 | KM853203 | KM853487 | KM853662 | mtgenome | mtgenome | |
| Tropidopolinae | Petamella prosternalis | OR560 | MG993412 | KM853343 | KM853349 | KM853525 | mtgenome | mtgenome | |
| Lentulidae | Lentulinae | Lentula callani | OR295 | NC_020774 | KM853234 | KM853456 | KM853632 | mtgenome | mtgenome |
| Lithidiidae | Lithidiinae | Lithidiopsis carinatus | OR316 | NC_020775 | KM853245 | KM853445 | KM853621 | mtgenome | mtgenome |
| Ommexechidae | Ommexechinae | Ommexecha virens | OR367 | NC_020778 | KM853269 | KM853421 | KM853597 | mtgenome | mtgenome |
| Pamphagidae | Thrinchinae | Prionotropis hystrix | OR151 | JX913764 | KM853180 | KM853509 | KM853684 | mtgenome | mtgenome |
| Pamphagodidae | Unplaced | Hemicharilaus monomorphus | OR540 | JX913773 | KM853337 | KM853355 | KM853531 | mtgenome | mtgenome |
| Romaleidae | Romaleinae | Romalea microptera | OR1000 | MG993392, MG993394, MG993454, MG993455, MG993456, MG993457 | MG888294 | MG888343 | MG888251 | mtgenome | mtgenome |
| Xyleus modestus | OR265 | NC_014490 | KM853221 | KM853469 | KM853644 | mtgenome | mtgenome | ||
| Tristiridae | Tristirinae | Tristira magellanica | OR204 | NC_020773 | KM853197 | KM853493 | KM853668 | mtgenome | mtgenome |
‘N/A’ means that the sequence data were not available. ‘mtgenome’ means that the corresponding sequences were derived from the available mtgenome data. For some taxa, multiple Genbank accession numbers are assigned for mtgenome, which indicates partial mtgenomes consisting of several fragments.
Taxonomic information and Genbank accession numbers for 142 taxa used in total evidence analysis
| Family . | Subfamily . | Species . | Voucher # (TAMUIC-IGC-#) . | mtgenome . | 18S . | 28S . | H3 . | COI . | COII . |
|---|---|---|---|---|---|---|---|---|---|
| Acrididae | Acridinae | Acrida willemsei | OR059 | NC_011303 | KM853177 | KM853512 | KM853687 | mtgenome | mtgenome |
| Calephorus compressicornis | OR192 | N/A | KM853192 | KM853498 | KM853673 | MG888076 | MG888143 | ||
| Coryphosima stenoptera | OR512 | N/A | MG888284 | MG888333 | MG888241 | MG888120 | MG888187 | ||
| Gymnobothrus sp | OR511 | N/A | MG888283 | MG888332 | MG888240 | MG888119 | MG888186 | ||
| Hyalopteryx rufipennis | OR240 | N/A | KM853210 | KM853480 | KM853655 | MG888088 | MG888155 | ||
| Keya capicola | OR514 | N/A | MG888286 | MG888335 | MG888243 | MG888122 | MG888189 | ||
| Orthochtha sp | OR513 | N/A | MG888285 | MG888334 | MG888242 | MG888121 | MG888188 | ||
| Phlaeoba albonema | N/A | NC_011827 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Truxalis sp | OR510 | N/A | KM853325 | KM853367 | KM853543 | MG888118 | N/A | ||
| Calliptaminae | Acorypha sp | OR195 | N/A | MG888254 | MG888300 | MG888208 | MG888078 | MG888145 | |
| Calliptamus italicus | OR193 | NC_011305 | KM853193 | KM853497 | KM853672 | mtgenome | mtgenome | ||
| Paracaloptenus caloptenoides | OR194 | N/A | KM853194 | KM853496 | KM853671 | MG888077 | MG888144 | ||
| Catantopinae | Apotropis vittata | OR493 | N/A | MG888277 | MG888325 | MG888233 | MG888106 | MG888173 | |
| Buforania sp | OR500 | N/A | MG888278 | MG888327 | MG888235 | MG888113 | MG888180 | ||
| Catantops sp | OR237 | N/A | KM853209 | KM853481 | KM853656 | MG888086 | MG888153 | ||
| Cedarinia sp | OR490 | N/A | MG888274 | MG888322 | MG888230 | MG888103 | MG888170 | ||
| Coryphistes ruricola | OR503 | MG993389, MG993390, MG993403, MG993406 | MG888281 | MG888330 | MG888238 | mtgenome | mtgenome | ||
| Ecphantus quadrilobus | OR495 | N/A | MG888295 | MG888326 | MG888234 | MG888108 | MG888175 | ||
| Gen nov. 46 sp. 1 | OR491 | N/A | MG888275 | MG888323 | MG888231 | MG888104 | MG888171 | ||
| Gen nov. 64 sp. 1 | OR489 | N/A | MG888273 | MG888321 | MG888229 | MG888102 | MG888169 | ||
| Goniaea vocans | OR502 | N/A | MG888280 | MG888329 | MG888237 | MG888115 | MG888182 | ||
| Kinangopa jeanneli | OR574 | N/A | KM853345 | KM853348 | KM853523 | N/A | MG888205 | ||
| Macrolopholia sp | OR235 | N/A | KM853208 | KM853482 | KM853657 | MG888085 | MG888152 | ||
| Macrotona sp | OR488 | N/A | MG888272 | MG888320 | MG888228 | MG888101 | MG888168 | ||
| Pezocatantops sp | OR505 | N/A | KM853321 | KM853372 | KM853548 | MG888117 | MG888184 | ||
| Phaeocatantops sp | OR504 | N/A | MG888282 | MG888331 | MG888239 | MG888116 | MG888183 | ||
| Porraxia sp | OR494 | N/A | KM853316 | KM853377 | KM853553 | MG888107 | MG888174 | ||
| Retuspia validicornis | OR496 | N/A | KM853317 | KM853376 | KM853552 | MG888109 | MG888176 | ||
| Rusurplia tristis | OR497 | N/A | KM853318 | KM853375 | KM853551 | MG888110 | MG888177 | ||
| Stenocatantops vitripennis | OR498 | N/A | KM853319 | KM853374 | KM853550 | MG888111 | MG888178 | ||
| Traulia szetschuanensis | N/A | NC_013826 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Typaya semicristata | OR492 | N/A | MG888276 | MG888324 | MG888232 | MG888105 | MG888172 | ||
| Urnisa guttulosa | OR501 | N/A | MG888279 | MG888328 | MG888236 | MG888114 | MG888181 | ||
| Urnisiella rubropunctata | OR499 | N/A | KM853320 | KM853373 | KM853549 | MG888112 | MG888179 | ||
| Xenocatantops brachycerus | OR236 | NC_021609 | MG888296 | MG888303 | MG888211 | mtgenome | mtgenome | ||
| Copiocerinae | Copiocera sp | OR333 | MG993384 | KM853250 | KM853440 | KM853616 | mtgenome | mtgenome | |
| Cyphacris sp | OR334 | N/A | KM853251 | KM853439 | KM853615 | MG888096 | MG888163 | ||
| Coptacrinae | Eucoptacra sp | OR509 | MG993445 | KM853324 | KM853368 | KM853544 | mtgenome | mtgenome | |
| Parepistaurus deses | OR508 | N/A | KM853323 | KM853369 | KM853545 | N/A | MG888185 | ||
| Cyrtacanthacridinae | Acanthacris ruficornis | OR183 | N/A | MG888253 | MG888299 | MG888207 | MG888071 | MG888138 | |
| Acridoderes sp | OR546 | N/A | MG888293 | MG888342 | MG888250 | MG888137 | MG888203 | ||
| Anacridium incisum | OR184 | N/A | KM853185 | KM853505 | KM853680 | MG888072 | MG888139 | ||
| Acrididae | Cyrtacanthacridinae | Austracris guttulosa | OR182 | MG993415 | MG888252 | MG888298 | MG888206 | mtgenome | mtgenome |
| Chondracris rosea | N/A | NC_019993 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Cyrtacanthacris tatarica | OR181 | MG993444 | KM853184 | KM853506 | KM853681 | mtgenome | mtgenome | ||
| Nomadacris septemfasciata | OR545 | N/A | KM853340 | KM853352 | KM853528 | MG888136 | N/A | ||
| Ornithacris sp | OR544 | N/A | KM853339 | KM853353 | KM853529 | MG888135 | MG888202 | ||
| Rhadinacris schistocercoides | OR547 | N/A | KM853341 | KM853351 | KM853527 | N/A | MG888204 | ||
| Schistocerca gregaria | OR185 | NC_013240 | KM853186 | KM853504 | KM853679 | mtgenome | mtgenome | ||
| Euryphyminae | Calliptamicus sp | OR313 | N/A | MG888261 | MG888308 | MG888216 | N/A | N/A | |
| Calliptamulus sp | OR311 | N/A | KM853241 | KM853449 | KM853625 | MG888094 | MG888161 | ||
| Euryphymus sp | OR314 | MG993388, MG993422, MG993436 | KM853243 | KM853447 | KM853623 | mtgenome | mtgenome | ||
| Pachyphymus sp | OR308 | N/A | MG888260 | MG888307 | MG888215 | MG888092 | MG888159 | ||
| Rhachitopis sp | OR312 | N/A | KM853242 | KM853448 | KM853624 | N/A | N/A | ||
| Eyprepocnemidinae | Cataloipus sp | OR218 | N/A | KM853201 | KM853489 | KM853664 | MG888079 | MG888146 | |
| Eyprepocnemis plorans | OR309 | MG993386, MG993418,MG993424, MG993425,MG993427, MG993433,MG993437, MG993450 | KM853239 | KM853451 | KM853627 | mtgenome | mtgenome | ||
| Heteracris sp | OR310 | N/A | KM853240 | KM853450 | KM853626 | MG888093 | MG888160 | ||
| Shirakiacris shirakii | N/A | NC_021610 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Tylotropidius sp | OR219 | N/A | MG888255 | MG888301 | MG888209 | MG888080 | MG888147 | ||
| Gomphocerinae | Arcyptera coreana | N/A | NC_013805 | N/A | N/A | N/A | mtgenome | mtgenome | |
| Aulocara elliotii | OR521 | N/A | KM853329 | KM853363 | KM853539 | MG888129 | MG888196 | ||
| Dichromorpha viridis | OR226 | N/A | KM853205 | KM853485 | KM853660 | MG888083 | MG888150 | ||
| Euchorthippus fusigeniculatus | N/A | NC_014449 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Gomphocerus sibiricus | N/A | NC_015478 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Mermiria intertexta | OR520 | N/A | KM853328 | KM853364 | KM853540 | MG888128 | MG888195 | ||
| Mesopsis sp | OR239 | N/A | MG888257 | MG888304 | MG888212 | MG888087 | MG888154 | ||
| Orinhippus tibetanus | N/A | NC_023467 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Pacris xizangensis | N/A | NC_023919 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Prorocorypha snowi | OR214 | MG993438, MG993452, MG993453 | KM853199 | KM853491 | KM853666 | mtgenome | mtgenome | ||
| Pseudogmothela sp | OR519 | N/A | MG888289 | MG888338 | MG888246 | MG888127 | MG888194 | ||
| Rhammatocerus schistocercoides | OR346 | N/A | KM853258 | KM853432 | KM853608 | MG888098 | MG888164 | ||
| Rhaphotittha sp | OR518 | N/A | MG888288 | MG888337 | MG888245 | MG888126 | MG888193 | ||
| Silvitettix sp | OR343 | N/A | MG888267 | MG888314 | MG888222 | N/A | N/A | ||
| Syrbula montezuma | OR227 | N/A | KM853206 | KM853484 | KM853659 | MG888084 | MG888151 | ||
| Acrididae | Hemiacridinae | Dirshacris aridus | OR305 | MG993398, MG993399, MG993410, MG993417, MG993420, MG993434, MG993435, MG993451 | MG888259 | MG888306 | MG888214 | mtgenome | mtgenome |
| Euroryma sp | OR302 | N/A | KM853236 | KM853454 | KM853630 | MG888090 | MG888157 | ||
| Hieroglyphus tonkinensis | N/A | NC_030587 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Leptacris sp | OR304 | MG993429 | KM853238 | KM853452 | KM853628 | mtgenome | mtgenome | ||
| Paulianiobia hirsuta | OR301 | N/A | MG888258 | MG888305 | MG888213 | MG888089 | MG888156 | ||
| Pristocorypha sp | OR303 | N/A | KM853237 | KM853453 | KM853629 | MG888091 | MG888158 | ||
| Leptysminae | Stenacris sp | OR342 | N/A | KM853255 | KM853435 | KM853611 | N/A | N/A | |
| Stenopola sp | OR220 | N/A | MG888256 | MG888302 | MG888210 | MG888081 | MG888148 | ||
| Tetrataenia surinama | OR338 | MG993385, MG993395, MG993396, MG993404, MG993407, MG993409, MG993432, MG993448 | KM853254 | KM853436 | KM853612 | mtgenome | mtgenome | ||
| Marelliinae | Marellia remipes | OR344 | MG993387, MG993423, MG993442, MG993447 | KM853256 | KM853434 | KM853610 | mtgenome | mtgenome | |
| Melanoplinae | Anapodisma miramae | OR356 | N/A | KM853265 | KM853425 | KM853601 | MG888100 | MG888166 | |
| Aptenopedes sphenarioides | OR516 | N/A | MG888287 | MG888336 | MG888244 | MG888124 | MG888191 | ||
| Bradynotes obesa | OR515 | N/A | KM853326 | KM853366 | KM853542 | MG888123 | MG888190 | ||
| Dichroplus sp | OR325 | N/A | KM853248 | KM853442 | KM853618 | MG888095 | MG888162 | ||
| Fruhstorferiola kulinga | N/A | NC_026716 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Hesperotettix viridis | OR517 | N/A | KM853327 | KM853365 | KM853541 | MG888125 | MG888192 | ||
| Jivarus ronderosi | OR328 | MG993400, MG993405 | KM853249 | KM853441 | KM853617 | mtgenome | mtgenome | ||
| Kingdonella bicollina | N/A | NC_023920 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Maeacris aptera | OR329 | N/A | MG888266 | MG888313 | MG888221 | N/A | N/A | ||
| Melanoplus bivittatus | OR245 | MG993426 | KM853211 | KM853479 | KM853654 | mtgenome | mtgenome | ||
| Ognevia longipennis | OR394 | NC_013701 | MG888297 | MG888319 | MG888227 | mtgenome | mtgenome | ||
| Ponderacris peruvianus | OR324 | N/A | MG888265 | MG888312 | MG888220 | N/A | N/A | ||
| Prumna arctica | OR395 | NC_013835 | KM853277 | KM853412 | KM853589 | mtgenome | mtgenome | ||
| Qinlingacris taibaiensis | N/A | NC_027187 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Oedipodinae | Acrotylus patruelis | OR190 | N/A | KM853190 | KM853500 | KM853675 | MG888075 | MG888142 | |
| Acrididae | Oedipodinae | Angaracris barabensis | N/A | NC_025946 | N/A | N/A | N/A | mtgenome | mtgenome |
| Bryodema miramae miramae | N/A | KP889242 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Ceracris kiangsu | N/A | NC_019994 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Chortoicetes terminifera | OR524 | N/A | MG888290 | MG888339 | MG888247 | MG888132 | MG888199 | ||
| Gastrimargus marmoratus | N/A | NC_011114 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Heteropternis sp | OR225 | N/A | KM853204 | KM853486 | KM853661 | MG888082 | MG888149 | ||
| Locusta migratoria | OR191 | NC_001712 | KM853191 | KM853499 | KM853674 | mtgenome | mtgenome | ||
| Psinidia fenestralis | OR522 | N/A | KM853330 | KM853362 | KM853538 | MG888130 | MG888197 | ||
| Pycnostictus seriatus | OR525 | N/A | MG888291 | MG888340 | MG888248 | MG888133 | MG888200 | ||
| Qualetta maculata | OR526 | N/A | MG888292 | MG888341 | MG888249 | MG888134 | MG888201 | ||
| Tomonotus ferruginosus | OR523 | N/A | KM853331 | KM853361 | KM853537 | MG888131 | MG888198 | ||
| Trilophidia annulata | N/A | NC_027179 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Trimerotropis sp | OR186 | N/A | KM853187 | KM853503 | KM853678 | MG888073 | MG888140 | ||
| Xanthippus sp | OR187 | N/A | KM853188 | KM853502 | KM853677 | MG888074 | MG888141 | ||
| Ommatolampidinae | Abracris sp | OR222 | MG993440 | KM853202 | KM853488 | KM853663 | mtgenome | mtgenome | |
| Anablysis teres | OR362 | N/A | MG888269 | MG888316 | MG888224 | N/A | N/A | ||
| Kyphiacris sp | OR363 | N/A | MG888270 | MG888317 | MG888225 | N/A | N/A | ||
| Locheuma brunneri | OR366 | N/A | KM853268 | KM853422 | KM853598 | N/A | N/A | ||
| Lysacris festae | OR365 | N/A | MG888271 | MG888318 | MG888226 | N/A | N/A | ||
| Ommatolampis quadrimaculata | OR364 | MG993443 | KM853267 | KM853423 | KM853599 | mtgenome | mtgenome | ||
| Pollostacris sp | OR322 | MG993391, MG993411, MG993413 | MG888263 | MG888310 | MG888218 | mtgenome | mtgenome | ||
| Psiloscirtus sp | OR348 | N/A | MG888268 | MG888315 | MG888223 | MG888099 | MG888165 | ||
| Syntomacrella sp | OR323 | N/A | MG888264 | MG888311 | MG888219 | N/A | N/A | ||
| Vilerna sp | OR336 | N/A | KM853252 | KM853438 | KM853614 | MG888097 | N/A | ||
| Xiphidiopteron sp | OR321 | N/A | MG888262 | MG888309 | MG888217 | N/A | N/A | ||
| Oxyinae | Kosciuscola tristis | OR396 | MG993402, MG993408, MG993414 | KM853278 | KM853411 | KM853588 | mtgenome | mtgenome | |
| Oxya chinensis | OR315 | NC_010219 | KM853244 | KM853446 | KM853622 | mtgenome | mtgenome | ||
| Pseudoxya diminuta | N/A | NC_025765 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Pauliniinae | Paulinia acuminata | OR345 | MG993401, MG993416, MG993419, MG993430, MG993431, MG993446 | KM853257 | KM853433 | KM853609 | mtgenome | mtgenome | |
| Proctolabinae | Coscineuta sp | OR249 | MG993441 | KM853212 | KM853478 | KM853653 | mtgenome | mtgenome | |
| Poecilocloeus napoana | OR368 | N/A | KM853270 | KM853420 | KM853596 | N/A | N/A | ||
| Rhytidochrotinae | Galidacris variabilis | OR371 | N/A | KM853271 | KM853419 | KM853595 | N/A | MG888167 | |
| Acrididae | Rhytidochrotinae | Paropaon sp | OR337 | MG993393, MG993397, MG993421, MG993428, MG993449 | KM853253 | KM853437 | KM853613 | mtgenome | mtgenome |
| Spathosterninae | Spathosternum nigrotaeniatum | OR224 | MG993439 | KM853203 | KM853487 | KM853662 | mtgenome | mtgenome | |
| Tropidopolinae | Petamella prosternalis | OR560 | MG993412 | KM853343 | KM853349 | KM853525 | mtgenome | mtgenome | |
| Lentulidae | Lentulinae | Lentula callani | OR295 | NC_020774 | KM853234 | KM853456 | KM853632 | mtgenome | mtgenome |
| Lithidiidae | Lithidiinae | Lithidiopsis carinatus | OR316 | NC_020775 | KM853245 | KM853445 | KM853621 | mtgenome | mtgenome |
| Ommexechidae | Ommexechinae | Ommexecha virens | OR367 | NC_020778 | KM853269 | KM853421 | KM853597 | mtgenome | mtgenome |
| Pamphagidae | Thrinchinae | Prionotropis hystrix | OR151 | JX913764 | KM853180 | KM853509 | KM853684 | mtgenome | mtgenome |
| Pamphagodidae | Unplaced | Hemicharilaus monomorphus | OR540 | JX913773 | KM853337 | KM853355 | KM853531 | mtgenome | mtgenome |
| Romaleidae | Romaleinae | Romalea microptera | OR1000 | MG993392, MG993394, MG993454, MG993455, MG993456, MG993457 | MG888294 | MG888343 | MG888251 | mtgenome | mtgenome |
| Xyleus modestus | OR265 | NC_014490 | KM853221 | KM853469 | KM853644 | mtgenome | mtgenome | ||
| Tristiridae | Tristirinae | Tristira magellanica | OR204 | NC_020773 | KM853197 | KM853493 | KM853668 | mtgenome | mtgenome |
| Family . | Subfamily . | Species . | Voucher # (TAMUIC-IGC-#) . | mtgenome . | 18S . | 28S . | H3 . | COI . | COII . |
|---|---|---|---|---|---|---|---|---|---|
| Acrididae | Acridinae | Acrida willemsei | OR059 | NC_011303 | KM853177 | KM853512 | KM853687 | mtgenome | mtgenome |
| Calephorus compressicornis | OR192 | N/A | KM853192 | KM853498 | KM853673 | MG888076 | MG888143 | ||
| Coryphosima stenoptera | OR512 | N/A | MG888284 | MG888333 | MG888241 | MG888120 | MG888187 | ||
| Gymnobothrus sp | OR511 | N/A | MG888283 | MG888332 | MG888240 | MG888119 | MG888186 | ||
| Hyalopteryx rufipennis | OR240 | N/A | KM853210 | KM853480 | KM853655 | MG888088 | MG888155 | ||
| Keya capicola | OR514 | N/A | MG888286 | MG888335 | MG888243 | MG888122 | MG888189 | ||
| Orthochtha sp | OR513 | N/A | MG888285 | MG888334 | MG888242 | MG888121 | MG888188 | ||
| Phlaeoba albonema | N/A | NC_011827 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Truxalis sp | OR510 | N/A | KM853325 | KM853367 | KM853543 | MG888118 | N/A | ||
| Calliptaminae | Acorypha sp | OR195 | N/A | MG888254 | MG888300 | MG888208 | MG888078 | MG888145 | |
| Calliptamus italicus | OR193 | NC_011305 | KM853193 | KM853497 | KM853672 | mtgenome | mtgenome | ||
| Paracaloptenus caloptenoides | OR194 | N/A | KM853194 | KM853496 | KM853671 | MG888077 | MG888144 | ||
| Catantopinae | Apotropis vittata | OR493 | N/A | MG888277 | MG888325 | MG888233 | MG888106 | MG888173 | |
| Buforania sp | OR500 | N/A | MG888278 | MG888327 | MG888235 | MG888113 | MG888180 | ||
| Catantops sp | OR237 | N/A | KM853209 | KM853481 | KM853656 | MG888086 | MG888153 | ||
| Cedarinia sp | OR490 | N/A | MG888274 | MG888322 | MG888230 | MG888103 | MG888170 | ||
| Coryphistes ruricola | OR503 | MG993389, MG993390, MG993403, MG993406 | MG888281 | MG888330 | MG888238 | mtgenome | mtgenome | ||
| Ecphantus quadrilobus | OR495 | N/A | MG888295 | MG888326 | MG888234 | MG888108 | MG888175 | ||
| Gen nov. 46 sp. 1 | OR491 | N/A | MG888275 | MG888323 | MG888231 | MG888104 | MG888171 | ||
| Gen nov. 64 sp. 1 | OR489 | N/A | MG888273 | MG888321 | MG888229 | MG888102 | MG888169 | ||
| Goniaea vocans | OR502 | N/A | MG888280 | MG888329 | MG888237 | MG888115 | MG888182 | ||
| Kinangopa jeanneli | OR574 | N/A | KM853345 | KM853348 | KM853523 | N/A | MG888205 | ||
| Macrolopholia sp | OR235 | N/A | KM853208 | KM853482 | KM853657 | MG888085 | MG888152 | ||
| Macrotona sp | OR488 | N/A | MG888272 | MG888320 | MG888228 | MG888101 | MG888168 | ||
| Pezocatantops sp | OR505 | N/A | KM853321 | KM853372 | KM853548 | MG888117 | MG888184 | ||
| Phaeocatantops sp | OR504 | N/A | MG888282 | MG888331 | MG888239 | MG888116 | MG888183 | ||
| Porraxia sp | OR494 | N/A | KM853316 | KM853377 | KM853553 | MG888107 | MG888174 | ||
| Retuspia validicornis | OR496 | N/A | KM853317 | KM853376 | KM853552 | MG888109 | MG888176 | ||
| Rusurplia tristis | OR497 | N/A | KM853318 | KM853375 | KM853551 | MG888110 | MG888177 | ||
| Stenocatantops vitripennis | OR498 | N/A | KM853319 | KM853374 | KM853550 | MG888111 | MG888178 | ||
| Traulia szetschuanensis | N/A | NC_013826 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Typaya semicristata | OR492 | N/A | MG888276 | MG888324 | MG888232 | MG888105 | MG888172 | ||
| Urnisa guttulosa | OR501 | N/A | MG888279 | MG888328 | MG888236 | MG888114 | MG888181 | ||
| Urnisiella rubropunctata | OR499 | N/A | KM853320 | KM853373 | KM853549 | MG888112 | MG888179 | ||
| Xenocatantops brachycerus | OR236 | NC_021609 | MG888296 | MG888303 | MG888211 | mtgenome | mtgenome | ||
| Copiocerinae | Copiocera sp | OR333 | MG993384 | KM853250 | KM853440 | KM853616 | mtgenome | mtgenome | |
| Cyphacris sp | OR334 | N/A | KM853251 | KM853439 | KM853615 | MG888096 | MG888163 | ||
| Coptacrinae | Eucoptacra sp | OR509 | MG993445 | KM853324 | KM853368 | KM853544 | mtgenome | mtgenome | |
| Parepistaurus deses | OR508 | N/A | KM853323 | KM853369 | KM853545 | N/A | MG888185 | ||
| Cyrtacanthacridinae | Acanthacris ruficornis | OR183 | N/A | MG888253 | MG888299 | MG888207 | MG888071 | MG888138 | |
| Acridoderes sp | OR546 | N/A | MG888293 | MG888342 | MG888250 | MG888137 | MG888203 | ||
| Anacridium incisum | OR184 | N/A | KM853185 | KM853505 | KM853680 | MG888072 | MG888139 | ||
| Acrididae | Cyrtacanthacridinae | Austracris guttulosa | OR182 | MG993415 | MG888252 | MG888298 | MG888206 | mtgenome | mtgenome |
| Chondracris rosea | N/A | NC_019993 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Cyrtacanthacris tatarica | OR181 | MG993444 | KM853184 | KM853506 | KM853681 | mtgenome | mtgenome | ||
| Nomadacris septemfasciata | OR545 | N/A | KM853340 | KM853352 | KM853528 | MG888136 | N/A | ||
| Ornithacris sp | OR544 | N/A | KM853339 | KM853353 | KM853529 | MG888135 | MG888202 | ||
| Rhadinacris schistocercoides | OR547 | N/A | KM853341 | KM853351 | KM853527 | N/A | MG888204 | ||
| Schistocerca gregaria | OR185 | NC_013240 | KM853186 | KM853504 | KM853679 | mtgenome | mtgenome | ||
| Euryphyminae | Calliptamicus sp | OR313 | N/A | MG888261 | MG888308 | MG888216 | N/A | N/A | |
| Calliptamulus sp | OR311 | N/A | KM853241 | KM853449 | KM853625 | MG888094 | MG888161 | ||
| Euryphymus sp | OR314 | MG993388, MG993422, MG993436 | KM853243 | KM853447 | KM853623 | mtgenome | mtgenome | ||
| Pachyphymus sp | OR308 | N/A | MG888260 | MG888307 | MG888215 | MG888092 | MG888159 | ||
| Rhachitopis sp | OR312 | N/A | KM853242 | KM853448 | KM853624 | N/A | N/A | ||
| Eyprepocnemidinae | Cataloipus sp | OR218 | N/A | KM853201 | KM853489 | KM853664 | MG888079 | MG888146 | |
| Eyprepocnemis plorans | OR309 | MG993386, MG993418,MG993424, MG993425,MG993427, MG993433,MG993437, MG993450 | KM853239 | KM853451 | KM853627 | mtgenome | mtgenome | ||
| Heteracris sp | OR310 | N/A | KM853240 | KM853450 | KM853626 | MG888093 | MG888160 | ||
| Shirakiacris shirakii | N/A | NC_021610 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Tylotropidius sp | OR219 | N/A | MG888255 | MG888301 | MG888209 | MG888080 | MG888147 | ||
| Gomphocerinae | Arcyptera coreana | N/A | NC_013805 | N/A | N/A | N/A | mtgenome | mtgenome | |
| Aulocara elliotii | OR521 | N/A | KM853329 | KM853363 | KM853539 | MG888129 | MG888196 | ||
| Dichromorpha viridis | OR226 | N/A | KM853205 | KM853485 | KM853660 | MG888083 | MG888150 | ||
| Euchorthippus fusigeniculatus | N/A | NC_014449 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Gomphocerus sibiricus | N/A | NC_015478 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Mermiria intertexta | OR520 | N/A | KM853328 | KM853364 | KM853540 | MG888128 | MG888195 | ||
| Mesopsis sp | OR239 | N/A | MG888257 | MG888304 | MG888212 | MG888087 | MG888154 | ||
| Orinhippus tibetanus | N/A | NC_023467 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Pacris xizangensis | N/A | NC_023919 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Prorocorypha snowi | OR214 | MG993438, MG993452, MG993453 | KM853199 | KM853491 | KM853666 | mtgenome | mtgenome | ||
| Pseudogmothela sp | OR519 | N/A | MG888289 | MG888338 | MG888246 | MG888127 | MG888194 | ||
| Rhammatocerus schistocercoides | OR346 | N/A | KM853258 | KM853432 | KM853608 | MG888098 | MG888164 | ||
| Rhaphotittha sp | OR518 | N/A | MG888288 | MG888337 | MG888245 | MG888126 | MG888193 | ||
| Silvitettix sp | OR343 | N/A | MG888267 | MG888314 | MG888222 | N/A | N/A | ||
| Syrbula montezuma | OR227 | N/A | KM853206 | KM853484 | KM853659 | MG888084 | MG888151 | ||
| Acrididae | Hemiacridinae | Dirshacris aridus | OR305 | MG993398, MG993399, MG993410, MG993417, MG993420, MG993434, MG993435, MG993451 | MG888259 | MG888306 | MG888214 | mtgenome | mtgenome |
| Euroryma sp | OR302 | N/A | KM853236 | KM853454 | KM853630 | MG888090 | MG888157 | ||
| Hieroglyphus tonkinensis | N/A | NC_030587 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Leptacris sp | OR304 | MG993429 | KM853238 | KM853452 | KM853628 | mtgenome | mtgenome | ||
| Paulianiobia hirsuta | OR301 | N/A | MG888258 | MG888305 | MG888213 | MG888089 | MG888156 | ||
| Pristocorypha sp | OR303 | N/A | KM853237 | KM853453 | KM853629 | MG888091 | MG888158 | ||
| Leptysminae | Stenacris sp | OR342 | N/A | KM853255 | KM853435 | KM853611 | N/A | N/A | |
| Stenopola sp | OR220 | N/A | MG888256 | MG888302 | MG888210 | MG888081 | MG888148 | ||
| Tetrataenia surinama | OR338 | MG993385, MG993395, MG993396, MG993404, MG993407, MG993409, MG993432, MG993448 | KM853254 | KM853436 | KM853612 | mtgenome | mtgenome | ||
| Marelliinae | Marellia remipes | OR344 | MG993387, MG993423, MG993442, MG993447 | KM853256 | KM853434 | KM853610 | mtgenome | mtgenome | |
| Melanoplinae | Anapodisma miramae | OR356 | N/A | KM853265 | KM853425 | KM853601 | MG888100 | MG888166 | |
| Aptenopedes sphenarioides | OR516 | N/A | MG888287 | MG888336 | MG888244 | MG888124 | MG888191 | ||
| Bradynotes obesa | OR515 | N/A | KM853326 | KM853366 | KM853542 | MG888123 | MG888190 | ||
| Dichroplus sp | OR325 | N/A | KM853248 | KM853442 | KM853618 | MG888095 | MG888162 | ||
| Fruhstorferiola kulinga | N/A | NC_026716 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Hesperotettix viridis | OR517 | N/A | KM853327 | KM853365 | KM853541 | MG888125 | MG888192 | ||
| Jivarus ronderosi | OR328 | MG993400, MG993405 | KM853249 | KM853441 | KM853617 | mtgenome | mtgenome | ||
| Kingdonella bicollina | N/A | NC_023920 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Maeacris aptera | OR329 | N/A | MG888266 | MG888313 | MG888221 | N/A | N/A | ||
| Melanoplus bivittatus | OR245 | MG993426 | KM853211 | KM853479 | KM853654 | mtgenome | mtgenome | ||
| Ognevia longipennis | OR394 | NC_013701 | MG888297 | MG888319 | MG888227 | mtgenome | mtgenome | ||
| Ponderacris peruvianus | OR324 | N/A | MG888265 | MG888312 | MG888220 | N/A | N/A | ||
| Prumna arctica | OR395 | NC_013835 | KM853277 | KM853412 | KM853589 | mtgenome | mtgenome | ||
| Qinlingacris taibaiensis | N/A | NC_027187 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Oedipodinae | Acrotylus patruelis | OR190 | N/A | KM853190 | KM853500 | KM853675 | MG888075 | MG888142 | |
| Acrididae | Oedipodinae | Angaracris barabensis | N/A | NC_025946 | N/A | N/A | N/A | mtgenome | mtgenome |
| Bryodema miramae miramae | N/A | KP889242 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Ceracris kiangsu | N/A | NC_019994 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Chortoicetes terminifera | OR524 | N/A | MG888290 | MG888339 | MG888247 | MG888132 | MG888199 | ||
| Gastrimargus marmoratus | N/A | NC_011114 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Heteropternis sp | OR225 | N/A | KM853204 | KM853486 | KM853661 | MG888082 | MG888149 | ||
| Locusta migratoria | OR191 | NC_001712 | KM853191 | KM853499 | KM853674 | mtgenome | mtgenome | ||
| Psinidia fenestralis | OR522 | N/A | KM853330 | KM853362 | KM853538 | MG888130 | MG888197 | ||
| Pycnostictus seriatus | OR525 | N/A | MG888291 | MG888340 | MG888248 | MG888133 | MG888200 | ||
| Qualetta maculata | OR526 | N/A | MG888292 | MG888341 | MG888249 | MG888134 | MG888201 | ||
| Tomonotus ferruginosus | OR523 | N/A | KM853331 | KM853361 | KM853537 | MG888131 | MG888198 | ||
| Trilophidia annulata | N/A | NC_027179 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Trimerotropis sp | OR186 | N/A | KM853187 | KM853503 | KM853678 | MG888073 | MG888140 | ||
| Xanthippus sp | OR187 | N/A | KM853188 | KM853502 | KM853677 | MG888074 | MG888141 | ||
| Ommatolampidinae | Abracris sp | OR222 | MG993440 | KM853202 | KM853488 | KM853663 | mtgenome | mtgenome | |
| Anablysis teres | OR362 | N/A | MG888269 | MG888316 | MG888224 | N/A | N/A | ||
| Kyphiacris sp | OR363 | N/A | MG888270 | MG888317 | MG888225 | N/A | N/A | ||
| Locheuma brunneri | OR366 | N/A | KM853268 | KM853422 | KM853598 | N/A | N/A | ||
| Lysacris festae | OR365 | N/A | MG888271 | MG888318 | MG888226 | N/A | N/A | ||
| Ommatolampis quadrimaculata | OR364 | MG993443 | KM853267 | KM853423 | KM853599 | mtgenome | mtgenome | ||
| Pollostacris sp | OR322 | MG993391, MG993411, MG993413 | MG888263 | MG888310 | MG888218 | mtgenome | mtgenome | ||
| Psiloscirtus sp | OR348 | N/A | MG888268 | MG888315 | MG888223 | MG888099 | MG888165 | ||
| Syntomacrella sp | OR323 | N/A | MG888264 | MG888311 | MG888219 | N/A | N/A | ||
| Vilerna sp | OR336 | N/A | KM853252 | KM853438 | KM853614 | MG888097 | N/A | ||
| Xiphidiopteron sp | OR321 | N/A | MG888262 | MG888309 | MG888217 | N/A | N/A | ||
| Oxyinae | Kosciuscola tristis | OR396 | MG993402, MG993408, MG993414 | KM853278 | KM853411 | KM853588 | mtgenome | mtgenome | |
| Oxya chinensis | OR315 | NC_010219 | KM853244 | KM853446 | KM853622 | mtgenome | mtgenome | ||
| Pseudoxya diminuta | N/A | NC_025765 | N/A | N/A | N/A | mtgenome | mtgenome | ||
| Pauliniinae | Paulinia acuminata | OR345 | MG993401, MG993416, MG993419, MG993430, MG993431, MG993446 | KM853257 | KM853433 | KM853609 | mtgenome | mtgenome | |
| Proctolabinae | Coscineuta sp | OR249 | MG993441 | KM853212 | KM853478 | KM853653 | mtgenome | mtgenome | |
| Poecilocloeus napoana | OR368 | N/A | KM853270 | KM853420 | KM853596 | N/A | N/A | ||
| Rhytidochrotinae | Galidacris variabilis | OR371 | N/A | KM853271 | KM853419 | KM853595 | N/A | MG888167 | |
| Acrididae | Rhytidochrotinae | Paropaon sp | OR337 | MG993393, MG993397, MG993421, MG993428, MG993449 | KM853253 | KM853437 | KM853613 | mtgenome | mtgenome |
| Spathosterninae | Spathosternum nigrotaeniatum | OR224 | MG993439 | KM853203 | KM853487 | KM853662 | mtgenome | mtgenome | |
| Tropidopolinae | Petamella prosternalis | OR560 | MG993412 | KM853343 | KM853349 | KM853525 | mtgenome | mtgenome | |
| Lentulidae | Lentulinae | Lentula callani | OR295 | NC_020774 | KM853234 | KM853456 | KM853632 | mtgenome | mtgenome |
| Lithidiidae | Lithidiinae | Lithidiopsis carinatus | OR316 | NC_020775 | KM853245 | KM853445 | KM853621 | mtgenome | mtgenome |
| Ommexechidae | Ommexechinae | Ommexecha virens | OR367 | NC_020778 | KM853269 | KM853421 | KM853597 | mtgenome | mtgenome |
| Pamphagidae | Thrinchinae | Prionotropis hystrix | OR151 | JX913764 | KM853180 | KM853509 | KM853684 | mtgenome | mtgenome |
| Pamphagodidae | Unplaced | Hemicharilaus monomorphus | OR540 | JX913773 | KM853337 | KM853355 | KM853531 | mtgenome | mtgenome |
| Romaleidae | Romaleinae | Romalea microptera | OR1000 | MG993392, MG993394, MG993454, MG993455, MG993456, MG993457 | MG888294 | MG888343 | MG888251 | mtgenome | mtgenome |
| Xyleus modestus | OR265 | NC_014490 | KM853221 | KM853469 | KM853644 | mtgenome | mtgenome | ||
| Tristiridae | Tristirinae | Tristira magellanica | OR204 | NC_020773 | KM853197 | KM853493 | KM853668 | mtgenome | mtgenome |
‘N/A’ means that the sequence data were not available. ‘mtgenome’ means that the corresponding sequences were derived from the available mtgenome data. For some taxa, multiple Genbank accession numbers are assigned for mtgenome, which indicates partial mtgenomes consisting of several fragments.
DNA-grade tissue samples used for this study were either collected by the authors or provided by collaborators. They were preserved in 100% ethanol and vouchered in the −80°C freezer in the Texas A&M University Insect Collection’s Insect Genomic Collection (TAMUIC-IGC). To generate 18S, 28S, H3, COI, and COII sequences, we followed standard protocols for DNA extraction, PCR, and Sanger sequencing, which we described in detail elsewhere (Mugleston et al. 2013, Song et al. 2015). To generate mtgenome sequences, we performed shotgun sequencing of genomic DNA using the Illumina platform. To extract high molecular weight DNA required for Illumina sequencing, we used a Gentra Puregene Tissue Kit (Qiagen) following the manufacturer’s guidelines. The quality and concentration of DNA extracts were initially measured using either Qubit Fluorometer (Thermo Fisher) or DeNovix Spectrophotometer, and more thoroughly analyzed using Fragment Analyzer (Advanced Analytical Technologies). We used a Nextera XT DNA Library Prep Kit for library preparation and performed either 150bp paired-end (PE) sequencing using NextSeq500 or 125bp PE sequencing using HiSeq2500.
Library preparation and next-generation sequencing (NGS) were conducted at either Georgia Genomic Facility (NextSeq500) or Texas A&M Genomics and Bioinformatics Service (HiSeq2500). The resulting raw reads were quality-trimmed in CLC Genomics Workbench 8 (Qiagen). We used the MITObim pipeline (Hahn et al. 2013) to assemble mtgenomes de novo from the NGS reads. All newly assembled mtgenomes were first uploaded as raw fasta files to MITOS (Bernt et al. 2013) to identify open reading frames (ORFs) and tRNAs. The initial MITOS annotation was used as a guideline to delimit gene boundaries, and start and stop codons of each protein-coding gene were manually identified in Geneious 10.0.9 (Biomatters), following the recommendation by Cameron (2014). DNA sequence data generated for this study were deposited in Genbank with accession numbers presented in Table 2.
Phylogenetic Analyses
For both mitochondrial and nuclear protein-coding genes, we aligned based on the conservation of reading frames by first translating into amino acids and aligning individually in MUSCLE (Edgar 2004) using default parameters in Geneious. All other genes were individually aligned in MUSCLE using default parameters, also in Geneious. All these individual alignments were concatenated into a single matrix using SequenceMatrix (Vaidya et al. 2011). We divided the data into a total of 66 data blocks (13 mitochondrial protein-coding genes divided into individual codon positions, 22 tRNAs, 2 mitochondrial rRNAs, 2 nuclear rRNAs, and 1 nuclear protein-coding gene). We then used PartitionFinder v.1.1.1 (Lanfear et al. 2012) using the ‘greedy’ algorithm (heuristic search) with branch lengths estimated as ‘linked’ to search for the best-fit scheme as well as to estimate the model of nucleotide evolution for each partition using the Bayesian Information Criterion (BIC).
We performed maximum likelihood (ML) and Bayesian (BA) analyses on two datasets that include both mitochondrial and nuclear loci: a backbone dataset (20,425 aligned bp and 58 taxa) and a total evidence dataset (20,425 aligned bp and 142 taxa). The backbone dataset had 11.1% missing data due to those 19 taxa that we obtained from Genbank without nuclear gene data, and the total evidence dataset had 46.2% missing data due to 83 taxa without sequenced mtgenomes. We compared the resulting topologies to examine the effect of missing data in phylogenetic reconstruction in the total evidence dataset. For the ML analyses, we used the best-fit partitioning scheme recommended by PartitionFinder with the GTRCAT model applied to each partition and analyzed using RAxML 7.2.8 (Stamatakis et al. 2008) on XSEDE (Extreme Science and Engineering Discovery Environment, https://www.xsede.org) through the CIPRES Science Gateway (Miller et al. 2011). Nodal support was evaluated using 1,000 replications of rapid bootstrapping implemented in RAxML. For the BA analyses, we used the best-fit partitioning scheme and partition-specific models recommended by PartitionFinder and analyzed using MrBayes 3.2.6 (Ronquist et al. 2012) also on CIPRES. We used default priors and ran four runs with four chains each for 100 million generations, sampling every 5,000 generations. We plotted the likelihood trace for each run to assess convergence in Tracer (Rambaut and Drummond 2003–2009), and discarded an average of 25% of each run as burn-in. The resulting trees were visualized using FigTree (Rambaut 2006–2009). Our aligned datasets and the resulting trees were deposited to Mendeley (doi:10.17632/3cgttymztk.1).
Divergence Time Estimate Analysis
In order to estimate timing and rates of divergence across major grasshopper lineages using fossil records, we performed a divergence time estimate analysis in a Bayesian framework using the BEAST2 package (Bouckaert et al. 2014). We used the Fossilworks database (Behrensmeyer and Turner 2013) to search for known fossils of Acrididae. Although there are more than 50 fossil acridid specimens, many of them were discovered from the same deposits, and, so, there were only a small number of calibration points available to use for the analysis. Furthermore, many of the known fossils represented either crown groups or extant species. As a result, we selected three fossil species representing stem groups of different lineages within Acrididae. The first fossil was Proschistocerca oligocaenica Zeuner 1937, known from the Eocene of the United Kingdom (37.2 to 33.9 million years ago [MYA]). This is the oldest definitive fossil of the subfamily Cyrtacanthacridinae and we used it to calibrate this monophyletic group (Song and Wenzel 2008). Although there is no modern lineage of Cyrtacanthacridinae occurring in northern Europe, the fossil deposition site suggests that this Old World subfamily (except for Schistocerca, which is discussed later) must have had a broader distribution in the Eocene than today. The second fossil was Tyrbula russelli Scudder, 1885, known from the Florissant Formation of the Eocene of the United States (37.2 to 33.9 MYA). A number of fragmentary fossils of Acrididae have been found in this formation and the one we chose is the most completely preserved specimen known, which characteristically resembles the North American gomphocerine Syrbula (Scudder 1890). In our analyses, Gomphocerinae was paraphyletic with Acridinae and Oedipodinae, although these three subfamilies as a whole formed a clade. In fact, they have been considered paraphyletic grades in previous studies (Chapco and Contreras 2011). Thus, we used T. russelli to calibrate the clade consisting of these three subfamilies. The final fossil was Menatacridium eocenicum Piton 1936 of the Paleocene of France (58.7 to 55.8 MYA), which is the oldest known fossil of Acrididae. However, we did not use this to calibrate the node at the base of Acrididae, but at a node of an internal clade consisting of clades B, C, and D (Fig. 3). The rationale for this is as follows: in all of the phylogenetic analyses, the earliest diverging clade (A in Fig. 3) within Acrididae consisted of subfamilies restricted to the Neotropics, suggesting a South American origin for Acrididae. Since this fossil is from France, which in the Paleocene was not far from its present-day location, we considered that it could not be the stem Acrididae, but the stem of the group that colonized the Old World. Therefore, we used it to calibrate this node.
For this analysis, we used the total evidence dataset following the partitioning scheme and the models of nucleotide evolution recommended by PartitionFinder. We created an XML file in BEAUti from the BEAST2 package, specifying the fossil priors and monophyly constraints. We used the relaxed clock lognormal model for the clock model, the birth-death model with a uniform distribution as a tree prior, and a lognormal distribution as a distribution prior for fossil calibration points following a general guideline discussed in Ho and Phillips (2009). To assess convergence across independent runs, we conducted two separate analyses each for 100 million generations, sampling every 2,500 generations. We inspected the results using Tracer (Rambaut and Drummond 2003–2009), discarded 25% of each run as burn-in, and combined the trees using LogCombiner (Rambaut and Drummond 2002–2013a). A maximum clade credibility tree was summarized in TreeAnnotator (Rambaut and Drummond 2002–2013b) and visualized in FigTree.
Biogeographic Analysis
We used the package BioGeoBEARS (Biogeography with Bayesian [and Likelihood] Evolutionary Analysis in R Scripts) (Matzke 2013) within R (R Core Team 2017) to infer biogeographical patterns during the diversification of different lineages within Acrididae. BioGeoBEARS performs different models of ancestral range estimation because different ancestral-area reconstructions have different assumptions. The input files were: 1) a dated phylogeny inferred from the BEAST analysis and 2) a file of geographical ranges indicating presence/absence of each taxon in each discrete area in the analysis. We defined six areas: Neotropical, Nearctic, Palearctic, Ethiopian, Oriental, and Australian following the most commonly used divisions of the biogeographical realms. We identified a distribution range for each terminal (treated at the genus level) from the distribution maps available from the OSF (Cigliano et al. 2018). Although this information from the OSF was not based on specimen-level databases, we were able to identify the distribution ranges with confidence. To cope with those genera with broad geographical distributions, we allowed a maximum of five areas for a given genus to occur within. We tested six models implemented in the program: 1) DEC (dispersal-extinction-cladogenesis) (Ree et al. 2005); 2) DEC+J (including founder-event speciation); 3) DIVALIKE, a likelihood version of DIVA (dispersal-vicariance) (Ronquist 1997); 4) DIVALIKE+J (including founder-event speciation); 5) BAYAREALIKE, a likelihood version of the Bayesian inference of historical biogeography for discrete areas (BayArea) (Landis et al. 2013); and, 6) BAYAREALIKE+J (including founder-event speciation). These six models included two parameters: d = dispersal and e = extinction. Likelihood values of these models were compared using a likelihood ratio test and we used the Akaike Information Criterion (AIC) to directly compare how well the different models fit the data (Matzke 2013, 2014).
Results
Phylogeny of Acrididae
We recovered monophyletic Acrididae with strong nodal support in both ML and BA analyses of the backbone dataset and the total evidence dataset. For the backbone dataset, the topologies recovered from both analyses were mostly congruent except the clade consisting of Copiocera Burmeister, 1838 (Copiocerinae) and Coscineuta Stål, 1873 (Proctolabinae) (Fig. 2). This clade grouped closely with Oxyinae and Spathosterninae in the ML tree, but with the earliest diverging lineage in the Bayesian tree. However, the nodal support values for the placements of this lineage were low in both analyses (Fig. 2). For the total evidence dataset, the resulting relationships were also largely congruent between the two analyses, but in the Bayesian tree (not shown, available at Mendeley), the positions of some of the smaller clades, which had low bootstrap support (<50) in the ML tree (Fig. 3), were not fully resolved. Between the backbone trees and the total evidence trees in both inference methods, the majority of backbone relationships were preserved in the total evidence trees and the taxa with lots of missing data (those without mtgenome sequenced) were placed in the expected positions in the backbone topology. The clade that had different positions between the inference methods in the backbone trees grouped with Melanoplinae in the total evidence trees. The relationships among major lineages, including the placement of Acrididae in relation to the outgroups, as well as the early divergence of the New World endemic subfamilies within Acrididae, were consistent among all analyses. In other words, there seemed to be no negative effect of missing data in this data combination strategy. Because the ML analysis resulted in a single fully resolved topology with the best likelihood score (Fig. 3), even though some of the relationships were supported with low bootstrap values, we base the following discussion on the relationships resolved by the ML total evidence analysis.
Phylogeny of Acrididae based on the backbone dataset inferred in ML (a) and Bayesian (b) framework. Shown in red is the relationships that are different between ML and Bayesian analyses. Green circles on the nodes represent 100% bootstrap values in the ML tree and 100% posterior probability values in the Bayesian tree. Values lower than 100 are shown.
Phylogeny of Acrididae inferred from ML analysis. The numbers on nodes indicate bootstrap support values. Values lower than 50 are not known. Asterisk indicates a paraphyletic group.
In terms of outgroup relationships, we found Acrididae to be most closely related to Ommexechidae, Romaleidae, and Tristiridae, all of which are the New World families. These three New World endemic families and Acrididae collectively formed a monophyletic group, which was sister to the South African endemic clade consisting of Lentulidae and Lithidiidae. Within Acrididae, we recovered four major clades, which we tentatively refer to as clades A, B, C, and D, although the nodal support values for these clades were low (Fig. 3). Across all analyses, the placement of clade A at the base of the Acrididae phylogeny was consistent, but the relationships among the remaining three clades were unclear. The earliest diverging clade (clade A, green in Fig. 3) consisted of the Central and South American endemic subfamilies: Marelliinae, Pauliniinae, Ommatolampidinae, Leptysminae, and Rhytidochrotinae. Ommatolampidinae was found to be largely paraphyletic. The two monotypic subfamilies, Marelliinae and Pauliniinae, formed a clade, which represented the earliest diverging lineage within clade A. The second clade (clade B, red in Fig. 3) consisted of the Old World subfamilies (one representative each of Hemiacridinae, Tropidopolinae, Coptacrinae [= Coptacridinae], one representative of Catantopinae) and the cosmopolitan subfamilies (Acridinae, Gomphocerinae, and Oedipodinae). Hemiacridinae and Catantopinae were paraphyletic because other members of these subfamilies were found in other clades. Acridinae, Gomphocerinae, and Oedipodinae collectively formed a monophyletic group, which was consistently found in all analyses, but each subfamily was paraphyletic within.
The third clade (clade C, blue in Fig. 3) consisted of three Old World subfamilies (Hemiacridinae, Oxyinae, and Spathosterninae), two Neotropical subfamilies (Copiocerinae and Proctolabinae), and one cosmopolitan subfamily (Melanoplinae). Hemiacridinae and Copiocerinae were paraphyletic. Spathosterninae was nested within Oxyinae, rendering Oxyinae paraphyletic. Proctolabinae and Melanoplinae were the only monophyletic groups in this clade and Melanoplinae was found to be closely related to Proctolabine and Copiocerinae. The fourth clade (clade D, orange in Fig. 3) consisted of the Old World subfamilies (Eyprepocnemidinae, Calliptaminae, Euryphyminae, and Catantopinae) and one cosmopolitan subfamily (Cyrtacantharidinae). Of these, Catantopinae was the only paraphyletic group, and all other subfamilies were found to be monophyletic. Eyprepocnemidinae, Calliptaminae, and Euryphyminae collectively formed a strongly supported clade.
Divergence Time Estimate and Biogeography
The time-calibrated tree estimated from the BEAST analysis (Fig. 4) indicated that Acrididae originated in the Paleocene of the Cenozoic period (59.3 MYA). The BioGeoBEARS analysis suggested BAYAREALIKE+J to be the best-fit model for our data (LnL = −307.0228), allowing us to infer that the diversification of the family can be characterized as a series of founder events with subsequent radiation (Fig. 5). We found that the common ancestor of the South American endemic families (Tristiridae, Romaleidae, and Ommexechidae) and Acrididae diverged from their African relatives (Pamphagidae, Pamphagodidae, Lentulidae, and Lithidiidae) in the late Cretaceous. Soon after the K/T boundary, Acrididae diverged from Ommexechidae and started to diversify in northern South America, giving rise to Marelliinae, Pauliniinae, Ommatolampidinae, Leptysminae, and Rhytidochrotinae (clade A). The analysis also suggested that the common ancestor of the remaining subfamilies colonized Africa in the late Paleocene, radiated throughout Africa and progressively colonized the Palearctic and Oriental regions. Subsequently, several lineages recolonized the New World multiple times.
A time-calibrated phylogeny of Acrididae based on three fossil calibration points using BEAST. Posterior probability values are shown as colored circles (yellow: 96–100%, orange: 90–95%, red: 80–89%, below 79% not shown). Green bar indicates 95% HPD for a time estimate. Asterisk indicates a paraphyletic group. (a) Marellia remipes Uvarov, 1929 (Marelliinae); (b) Paulinia acuminata (De Geer, 1773) (Pauliniinae); (c) Tucayaca parvula Roberts, 1977 (Leptysminae); (d) Trichopaon tatei (Hebard, 1924) (Rhytidochrotinae); (e) Ommatolampis perspicillata (Johannson, 1763) (Ommatolampidinae); (f) Homoxyrrhepes punctipennis (Walker, 1870) (Tropidopolinae); (g) Eucoptacra anguliflava (Karsch, 1893) (Coptacrinae); (h) Acanthoxia gladiator (Westwood, 1841) (Hemiacridinae); (i) Uvarovium dirshi Uvarov, 1933 (Hemiacridinae); (j) Truxalis robusta (Uvarov, 1916) (Acridinae); (k) Mermiria picta (Walker, 1870) (Gomphocerinae); (l) Amblytropidia mysteca (Saussure, 1861) (Gomphocerinae); (m) Oedipoda miniata (Pallas, 1771) (Oedipodinae); (n) Oxya japonica (Thunberg, 1815) (Oxyinae); (o) Spathosternum pygmaeum Karsch, 1893 (Spathosterninae); (p) Adimantus ornatissimus (Burmeister, 1838) (Copiocerinae); (q) Proctolabus brachypterus Bruner, 1908 (Proctolabinae); (r) Melanoplus sanguinipes (Fabricius, 1798) (Melanoplinae); (s) Eyprepocnemis plorans (Charpentier, 1825) (Eyprepocneminae); (t) Calliptamus italicus (Linnaeus, 1758) (Calliptaminae); (u) Euryphymus haematopus (Linnaeus, 1758) (Euryphyminae); (v) Oxycatantops spissus (Walker, 1870) (Catantopinae); (w) Cyrtacanthacris tatarica (Linnaeus, 1758) (Cyrtacanthacridinae); (x) Eumecistes gratiosus Brančik, 1896 (Catantopinae).
Biogeographical histories of different lineages of Acrididae as inferred by BioGeoBEARS. Each terminal is a genus, and the colored squares indicate where the species in the genus are currently distributed, as defined by six biogeographical realms. The colored circles on the nodes represent the probabilities of each possible geographical range just before and after each speciation event. Some of the colored circles do not match with one of the six pre-defined colors for the six biogeographical realms, which show ambiguity in the ancestral distribution. Orange represents either Ethiopian (yellow) or Oriental (red), and olive green represents either Ethiopian (yellow), Oriental (red), or Palearctic (green). Major biogeographical events are indicated by the arrows on specific nodes, and the recolonization of the New World (NW) from the Old World (OW) by various lineages is indicated by OW→NW. The colored maps next to each clade show general hypothesized patterns of dispersals and colonization events for the subfamilies within each clade. A star represents the origin of Acrididae in South America. Black circles represent a likely area where the most recent common ancestor of each clade could have originated. Thick arrows indicate likely paths of colonization by major lineages, and thin arrows represent likely paths of colonization by lower taxonomic units (e.g., genus). Dotted arrows indicate possible dispersal events between the Old World and the New World.
Discussion
Phylogenetic Relationships of Acrididae to Other Families
Our study represents the first modern phylogenetic hypothesis of Acrididae based on a large taxon and molecular character sampling. While we recovered the monophyly of Acrididae as well as several well-characterized subfamilies with strong support, we found that paraphyly is rampant across many subfamilies. This finding highlights that the current classification has been affected by inadequate subfamily definitions and calls for the urgent need to revise the taxonomy of Acrididae as a whole.
Although the family concept of Acrididae has been used since the 19th century, the formal definition of what constitutes the family has never been clearly made because different authors relied on different characters for recognizing the family (Dirsh 1975, Xia 1994, Eades 2000). In fact, there are no external characters whose presence define the family (Rowell 2013). As mentioned previously, the only morphological character that has been suggested to unite all family members is the presence of a well-developed arch sclerite in the male phallic complex (Eades 2000), which is a difficult character to observe for untrained eyes and without careful dissection. Nevertheless, the fact that we have recovered the monophyletic Acrididae strongly suggests that this obscure genital character may indeed be a synapomorphy for the family. Amedegnato (1977) also suggested the preapical diverticulum of the female spermatheca could be a diagnostic feature for Acrididae, but because her work focused only on the Neotropical taxa, it is not clear how applicable this trait might be to other Old World taxa.
Due to the lack of obvious morphological characters that can be used to distinguish Acrididae from other related families, several currently recognized families were previously considered as members of Acrididae. For example, Dirsh (1975) considered the South American endemic Tristiridae and the South African endemic Lithidiidae to be closely related to the acridid subfamily Hemiacridinae (Hemiacrididae in his concept). These two families, however, have since been shown to be quite divergent from Acrididae and represent lineages that diverged much earlier than Acrididae (Leavitt et al. 2013, Song et al. 2015). The family Romaleidae is still sometimes regarded as part of Acrididae (Capinera et al. 2004, Johnson and Triplehorn 2005), but our study finds that it is a closely related but distinctly different family. Romaleidae can be morphologically distinguished from Acrididae by the sclerites of the aedeagus, which are derived from posterior prolongations of the endophallic plates, and the absence of the arch sclerite of the male phallic complex (Amedegnato 1977, Eades 2000, Rowell 2013).
The two most recent studies that investigated the phylogenetic relationships of Acrididae to other families used mtgenome data (Leavitt et al. 2013, Song et al. 2015) and our present phylogeny essentially used the same outgroup taxon sampling. Regardless of the scope of each study, the consistent pattern resulting from the mtgenome data is that Acrididae forms a monophyletic group with Ommexechidae and Romaleidae, which in turn is sister to Tristiridae. This relationship is significant because the closest living relatives of Acrididae are all endemic to South America, which supports the possibility of Acrididae originating in this region. We expand on this finding in the next section on biogeography. The common ancestor of Acrididae, Ommexechidae, and Romaleidae diverged from the ancestral Tristiridae in the late Cretaceous period. The common ancestor of these four families diverged from the Old World endemic lineage, including Lentulidae and Lithidiidae, probably due to variance.
Major Clades Within the Phylogeny of Acrididae
Although there have been several phylogenetic hypotheses of acridid grasshoppers at the subfamily level (Chapco et al. 2001, Litzenberger and Chapco 2001, Amédégnato et al. 2003, Litzenberger and Chapco 2003, Rowell and Flook 2004, Bugrov et al. 2006, Contreras and Chapco 2006, Fries et al. 2007, Song and Wenzel 2008, Chapco and Contreras 2011, Chintauan-Marquier et al. 2011, Li et al. 2011, Nattier et al. 2011), our study represents the first attempt to elucidate phylogenetic relationships among major subfamilies across the entire family. We found eight out of 21 subfamilies included in this analysis to be paraphyletic: Acridinae, Catantopinae, Copiocerinae, Gomphocerinae, Hemiacridinae, Oedipodinae, Ommatolampidinae, and Oxyinae. Most of these paraphyletic groupings are to be expected because these subfamilies have either not been clearly defined or used as taxonomic dumping grounds by previous taxonomists (Dirsh 1975, Grunshaw 1996, Eades 2000, Chapco and Contreras 2011, Rowell and Hemp 2017). Although more than one-third of the acridid subfamilies are paraphyletic, we have nevertheless recovered four major clades, which we tentatively call clades A, B, C, and D (Figs. 3 and 4). The relationships within each of these clades corroborate well with morphology and previous studies focusing on subfamilies (Amedegnato 1977, Eades 2000). One caveat is that the nodal support values for clades B, C, and D, especially the bootstrap support values from the ML analysis, are not strong (below 50), which we think is related to our character sampling that included a large number of missing data for those taxa without sequenced mtgenomes, although this did not have any effect on phylogenetic reconstruction. Thus, we discuss the higher relationships among these clades very little, and the following discussions should be viewed with some caution. Below, we present an in-depth discussion of the recovered relationships, along with commentary about our study’s implications for the evolution of grasshoppers in light of our phylogenetic analyses (Fig. 3) and the time-calibrated tree (Fig. 4). Because many of the subfamilies may be unfamiliar to the average readers, we also take this opportunity to highlight the diverse biology and ecology of each subfamily.
Clade A
The earliest diverging lineage within Acrididae is clade A (green in Figs. 3 and 4), which consists of members of two monotypic subfamilies, Marelliinae and Pauliniinae, two monophyletic subfamilies, Leptysminae, and Rhytidochrotinae, and the paraphyletic Ommatolampidinae. This clade represents the first radiation within the Neotropical region, although a few species of Leptysminae have reached the southern United States (such as Leptysma marginicollis (Serville, 1838) and Stenacris vitreipennis (Marschall, 1836)). The members of this clade are ecologically diverse and quite aberrant compared to typical grasshoppers. The most unusual case can be demonstrated with two aquatic species in Marelliinae (Marellia remipes Uvarov, 1929) and Pauliniinae (Paulinia acuminata (De Geer, 1773)). These species live on broad, floating leaves of aquatic plants, feeding and ovipositing on them, and their entire life cycle takes place on these plants. Their hind femora are flat and dilated, which help them swim underwater (Carbonell 1957). As reviewed in Carbonell (2000), the literature on Paulinia and Marellia suggests an uncertainty by authors regarding their relationships with other acridids and their taxonomic placement. Initially, these two taxa were treated as the only members of the family Pauliniidae, which Dirsh (1956) considered to be sufficiently different from Acrididae based on the male phallic complex. However, Dirsh (1961) later commented that Marellia probably did not belong to the same group as Paulinia and Carbonell (2000) conducted a detailed investigation of the internal and external morphology of both groups to suggest that Pauliniidae is an artificial group at best. He further commented that the external similarities between Paulinia and Marellia could be due to parallel adaptations to the aquatic habitat and not their shared ancestry. Eventually, Eades (2000) erected a new subfamily, Marelliinae, to accommodate Marellia, but its affinity to other acridid lineages has remained unclear. Our study found that Paulinia and Marellia form a well-supported clade, which instead suggests that their similar external morphology and unusual biology are, in fact, due to shared ancestry. However, we also found that the two lineages diverged in the Eocene, which means that there was ample time for each lineage to accumulate species-specific traits that might have confused earlier taxonomists.
Prior to 1974, all grasshopper species with the prosternal process were artificially grouped into Catantopinae, which was occasionally elevated to the level of family (Dirsh 1961). This structure is, however, present in all known subfamilies except Acridinae, Gomphocerinae, and Oedipodinae, and also present in Romaleidae, Ommexechidae, and Tristiridae, as well as other Old World families. This suggests that it is a plesiomorphic character and cannot be used as a diagnostic character for any one group within Acrididae. Amedegnato (1974) reclassified Neotropical grasshoppers based on a detailed examination of male and female genitalia and recognized six subfamilies that used to be classified as Catantopinae. Of these six subfamilies, she considered Leptysminae, Rhytidochrotinae, and Ommatolampidinae to form a clade (Amedegnato 1977), and our study also recovered these three subfamilies as a clade, although Ommatolampidinae was found to be paraphyletic. While Leptysminae and Rhytidochrotinae are defined by vertically hooked male cerci (Roberts and Carbonell 1980) and by a short pronotum and reduced aedeagus, respectively (Descamps and Amédégnato 1972), Ommatolampidinae is only defined as one of the Neotropical Acrididae with the prosternal process and a simple spermatheca (both of which the other two subfamilies have too), and without the defining features of Leptysminae and Rhytidochrotinae (Rowell 2013). Therefore, without any unique morphological characters of its own, it is not surprising that Ommatolampidinae was rendered paraphyletic in our phylogeny.
Leptysminae is a relatively small group with 21 genera and 79 valid species, well-characterized by the vertically hooked male cerci, angular lower external lobe of the hind knee, very short second tarsal segment of the hind legs, and the distal portion of the endophallic apodemes of the aedeagus that lies in the horizontal plane (Rowell 2013). A series of revisionary work by Roberts and Carbonell in the 1970s placed Leptysminae as one of the best-studied Neotropical acridid subfamilies (Roberts 1975, 1978; Roberts and Carbonell 1979, 1980). We included members of two major tribes, Leptysmini and Tetrataeniini, in our taxon sampling, and recovered the subfamily as monophyletic, which suggests the taxonomic stability of this group. Leptysmines are highly adapted to semi-aquatic habitats, preferring marshes or swampy areas, and feed on grasses, sedges, or broad-leaved monocots that thrive in these habitats (Rowell 2013). Like many grass-feeding grasshoppers, these insects have evolved an elongated body form (Rowell 2013) and many species even have the ability to swim. Unlike typical grasshoppers that lay eggs underground, leptysmines are known to engage in endophytic oviposition, in which the female first bites a hole in the surface of a stem and then inserts the toothed ovipositor to oviposit inside the plant (Braker 1989).
Rhytidochrotinae is also a small group, with 20 genera and 47 valid species, mainly distributed from Costa Rica to northern Brazil, with the highest diversity in Colombia (Descamps and Amédégnato 1972). Except for a few genera, rhytidochrotines live in montane forest (Rowell 2013). Most rhytidochrotines are completely apterous, often without tympana, and have a short pronotum with exposed metathoracic tergites (Descamps and Amédégnato 1972). Many species are also brilliantly colored. Unlike most other grasshopper groups, the male genitalia of rhytidochrotines are known to be homogeneous across species and genera with a reduced aedeagus (Amedegnato 1977, Rowell 2013). The lack of tympana and the lack of species-specific genitalia collectively suggest that these insects probably rely on non-acoustic and non-genital courtships. Judging from their protuberant eyes, bright body coloration, and varied patterning, it is likely that visual courtship might be the main mode of species recognition in this group. Rhytidochrotines are also known to feed on ferns (Rowell et al. 1983, Rowell 2013), which is quite unusual among grasshoppers. Another acridid subfamily that mainly feeds on aquatic ferns is Pauliniinae (Carbonell 2000) while other non-acridid fern-feeders belong to Eumastacoidea (Rowell 2013), an ancient lineage that originated in the Jurassic (Song et al. 2015). The association between eumastacoids and ferns makes sense due to their antiquity, but the association of rhytidochrotines (and pauliniines) with ferns must have evolved more recently because modern grasshoppers essentially diversified in the Cenozoic. Rowell et al. (1983) tested the feeding preference of a rhytidochrotine, Hylopedetes nigrithorax Descamps & Rowell, 1978, from Costa Rica, and found that it specializes on several species of ferns regardless of the secondary chemicals produced by the ferns. So, this is an interesting case of a young insect lineage associating with an old plant lineage. Whether these species can metabolize the plant chemicals (phenols and tannins) to use for their protection is unknown.
Ommatolampidinae is the largest subfamily in clade A with 114 genera and 292 valid species and it is rendered paraphyletic in our study. It is a heterogeneous Neotropical group distributed from Mexico to South America and Hispaniola. This subfamily includes morphologically and ecologically diverse species, such as the cryptic and geophilous Vilerna Stål, 1873, moss-mimicking Nicarchus Stål, 1878, leaf-litter-inhabiting Microtylopteryx Rehn, 1905, canopy-dwelling Anablysis Gerstaecker, 1889, as well as typical-looking Abracris Walker, 1870. Accordingly, there is great diversity in feeding habits and mode of oviposition among ommatolampidines (Rowell 2013). There are several well-characterized tribes within this subfamily, but the relationships among them are unclear. Our analysis included members of Syntomacrini, Abracrini, and Ommatolampidini, and recovered monophyly for the first two tribes. However, these tribes do not form a clade, but, rather, are intermingled with Leptysminae and Rhytidochrotinae. In fact, Amédégnato and Poulain (1998) speculated that Leptysminae and Rhytidochrotinae could have diverged from one stem of the Ommatolampidinae, which seems to fit the pattern we have recovered. Thus, it may be possible in the future to erect more groupings that are based on our phylogeny, or synonymize Leptysminae and Rhytidochrotinae with Ommatolampidinae.
Clade B
Clade B (red in Figs. 3 and 4) consists of members of the Old World subfamilies: Catantopinae, Coptacrinae, Hemiacridinae, Tropidopolinae, and a clade consisting of three cosmopolitan subfamilies without the prosternal process: Acridinae, Gomphocerinae, and Oedipodinae. Out of the four major clades recovered in our study, this clade seems to be the most unstable taxonomically since paraphyly is quite rampant. Because our taxon sampling for the Old World subfamilies in this clade was limited, it is difficult to make too much inference about them at this point, other than their placement in this clade. Both Hemiacridinae (38 genera, 121 species) and Tropidopolinae (11 genera, 34 species) are taxonomically ill-defined. Hemiacridinae, in particular, currently includes morphologically diverse genera that have been placed in this subfamily without a clear justification (Rowell and Hemp 2017). For example, some hemiacridine genera have elongated body forms characteristic of grassland-adapted ecomorphs (such as Leptacris Walker, 1870) and some have compressed body forms with forward-facing mandibles characteristic of arboreal ecomorphs (such as Pristocorypha Karsch, 1896), while others are flightless and small (such as Dirshacris Brown, 1959). As such, it is difficult to characterize what a typical hemiacridine is.
Most species currently classified as Tropidopolinae prefer long grass savanna in Africa and have elongated body forms mimicking grasses (Rowell and Hemp 2017). In our phylogeny, the single representative of tropidopoline Petamella Giglio-Tos, 1907 grouped with the hemiacridine Leptacris, which suggests that there is a need to re-classify these two subfamilies. Near the base of clade B, we recovered Coptacrinae (20 genera, 116 species) as monophyletic, which is characterized by the presence of furculae, an elongated and tapered supra-anal plate, and subgenital plate with a transverse fold (Rowell and Hemp 2017). The members of this subfamily prefer open savanna, savanna woodlands, and forest edges, and are known to feed on herbaceous plants, with some species specializing on Asteraceae (Johnsen 1982, Rowell and Hemp 2017). Not much is known about the biology of this group but many species are brachypterous and quite colorful, reminiscent of the Ommatolampidinae in the New World.
Traulia Stål, 1873 is the only member of Catantopinae represented in clade B and all other catantopines included in this analysis are clustered in clade D (orange clade in Figs. 3 and 4), which makes its position questionable. This genus has been classified as Catantopinae because of the presence of the prosternal process, but previous phylogenetic studies that included Traulia consistently found that this genus did not cluster with other catantopines. For example, Liu et al. (2008) found this genus to group with Acridinae and Gomphocerinae based on mitochondrial rRNA and Chapco (2013) suggested that the phylogenetic position of Traulia was unclear based on his analysis using four mitochondrial genes. Morphologically, Eades (2000) questioned the placement of Traulia in Catantopinae because its phallic structures show resemblance to members of Oedipodinae rather than other catantopines. In our ML tree, Traulia is at the base of the clade consisting of Acridinae, Gomphocerinae, and Oedipodinae. Therefore, all evidence seems to suggest that this genus needs to be reclassified and a more in-depth study is necessary to resolve the relationship between Traulia and these three subfamilies, which do not possess the prosternal process.
The clade consisting of Acridinae, Gomphocerinae, and Oedipodinae collectively represents the largest radiation within Acrididae, including more than 2,500 valid species (over 37% of total acridid diversity) (Cigliano et al. 2018). The members of this clade can produce sound either by stridulation (produced by rubbing their hind legs against the forewings) or crepitation (produced by snapping wings when they fold and unfold) (Otte 1981). Their high numbers of species, combined with worldwide distribution and common association with grasslands, is why they are the most familiar members of Acrididae to the general public. Simultaneously, though, they are also the most controversial group taxonomically as numerous authors have struggled to define what constitutes each subfamily (Rehn and Grant 1960, Jago 1971). During the early half of the 20th century, the only distinguishing feature between Acridinae and Oedipodinae (the concept Gomphocerinae was formalized much later) was the degree of facial angle, which is at an oblique angle in acridines and a vertical angle in oedipodines (Uvarov 1941).
However, Uvarov (1941) expressed an opinion that the degree of facial angle is highly variable and is not a suitable trait to define a subfamily, instead suggesting that the morphological adaptations that allow sound production should be the main characters to distinguish these groups. Uvarov (1941) observed that Acridinae could be characterized by a series of pegs on a special ridge of the hind femur that work against the raised second radial vein, while Oedipodinae lacks this trait. Following this observation, Dirsh (1951[1950]) proposed several tribes based on this stridulatory mechanism, such that Truxalini was defined by the presence of the stridulatory pegs along the inside of the hind femur coupled with the raised medial and radial vein, and Acridini was defined by the absence of these pegs and raised veins. Although Dirsh (1951[1950]) observed that the species classified as Oedipodinae could be characterized by the presence of stridulatory intercalary veins, he nevertheless did not recognize Oedipodinae as a separate subfamily and treated it under Acridinae.
Rehn and Grant (1960) criticized the overemphasis on the stridulatory structures as a main taxonomic character and documented numerous exceptions where the strict adherence to this character would cause conflicts in taxonomy, which led to their conclusion in recognizing only the single subfamily of Acridinae. However, Dirsh (1965) ignored this criticism and placed all species with a stridulatory file consisting of a row of pegs along the inside of the hind femora in either Eremogryllinae (a small subfamily in North Africa, consisting of two genera and five species that we did not include) or Truxalinae. All other species were placed in Egnatiinae (another small subfamily in the Middle East, consisting of nine genera and 36 species that we did not include) or Acridinae (which included Oedipodinae). Uvarov (1966) and Jago (1971) adopted a different view and recognized Truxalinae sensu Dirsh that have the stridulatory file consisting of a series of peglike hairs as a separate subfamily, Gomphocerinae. Consequently, the definition of Truxalinae was restricted to only those species with the stridulatory file consisting of unmodified hairs lying between peglike cuticular expansions. Uvarov (1966) reintroduced the definition of Oedipodinae as those without the stridulatory files on hind femora, but with a stridulatory file on a raised intercalary vein on the tegmina and that have a rounded-head profile. Otte (1981, 1984) took a similar approach in distinguishing the three subfamilies in North America, but also noted that the loss of stridulatory pegs appeared to be quite common, making it difficult to separate Gomphocerinae from Acridinae. He classified the Nearctic Acridinae as those with the forewings that are obliquely truncated at apex and male hind wings with enlarged cells near leading edges. Later, Jago (1996) synonymized Truxalinae (which has the stridulatory files) under Acridinae based on the morphology of the male epiphallus, which further downplayed the taxonomic importance of the stridulatory file.
Our phylogeny reflects this taxonomic controversy as none of the three subfamilies in question were recovered as monophyletic. This lack of monophyly has been suspected for many decades because there are many intermediate groups that do not neatly fit into any of the three subfamilies, such as the tribe Hyalopterygini in the New World (Rehn and Grant 1960). Chapco and Contreras (2011) reported the same pattern from a molecular phylogenetic study based on a dense taxon sampling of these three subfamilies. They suggested that while these subfamilies share a common ancestor, the current taxonomy does not correspond with the phylogeny, thereby labeling these subfamilies ‘fuzzy sets’. This pattern also extends to tribal levels. For example, Fries et al. (2007) presented a molecular phylogeny of Oedipodinae based on mitochondrial genes in which they found that many tribes were paraphyletic and suggested that the current taxonomic groupings appeared to be based on largely convergent characters. Nattier et al. (2011) studied the phylogeny of Gomphocerinae based on molecular data and also found many paraphyletic tribes. The relationships we have recovered in our study highlight the taxonomic instability at all levels and a thorough taxonomic and phylogenetic investigation of this clade is urgently needed, especially because this clade contains dozens of agriculturally important pest species and is also biologically fascinating.
As mentioned above, Acridinae (141 genera, 484 species) is a heterogeneous group, which is currently defined by negative characters, such as the lack of the stridulatory file. In our phylogeny, Acrida Linnaeus, 1758 (type genus of Acridinae) and Truxalis Fabricius, 1775 (type genus of Truxalinae) were recovered as sisters. The close relationship between these two genera was also recovered by Chapco and Contreras (2011) as well. These findings also support Jago’s (1996) synonymy of Truxalinae under Acridinae. In fact, both genera have a highly slanted head, an elongated body, and strongly ensiform antennae, making them well-adapted for inhabiting grasslands. It appears that the lack of the stridulatory file in Acrida is a loss of character. Otte (1981) considered there to be only one tribe of Acridinae in the New World, Hyalopterygini. In our phylogeny, Hyalopteryx Charpentier, 1845, the type genus for the tribe, forms a clade with Dichromorpha Morse, 1896 and Syrbula Stål, 1873, which are currently classified as Gomphocerinae. In Chapco and Contreras (2011), several species of Hyalopterygini formed a clade with Dichromorpha, Orphulella Giglio-Tos, 1894, and Orphulina Giglio-Tos, 1894, all of which are currently considered gomphocerines. These findings also collectively suggest that the stridulatory file has been lost and regained many times throughout the diversification of Acridinae and Gomphocerinae, and the current definition of each subfamily is not adequate.
Gomphocerinae (192 genera, 1,273 species) is the largest subfamily within Acrididae and we found it to be a highly paraphyletic group intermingled with Acridinae and Oedipodinae. We have shown that the original definition of Gomphocerinae based on the presence of stridulatory file is no longer valid. Nevertheless, the species of typical Gomphocerinae are among the best-studied grasshoppers in terms of mating behavior (Faber 1929, Jacobs 1953, Otte 1970, Eisner 1974). Many species show elaborate pre-copulatory courtship behaviors using a combination of acoustic, vibrational, and even visual signals sometimes. Nattier et al. (2011) demonstrated, based on a phylogenetic study of gomphocerine songs, that there are clades (such as Chorthippus Fieber, 1852) that include males that produce highly complex and unique calling songs, and whose courtship songs are very similar to calling songs. There are other clades in which males produce relatively simple and non-species-specific calling songs, but highly elaborate and multi-modal courtship songs and displays. These patterns show dynamic and rapid evolution of mating signals and behaviors, likely driven by sexual selection.
Oedipodinae (137 genera, 794 species) is commonly known as band-winged grasshoppers in North America as most species have hind wings that are usually banded or brightly colored with yellow, orange, red, or blue hues, often overlaid with black or smoky bands. However, not all species have this characteristic and there are many other species outside of Oedipodinae that have convergently evolved this trait. Many oedipodine grasshoppers exhibit acoustic communication by crepitation, but a more prominent mode of communication is visual, which involves hind wing color display during flight and various types of hind leg signaling (Otte 1970), although the function of hind wing color display might also be related to defense against visual predators. Many oedipodines are cryptically colored to mimic substrate coloration and prefer sandy habitats. Nymphs of oedipodines are known to express homochromy, the ability to change cuticular coloration to match background color during development (Rowell 1971, Edelaar et al. 2017, Peralta-Rincon et al. 2017).
Clade C
Clade C (blue in Figs. 3 and 4) includes the Old World subfamilies, Hemiacridinae and Oxyinae, both of which are recovered as paraphyletic, and Spathosterninae, represented by a single taxon, and the New World endemic subfamilies Copiocerinae and Proctolabinae, as well as the large subfamily Melanoplinae, which is found throughout the New World and parts of Eurasia. Many species in this clade are associated with herbaceous plants and some groups, such as Oxyinae, Copiocerinae, and Proctolabinae are adapted to rainforest habitats (Rowell 2013, Rowell and Hemp 2017) while other groups within Oxyinae and Melanoplinae appear to be secondarily associated with grasslands. Brachyptery appears to be common in this clade and many species have independently adapted to alpine habitats (Knowles 2001, Cigliano and Amédégnato 2010, Tatarnic et al. 2013, Pocco et al. 2015). As mentioned above, Hemiacridinae is an artificial group at best. While it is not a natural group, previous taxonomists have suggested that some species classified under Hemiacridinae may be closely related to Oxyinae (37 genera, 307 species) (Rowell and Hemp 2017). The presence of a radial area bearing a series of transverse stridulatory veinlets in the tegmina of males has been used as a main feature for characterizing Hemiacridinae (Dirsh 1975). Some species in Oxyinae also have this modification in wing venation (Rowell and Hemp 2017) and some hemiacridine species have the bridge of epiphallus split in half, a condition that is found in all Oxyinae members (Hollis 1975). Spathosterninae (three genera, 12 species), a small subfamily with three known genera, has also been attached to Oxyinae or Hemiacridinae because it has similar stridulatory veinlets on the tegmina (Dirsh 1975). Thus, it appears that at least some members of Hemiacridinae, Spathosterninae, and Oxyinae might collectively form a natural group.
Copiocerinae, Proctolabinae, and Melanoplinae collectively form a clade, although Copiocerinae was found to be paraphyletic in our study. The close relationship of these three subfamilies was originally postulated by Amedegnato (1977) who established these subfamilies based on shared morphological characters, such as a long vermiform preapical diverticulum of the spermatheca, laterally compressed anterior sclerites of the endophallus, and a cup-like male subgenital plate. Although our taxon sampling was small, especially for Copiocerinae and Proctolabinae, the resulting topology is congruent with the hypothesis based on morphology. Amedegnato (1977), as well as Rowell and Flook (2004), suggested that Copiocerinae is more closely related to Proctolabinae than Melanoplinae, and our analyses confirm this idea. Copiocerinae (21 genera, 82 species) is a Neotropical subfamily found from Mexico to South America, but the subfamily is not well-defined and does not seem to have a distinguishing characteristic other than a transverse groove that separates the vertex from the fastigium on the head. Rowell (2013) commented that the boundaries of the subfamily are not clear, hinting at the possibility of paraphyly. Ecologically, copiocerines are known to specialize on rainforest palms, which is quite unusual for grasshoppers (Rowell 2013). Proctolabinae (29 genera, 214 species), on the other hand, is a more distinct subfamily, characterized by a thickened transverse ridge bounding the tip of the fastigium, elongated second tarsal segment on hind legs, and several characters from the male genitalia (Amedegnato 1977), and its monophyly was well-supported by a previous molecular study (Rowell and Flook 2004) as well as hypotheses based on morphology (Descamps 1976, Amedegnato 1977). Proctolabines live in wet forest and many are arboreal while others, mostly brachypterous species, are found on woody forbs and shrubs. Many proctolabines are specialist herbivores on Solanaceae and Asteraceae in the Neotropics and engage in endophytic or epiphyllic oviposition on host plants (Rowell 2013).
Melanoplinae (146 genera, 1,172 species) is the largest subfamily in the New World, which is well-characterized by a thick pallium and species-specific male cerci and phallic complex. Of all grasshopper subfamilies, Melanoplinae has received the most attention in terms of phylogenetics, and all previous studies have consistently recovered it as a monophyletic group (Chapco et al. 2001, Amédégnato et al. 2003, Chintauan-Marquier et al. 2011, Chintauan-Marquier et al. 2014, Woller et al. 2014). Among melanoplines, there is an enormous amount of interspecific differences in genital morphology (Hubbell 1932, Cohn and Cantrall 1974), primarily in males, but females in some genera also show species-specific differences in genitalia (Cigliano and Ronderos 1994). Melanoplines do not exhibit any pre-copulatory courtship behavior, such as visual or acoustic. Instead, mating behavior appears to be coercive, in which males stealthily approach and jump onto females to initiate copulation (Otte 1970). A recent study by Woller and Song (2017) examined the internal morphology of copulation in Melanoplus rotundipennis (Scudder, 1878) using micro-CT technology and clarified the function of many genital components in both males and females. Most species in the subfamily live in grasslands and compete with grazing livestock, and some species have considerable economic importance in North American rangelands and South American grasslands (Pfadt 1988, Cigliano et al. 2002). Many species of melanoplines are also adapted to alpine habitats in South America in the Andes (Cigliano and Amédégnato 2010, Pocco et al. 2015, Scattolini et al. 2018) and North America in the Rockies (Knowles 2001).
Clade D
Clade D (orange in Figs. 3 and 4) represents a group of morphologically well-differentiated subfamilies that have diversified in the Old World. Except for the paraphyletic Catantopinae, the four other subfamilies included in this clade were shown to be monophyletic with strong nodal support. In our phylogeny, this clade is divided into two smaller clades: 1) Eyprepocnemidinae, Calliptaminae, and Euryphyminae; 2) Cyrtacanthacridinae and Catantopinae.
Eyprepocnemidinae, Calliptaminae, and Euryphyminae have been consistently shown to have affinities to each other based on morphology (Dirsh 1975, Rowell and Hemp 2017), but our study is the first to clearly propose that they constitute a monophyletic lineage. Not much is known about the biology of the species belonging to these subfamilies except for agriculturally important species, such as the Italian locust (Calliptamus italicus (Linnaeus, 1758)) and several minor pest species in Eyprepocnemidinae (COPR 1982). Most species appear to be associated with shrublands or woodlands, and several species can be characterized as geophilous, often being associated with bare soil. Although each subfamily has a distinct set of morphology that defines them, the grasshoppers in this group have somewhat enlarged eyes, hind wings that are often colored, and hind legs that are often brightly colored (at least on the inner portion). In our phylogeny, Calliptaminae and Euryphyminae are shown to be more closely related to each other than to Eyprepocnemidinae. Both calliptamines and euryphymines are small in size and cryptic in coloration, resembling soil, and often feed on woody plants. In a way, their general coloration is quite reminiscent of many oedipodines. Calliptaminae (12 genera, 93 species) is easily characterized by the pincer-like male cerci, which are elongated and strongly incurved. The function of these exaggerated cerci is unknown, but they are possibly used for holding females during copulation. Interestingly, the epiphallus in calliptamines does not possess lophi, which function as grasping organs in other grasshopper species (Randell 1963, Dirsh 1973, Woller and Song 2017). It is possible that the elaboration of male cerci corresponds with the reduction of lophi. The closely related Euryphyminae (23 genera, 87 species) is mostly confined to southern Africa and is characterized by male cerci with large basal articulation and strongly sclerotized supra-anal plate. Unlike calliptamines, euryphymines have well-developed lophi. Furthermore, the epiphallus is divided in the middle, similar to those observed in Oxyinae (Dirsh 1973, Rowell and Hemp 2017). Eyprepocnemidinae (26 genera, 159 species) is the most diverse group among the three subfamilies, and is characterized by a flat dorsum of pronotum, downcurved male cerci, and articulated ancorae of epiphallus. Most eyprepocnemidines are associated with woodlands or forests, but some species are known to be pests of economic importance (COPR 1982).
The subfamily Catantopinae (341 genera, 1,077 species) used to be a catch-all group to include any grasshopper species with the prosternal process, but now its definition has been reduced to any Old World and Australian acridid species with the prosternal process that do not fit well with other subfamilies. Many of the catantopine genera are monotypic or include a small number of species. Because its subfamily definition is not based on any distinct morphological feature, Catantopinae was expected to be paraphyletic, which is what we recovered, although our taxon sampling beyond Australian taxa was quite sparse. All catantopines from Australia, except Stenocatantops Dirsh, 1953 and Xenocatantops Dirsh, 1953 form a monophyletic group with strong nodal support (Figs. 3 and 4). About 85% of Australian acridid fauna (~300 sp.) belongs to Catantopinae and there is an incredible amount of morphological diversity among these Australian catantopines (Key and Colless 1993, Rentz et al. 2003). The fact that morphologically diverse species confined within Australia form a clade suggests major adaptive radiation, equivalent to the radiation of marsupials.
The subfamily Cyrtacanthacridinae (37 genera, 165 species) includes some of the largest grasshoppers, which also possess some of the strongest flying capabilities, giving them their common name of bird-wing grasshoppers. The group is well-defined by rectangular mesosternal lobes and its monophyly has been supported previously based on a morphological phylogeny (Song and Wenzel 2008), as well as the current study. This subfamily includes some of the most economically important locust species in the world, such as the desert locust (Schistocerca gregaria (Forskål, 1775)), the Central American locust (Schistocerca piceifrons (Walker, 1870)), the South American locust (Schistocerca cancellata (Serville, 1838)), and the red locust (Nomadacris septemfasciata (Serville, 1838)).
Historical Biogeography of Acrididae
The family Acrididae as a whole has a cosmopolitan distribution. Historically, many orthopterists assumed the origin of Acrididae to be Africa because there is a large diversity of other related families there, such as Pneumoridae, Pamphagidae, Pyrgomorphidae, and Lentulidae (Carbonell 1977, Jago 1979, Amedegnato 1993). The fact that the South American grasshopper fauna was not studied in-depth until the 1970s and that most European orthopterists initially focused on the Old World fauna probably contributed to this thinking as well. For example, Amedegnato (1993), who extensively studied South American grasshoppers, maintained that Acrididae originated in the Old World and diverged from Romaleidae due to the separation of Africa and South America. However, our biogeographical analysis, along with divergence time estimates, show a much more dynamic pattern of diversification and radiation, and sheds interesting new light on the evolution of these grasshoppers.
Our study suggests the origin of Acrididae to be South America, which is a novel hypothesis (Fig. 5). We infer this based on the monophyletic group consisting of Tristiridae (endemic to South America), Romaleidae (widely distributed in South America, extending to Central and North America), Ommexechidae (endemic to South America), and Acrididae. The other outgroups that we included in this study are all restricted to the Old World and none of the species belonging to these Old World families shows any morphological affinity to Acrididae. The earlier study by Flook and Rowell (1997) that grouped Proctolabinae (New World endemic subfamily of Acrididae) with the Old World family Pamphagidae is most likely due to the small molecular character sampling (930 bp from mitochondrial rRNAs) and sparse taxon sampling that did not include any of the New World endemic families. Even the authors mentioned that this relationship was very unexpected because the relationship did not make sense in terms of male genital morphology.
Our biogeographic analysis using BioGeoBEARS (Fig. 5) suggests that the common ancestor of Tristiridae, Romaleidae, Ommexechidae, and Acrididae diverged from its Old World relatives in the late Cretaceous due to vicariance when the South American continent separated from Africa. This common ancestor gave rise to current families within South America, which was essentially isolated until the emergence of the Isthmus of Panama, which is now estimated to have taken place about 20 MYA (Montes et al. 2015, Bacon et al. 2016), although this older date is not universally accepted (O’Dea et al. 2016). We estimate that the Acrididae originated in the early Paleogene in South America.
The earliest diverging lineage within Acrididae includes the South American endemic subfamilies Marelliinae, Pauliniinae, Ommatolapidinae, Leptysminae, and Rhytidochrotinae. With the exception of Rhytidochrotinae, which is mostly distributed in the montane forests of Colombia (Descamps and Amédégnato 1972), the remaining subfamilies are widely distributed in the Amazon basin and northern South America, with some later expanding their range up to Central America. This distribution appears to be related to what has been called the ‘pan-Amazonian’ region (Hoorn et al. 2010), a large region that included the present-day Amazon, Orinoco, Magdalena drainage basins, and Paraná river during the Paleogene. This vast area was characterized by the diverse fauna that existed there, elements of which are now restricted to Amazonia. Furthermore, the diversification of these subfamilies could be also associated with the large wetland of shallow lakes and swamps that developed in western Amazonia, originating in parallel with the intensified uplift of the Andes (late middle Miocene) (Hoorn et al. 2010). These new aquatic environments, the ‘Pebas system’, may have favored the diversification of those subfamilies associated with marshy/swamp habitats, wet forests, and canopies. It is interesting that these grasshoppers continue to be associated with the niches that their ancestors may have evolved in, rather than more typical terrestrial and grassland habitats that most other acridids favor, which may suggest conservation of the ancestral ecological niche within this lineage.
During the initial diversification period within South America, there appears to have been a single transatlantic colonization to Africa by the common ancestor of clades B, C, and D (Fig. 5), which took place in the early Paleogene (~57 MYA). Although South America and Africa were already separated at that point, these two continents were closer compared to today’s configuration and dispersal across the narrowest point between the two continents could have been possible. In fact, there are a number of organisms, such as amphibians and reptiles, that display similar patterns (George and Lavocat 1993). At this point in time, northern Africa was covered with tropical rainforests, not too different from the original habitats that the ancestral grasshoppers experienced in northern South America. Once in Africa, these ancestral acridids could have quickly radiated, giving rise to numerous lineages, which eventually differentiated into what we now recognize as different subfamilies. However, we currently have several more subfamilies in the New World scattered throughout the phylogeny in addition to the five subfamilies in clade A (green in Fig. 5). This pattern suggests that there must have been some recolonization events occurring from the Old World back to the New World. Our study hypothesizes that there were at least three distinct waves of recolonization throughout the diversification of Acrididae.
The first wave of recolonization of the New World was likely to be a westward transatlantic colonization by the common ancestor of Copiocerinae, Proctolabinae, and Melanoplinae that took place in the early Eocene. It is not clear how this common ancestor could have recolonized South America from Africa, but the distance between the two continents was still narrow enough for an unusual westward dispersal to have taken place. Upon arrival, probably to northern South America, this common ancestor gave rise to what would eventually become the three subfamilies. Many proctolabines are known to be associated with the plant family Solanaceae (Rowell 1978, 2013). The origin of Solanaceae has recently been hypothesized to be in the Eocene (Särkinen et al. 2013) and this family has great diversity in the Neotropics, which provides some support for the diversification of Proctolabinae. Compared to Copiocerinae and Proctolabinae, which are relatively small endemic subfamilies confined to the Neotropics, Melanoplinae is a much more diverse subfamily that radiated throughout South, Central, and North America, and extends to Eurasia. Many of its species are associated with either grasslands or alpine habitats. Amédégnato et al. (2003) conducted a molecular phylogenetic analysis of Melanoplinae and included the tribes Melanoplini (North America), Podismini (Eurasia), Dichroplini (South America), and Jivarini (South America). They recovered basal placement of the South American tribes and put forth a biogeographical hypothesis suggesting that the center of origin for this subfamily was South America, after which it diversified through North America and then to Eurasia. Chintauan-Marquier et al. (2011) found a similar biogeographical pattern, but estimated the divergence date of the subfamily to be 69 MYA, which predates our estimate of the origin of Acrididae. Woller et al. (2014) included several members of Dactylotini (Central America) that were not well-represented in the previous studies and recovered the basal placement of Jivarini, followed by Dichroplini. Our study confirms the South American origin of Melanoplinae and furthers the inference by suggesting that the ancestral stock that gave rise to Melanoplinae and the two other related subfamilies actually originated from Africa. We estimate that the common ancestor of Melanoplinae diverged from the other two subfamilies in the early Eocene (~43 MYA), giving rise to Jivarini and Dichroplini. Then, this ancestor expanded northward to give rise to Dactylotini and Melanoplini in North America, and further expanded westward through the Behring land bridge to give rise to Podismini in East Eurasia. Several members of Podismini have reached Europe and speciated in the mountain ranges (Kenyeres et al. 2009), similar to how Melanoplus speciated in the Rocky Mountains in North America (Knowles 2001). The origin of the Jivarini, which is exclusively distributed in the Central Andes (Cigliano and Amédégnato 2010), coincides with the first uplift of this geological feature, which slowly developed from the mid- Eocene and reached a peak in the Late Oligocene and Early Miocene (~23 MYA) (Gregory-Wodzicki 2000, Garzione et al. 2008, Hoorn et al. 2010). So, the highlands of the Andes may have served as a migration route for Melanoplinae toward North America.
The second wave of recolonization of the New World was achieved by several lineages within the Acridinae-Gomphocerinae-Oedipodinae complex. This clade is specifically associated with graminivory (Uvarov 1966, Pfadt 1988) and its diversification closely corresponds to the evolution and expansion of grasslands and open habitats (Song et al. 2015). Based on pollen fossils and phytoliths, Strömberg (2011) estimated that grasslands became abundant in western Eurasia, North America, and southern South America during the Eocene. Although the Acridinae-Gomphocerinae-Oedipodinae clade as a whole is cosmopolitan, more than 70% of its diversity (~1,790 spp.) occurs in the Palearctic (northern Africa, Europe, and temperate Asia) and the Ethiopian (sub-Saharan Africa) region, with the former region containing more than 48% of the diversity. This pattern of diversity, coupled with our biogeographical analysis and the diversification of grasslands, strongly suggest that this group originated in the Palearctic region, from which different lineages expanded their ranges, colonizing new regions. Furthermore, sexual selection on songs produced by stridulation or crepitation could have played another important role in promoting rapid speciation in this clade. Our analysis indicated that there were numerous recolonization events from the Old World to the New World throughout the diversification of this clade (Fig. 5). The relative paucity of this clade in the Neotropics (only 6.5% of the diversity) seems to suggest that the main routes of recolonization were probably from Eurasia to North America, either through the Thulean route (or dispersal flights across Greenland) or through Beringia from eastern Eurasia to North America. Additionally, there could have been ecological factors, such as the unsuitability of the habitats and tropical climates, which might have prevented the colonization of the Neotropics.
The third wave of recolonization of the New World occurred in the Old World subfamily Cyrtacanthacridinae. The genus Schistocerca Stål, 1873 is the only genus in this subfamily that has representatives in both the Old and New Worlds, while all other genera occur solely in the Old World (Amedegnato 1993, Song 2004). Furthermore, within Schistocerca, the infamous desert locust (S. gregaria) is the only species that occurs in Africa while the rest of the genus is found throughout North, Central, and South America. Recent molecular studies have consistently placed the desert locust at the base of the phylogeny of Schistocerca (Lovejoy et al. 2006, Song et al. 2013, Song et al. 2017), which indicates that the genus originated in Africa, and its current diversity is a result of a spectacular transatlantic colonization followed by rapid radiation. Song et al. (2017) estimated that the genus diverged from its relatives about 6–7 MYA, when the distance between Africa and South America was essentially identical to what it is today. In 1988, there was a large swarm of desert locusts that successfully crossed the Atlantic Ocean from western Africa to the West Indies (Kevan 1989, Rosenberg and Burt 1999), which suggests that such a long-distance flight could have been possible in the past. Taxonomically, there are two other genera in the Cyrtacanthacridinae in the New World: Halmenus Scudder, 1893 in the Galapagos and Nichelius Bolívar, 1888 in Cuba. The former is a small brachypterous genus with four species (Snodgrass 1902), but recent phylogenetic studies found the genus to be closely related to two other fully winged Schistocerca species in the Galapagos (Lovejoy et al. 2006, Song et al. 2013, Song et al. 2017). This suggests that Halmenus is simply a brachypterous Schistocerca, and currently, we are planning a taxonomic revision to synonymize Halmenus with Schistocerca to reflect this finding. Nichelius is only known from the type series of three specimens and has not been collected during the past 100 yr (Amedegnato 1993). The type specimens look suspiciously similar to Schistocerca, so it is quite possible that it might be an aberrant member of Schistocerca. Therefore, it is possible to postulate that there was indeed a single transatlantic colonization by the ancestral Schistocerca, which gave rise to the current diversity in the New World.
In Australia, we find perhaps the most dramatic adaptive radiation among all grasshopper lineages. Our biogeographical analysis suggests that there was a single colonization event by the common ancestor of a lineage within Catantopinae that entered Australia in the mid-to-late Eocene. By the middle Eocene, Australia was already an isolated island without any major connections to other landmasses. When the ancestral catantopine arrived in Australia, it must have found vast areas of complex habitats where no other acridids had entered before. The Australian catantopines have diversified in deserts, grasslands, shrublands, tropical rainforests, and alpine habitats. It is also possible to find numerous species in this lineage that have converged to resemble the members of other worldwide acridid subfamilies (Rentz et al. 2003), strongly mirroring the diversification of marsupials from placental mammals. Australia was later colonized by Acridinae, Oedipodinae, Cyrtacanthacridinae, and Oxyinae, but these collectively represent only a small percentage of grasshopper diversity (68 spp.) on the continent compared to Catantopinae (300+ spp.).
Concluding Remarks
Despite the familiarity and economic importance of the Acrididae, a large-scale phylogeny of this family has never been proposed until now. Our study represents a crucial step towards understanding the evolution and diversification of these insects. We have recovered monophyletic Acrididae and proposed that the family originated in South America based on substantial evidence, contrary to a popular belief that its center of origin is Africa. We have also shown that Acrididae diverged in the early Cenozoic and represents one of the most recently diverged lineages within Orthoptera, which originated in the early Permian (Song et al. 2015). Because of the relatively young age of Acrididae, we hypothesize that its current cosmopolitan distribution was largely achieved by dispersal followed by relatively rapid radiation in many cases. We estimate that there have been a number of colonization and recolonization events (three major waves) between the New World and the Old World throughout the diversification of Acrididae, and, thus, the current diversity in any given region is a reflection of this complex history. Although we discovered several intriguing patterns, our phylogeny is admittedly based on less than 2% of the current diversity of Acrididae. Therefore, there is still a lot more to discover by further resolving the phylogeny of Acrididae using more taxon and character sampling in the future. We hope that our study can be used as a solid foundation to understand the evolution of these fascinating grasshoppers and that it will be the basis for future taxonomic studies.
Supplementary Data
Supplementary data are available at Insect Systematics and Diversity online.
Acknowledgments
We thank numerous collaborators who provided valuable specimens used in this study: the late Christiane Amedegnato, Corinna Bazelet, Antoine Foucart, Claudia Hemp, Taewoo Kim, Kate Umbers, and Michael Whiting. We also thank several colleagues who provided logistic support and expertise during our field expeditions: Corinna Bazelet, Stephen Cameron, Joey Mugleston, Michael Samways, and You Ning Su. A large number of students have contributed to molecular data generation throughout the duration of this project: Gabriella Alava, Grace Avecilla, Beka Buckman, James Leavitt, Matthew Moulton, Tyler Raszick, and Steve Gotham. We thank Charlie Johnson and Richard Metz at Texas A&M AgriLife Research Genomics and Bioinformatics Service for the NGS data generation and data processing. The comments by two anonymous reviewers significantly improved the text. Fieldwork to South Africa and Australia was conducted by the senior author under permit numbers CRO 177/08CR and CRO 178/08CR (Eastern Cape) and a license number SF007010 (Western Australia). This work was supported by the National Science Foundation (grant numbers DEB-1064082 and IOS-1253493 to H.S.) and the US Department of Agriculture (Hatch Grant TEX0-1-6584 to H.S).
References Cited
R Core Team.