-
PDF
- Split View
-
Views
-
Cite
Cite
B. P. Mallikarjuna Swamy, K. Kaladhar, N. Shobha Rani, G. S. V. Prasad, B. C. Viraktamath, G. Ashok Reddy, N. Sarla, QTL Analysis for Grain Quality Traits in 2 BC2F2 Populations Derived from Crosses between Oryza sativa cv Swarna and 2 Accessions of O. nivara, Journal of Heredity, Volume 103, Issue 3, May-June 2012, Pages 442–452, https://doi.org/10.1093/jhered/esr145
Close - Share Icon Share
Abstract
The appearance and cooking quality of rice determine its acceptability and price to a large extent. Quantitative trait loci (QTLs) for 12 grain quality traits were mapped in 2 mapping populations derived from Oryza sativa cv Swarna × O. nivara. The BC2F2 population of the cross Swarna × O. nivara IRGC81848 (population 1) was evaluated during 2005 and that from Swarna × O. nivara IRGC81832 (population 2) was evaluated during 2006. Linkage maps were constructed using 100 simple sequence repeat (SSR) markers in population 1 and 75 SSR markers in population 2. In all, 21 QTLs were identified in population 1 (43% from O. nivara) and 37 in population 2 (38% QTLs from O. nivara). The location of O. nivara–derived QTLs mp1.2 for milling percent, kw6.1 for kernel width, and klac12.1 for kernel length after cooking coincided in the 2 populations and appear to be useful for Marker Assisted Selection (MAS). Four QTLs for milling percent, 1 QTL each for amylose content, water uptake, elongation ratio, 2 QTLs for kernel width, and 3 QTLs for gel consistency, each explained more than 20% phenotypic variance. Three QTL clusters for grain quality traits were close to the genes/QTLs for shattering and seed dormancy. QTLs for 4 quality traits were associated with 5 of the 7 major yield QTLs reported in the same 2 mapping populations. Useful introgression lines have been developed for several agronomic traits. It emerges that 40% O. nivara alleles were trait enhancing in both populations, and QTLs for grain quality overlapped with yield meta-QTLs and QTLs for dormancy and seed shattering.
Rice grain yield needs to be continuously increased for food security as rice is the most important staple food, but grain quality is increasingly becoming important from the commercial point of view. The consumer preference for rice is determined mainly by appearance and cooking–eating qualities. In addition, processing qualities such as milling percent (mp) and head rice recovery (hrr) impact the market value (Rodante et al. 2009; Hao et al. 2010; Samuel et al. 2010; Nelson et al. 2011). Nutritional quality is also gaining importance. Appearance is specified by kernel length (kl), width (kw), length–width ratio (lbr), color, and translucency of polished grains. The cooking and eating qualities are reported to be largely governed by amylose content (ac), gel consistency (gc), gelatinization temperature (gt or asv), and pasting properties of starch. The other important traits influencing quality of cooked rice are kernel length after cooking (klac), volume expansion ratio (ver), elongation ratio (er), and water uptake (wup). Most of the quality traits are complex and follow quantitative inheritance with considerable influence of genotype, environment, and their interactions (Li et al. 2003; Zhou et al. 2003; Ge et al. 2005; Wang et al. 2007; Yongmei et al. 2007; Amarawathi et al. 2008; Zheng et al. 2008; Nelson et al. 2011). Obtaining the right combination of these traits by phenotype-based classical breeding is very difficult, time consuming, and laborious. However, several recent developments have increased our understanding of the genes, pathways, and molecular mechanisms determining over all quality traits in rice (Fitzgerald et al. 2009).
Mapping of quantitative trait loci (QTLs) for grain quality traits in rice is a forward genetic approach to dissect and use them in marker-assisted selection and also for gene discovery. In rice, many quality traits have been mapped and tagged in different genetic backgrounds using molecular markers (Tan et al. 1999;,Aluko et al. 2004; Wan et al. 2006; Wang et al. 2007; Yongmei et al. 2007; Zheng et al. 2008; Zhou et al. 2009; Shao et al. 2010; Karim et al. 2011; Nelson et al. 2011). Several previous QTL mapping studies involving mapping populations obtained from intraspecific and interspecific crosses revealed that quality traits are quite complex, and several chromosomal locations are associated with the expression of a given phenotype. Redona and Mackill (1998) detected 2 QTLs for kl on chromosome 3, explaining 10.4% and 20.9% of the phenotypic variation (PV), respectively. Aluko et al. (2004) also mapped a QTL for grain length on chromosome 3, explaining 12.5% of the PV. Wan et al. (2006) fine mapped a QTL for grain length within a physical interval of 87.5 kb on chromosome 3. Amarawathi et al. (2008) mapped QTLs for grain length on chromosomes 1 and 7 explaining 10% and 7% PV. Bao et al. (2000) consistently detected a major QTL for ac near waxy locus in a doubled haploid population grown over 3 seasons. A QTL for ac located at RM3–RM217 on the short arm of chromosome 6 explained 39.6% of the PV (Amarawathi et al. 2008). Lanceras et al. (2000) reported an ac QTL on chromosome 5 with 58% PV and 4.48% additive effect. Aluko et al. (2004) reported ac QTL on chromosome 6 with a PV of 73% and had an additive effect of 2.6. Redona and Mackill (1998) mapped a major locus explaining 22% PV on chromosome 7. Tan et al. (2000) detected a QTL for grain width on chromosome 5 and a minor locus on chromosome 6 in 2 different populations. Jue et al. (2009) detected a pleiotropic main effect QTL on chromosome 3 influencing grain length, length and width ratio, and head rice ratio. A minor QTL qGL-7 with pleiotropic effects on grain length, grain width, and grain weight was identified in a Recombinant Inbred Lines (RIL) population (Bai et al. 2010). Shao et al. (2010) detected a grain length QTL GL-7 on chromosome 7 and narrowed it to a 278-kb region. Li et al. (2004) identified 2 QTLs, lwr3.1 from Oryza sativa and lwr12.1 from O. glaberrima, for length/width ratio. Some of these major effect QTLs can be used for marker-aided selection of target traits and/or exclusion of nondesirable traits in segregating populations. Marker-assisted selection has been successfully used to improve grain quality traits of a male sterile line and maintainer line widely used in hybrid rice breeding in China (Zhou et al. 2003; Jin et al. 2010) and to improve cooking quality traits in Myanmar rice cultivar Manawthukha (Myint et al. 2009).
Wild species may not be superior to cultivated rice varieties with respect to yield and grain quality traits. However, the hidden cryptic variations in the wild accessions and transgressive segregants that appear in the wide crosses provide an opportunity to further improve the yield and grain quality traits and to broaden the genetic base of the popular rice varieties. Most of the quality traits have been mapped in intraspecific crosses. Very few wild species have been explored for mapping and use of quality traits. There are only 2 reports on mapping of QTLs for grain quality traits from O. rufipogon (Septiningsih et al. 2003; Yuan et al. 2010) and none using O. nivara.
Oryza nivara (Sharma et Shastry) is one of the immediate wild progenitors of Asian cultivated rice O. sativa. Even though it is wild, its seeds are collected and cooked for traditional days of fasting in many parts of India, indicating its nutritional value (www.agri-history.org). The O. nivara accessions from different parts of India and abroad reveal abundant genetic diversity (Joshi et al. 2000; Sarla et al. 2003; Juneja et al. 2006) and are likely to have new alleles for improving grain quality traits.
Two accessions of O. nivara IRGC81848 (Uttar Pradesh) and IRGC81832 (Bihar) were used for developing mapping populations in the background of popular cultivated rice variety Swarna. These 2 accessions showed moderate molecular genetic diversity from Swarna, and the 2 states, where they belong to, have abundant genetic diversity for O. nivara accessions. The 2 accessions were also similar for grain quality traits, such as kw, lbr, asv, ac, and gc. However, the values were different for kl, klac, and er.
Two BC2F2 populations were developed from crosses between O. sativa cv Swarna and 2 different accessions of O. nivara. The population derived from Swarna × IRGC81848 is designated as population 1 and the population derived from Swarna × IRGC81832 is designated as population 2. The BC2F3 seeds from these 2 populations were used for mapping QTLs for grain quality traits. Population 1 and population 2 consisted of 140 and 146 BC2F3 families, respectively.
The main objectives of the present study were to map QTLs for 12 grain quality traits in BC2F3 seeds derived from 2 mapping populations, to know the proportion of trait-enhancing alleles that can be obtained from 2 O. nivara accessions and their coincidence in the 2 populations, and to know the colocation of QTLs for quality with QTLs mapped for yield and yield-related traits in the 2 O. nivara-derived populations.
Materials and Methods
Plant Material
The 2 accessions of O. nivara used were obtained from IRRI but collected from Uttar Pradesh (IRGC81848) and Bihar (IRGC81832), 2 Northern States of India. These 2 wild accessions are quite diverse from other accessions of O. nivara from 9 other States in India (Sarla et al. 2003). The popular rainfed lowland rice cultivar Swarna was used as the recipient. An advanced backcross strategy as described by Thomson et al. (2003) was followed to develop 2 mapping populations. Crosses were made using O. nivara accessions as a male parent and Swarna as a female parent to generate F1s. BC1F1s and BC2F1s were generated using Swarna as a male parent. BC2F2 families were developed by selfing BC2F1s. Two mapping populations, population 1 (Swarna × IRGC81848) and population 2 (Swarna × IRGC81832) consisted of 245 and 227 BC2F2 families, respectively. All the families from both the populations were genotyped and used for mapping yield and yield component traits. However, only 140 families from the population 1 and 146 families from the population 2 were used for grain quality analysis and QTL mapping.
Field Evaluation of the Mapping Populations
The experiments were conducted at the farm of Directorate of Rice Research , Hyderabad in South India. The soil type at the experimental site was clay Vertisol. During the field evaluation from July to November in wet season 2005, the average temperature was 30 °C, and total rainfall received was 508 mm. Whereas, during 2006, wet season experiment, the average temperature was 30.5 °C, and the total rainfall was 638 mm. Population 1 was evaluated during wet season 2006 and population 2 was evaluated during wet season 2005. Two populations were grown in 2 replications in an augmented block design. Each backcross family and the control (Swarna) consisted of 30 plants planted in 3 rows of 10 plants each adopting a uniform spacing of 20 cm between rows and 15 cm between plants. Standard agronomic practices and need-based plant protection measures were adopted uniformly to raise the crop.
Phenotyping for Quality Traits
Two-month-old seed samples after harvest with 14% moisture were used for phenotyping quality traits. One hundred grams of well-dried seeds were used for dehusking using Satake dehusker (THU35A). The dehusked and cleaned brown rice was polished to remove 8–10% bran using Satake polisher (TM05). The polished white raw rice was used for analyzing the cooking and eating quality traits.
Milling percent (mp): The ratio of weight of polished grain to weight of paddy expressed in percentage. Head rice recovery (hrr): The ratio of weight of the whole polished grains to the weight of paddy used for polishing expressed in percentage. Kernel length (kl): It is measured as average length of 10 grains in millimeters. Kernel width (kw): It is measured as average length of 10 grains in millimeters. Length/width ratio (lwr): Ratio of mean kernel length to mean kernel breadth for 10 grains. Alkali spreading value (asv): A set of 6 polished grains from each line was immersed in a freshly prepared 1.7% KOH solution and incubated at 27–30 °C for 23 h, and spreading of the rice grain was recorded by visual observation in a scale of 1–7, where score 1 is unaffected and 7 is completely dissolved based on Little et al. 1958. Amylose content (ac): It was estimated using the procedure of Juliano (1971) and expressed in percentage. Gel consistency (gc): GC was measured in millimeters according to the method of Cagampang et al. (1973). Volume expansion ratio (ver): Five grams of rice sample was taken in a test tube and soaked in 15 ml of water for 10 min and increase in volume noted. It was then cooked for 20 min. The cooked rice was dipped in 50 ml water taken in 100 ml measuring cylinder and increase in volume noted. The volume expansion ratio is estimated as ratio of increase in volume after cooking to the volume before cooking. Elongation ratio (er): It is the ratio of mean of the kernel length after cooking to mean of kernel length before cooking. Kernel elongation after cooking (klac): Mean length of 10 cooked rice kernels in millimeters. Water uptake (wup): Water absorption was calculated as ratio of weight of water absorbed on cooking (cooked rice kernel minus dry kernel) to the milled grain weight.
Trait Correlations
Trait correlations for 12 grain quality traits were obtained using pairwise Pearson correlations using Crop Stat v7.3.2.
Genotyping and QTL Analysis
Leaf samples were collected on 2-month-old seedlings from the parents and the 2 mapping populations. In the mapping populations, leaves were collected from 5 plants in the middle row and bulked. DNA was extracted following the method of Zheng et al. 1995. A set of 250 randomly selected microsatellite markers from all 12 chromosomes were used to select polymorphic loci between Swarna and O. nivara accessions (IRGC81848 and IRGC81832). A total of 100 polymorphic microsatellite markers separated by an average distance of 15 cM were used to analyze the segregation in 227 BC2F2 families of population 1 and 75 markers in 245 BC2F2 families of population 2. Among the simple sequence repeat (SSR) markers used for genotyping, 45 were common between the 2 studies. The PCR reaction for SSR primers was performed with 15 μl final volume containing, 45 ng of genomic DNA, 10X buffer, 0.125 mM final concentration of each dNTPs, 0.2 μM each of forward and reverse primer, 2% formamide, and 1 U of Biogene Taq DNA Polymerase. PCR amplification was performed under the following conditions: Initial denaturation at 94 °C for 5 min, followed by 35 cycles of denaturation at 94 °C for 1 min, annealing at 55 °C for 1 min, extension at 72 °C for 2 min, followed by the final extension 72 °C for 5 min. The amplified products were checked for polymorphism or marker segregation on an agarose gel (3%) and scored for segregating bands.
Linkage and QTL Analysis
A linkage map was constructed from the genotypic data of 100 markers in population 1 and 75 markers in population 2 using Join Map 3.0. Assignment of linkage groups to the respective chromosomes was done based on the rice maps developed at Cornell University. A chi-square test (P < 0.01) was used to identify markers with distorted segregation. A LOD value of 2.5 and map function Kosambi were used for the estimation of map distances. QTLs were identified separately for the 2 populations using QTL Cartographer 2.5 (http://statgen.ncsu.edu/qtlcart/WQTLCart.html). The QTLs were detected by interval mapping (IM) and composite interval mapping (CIM) (Zeng, 1994) with a threshold LOD of 3.0. A window size of 10 cM and walk speed of 1 cM were used in the genome scan. Digenic QTL interactions were identified using QTL network 2.0.
Results
Trait Analysis
The appearance of transgressive segregants in the progenies of wide crosses is a common phenomenon. This may be due to the accumulation of superior alleles from the 2 distinct parents in the segregating progenies and also because of the different types of interactions among them. In the present investigation, transgressive segregants were observed for all the grain quality traits except asv in both the populations (Table 1). As many as, 75% of all the families showed at least 5% increase over Swarna for the traits wup, er, and klac. One family each namely 83, 229, and 69 showed an increase of up to 120%, 37%, and 40% for wup, er, and klac, respectively. Twenty-nine families showed an increase of 11% in ac and 12 families showed an increase of 23% in gc. The increase in trait value was up to 10% in the remaining traits. The frequency of almost all the traits showed near normal distribution, and the distribution patterns in populations 1 and 2 were similar. Specifically, all the traits except ac and ver showed a near normal distribution (Supplementary Figure 1).
Variation for 12 quality traits in 3 parents and 2 populations derived from Swarna × Oryza nivara (IRGC81848 and IRGC81832) crosses
| Sl No. | Trait | Swarna mean | O. nivara mean | Range in BC2F2 families | No. of families showing >5% increase over Swarna |
| 1 | Milling (%) | 66.8 | — | 41.5–76.9 (41.5–76.9) | 31 (32) |
| 2 | Head rice recovery (%) | 60.5 | — | 12.2–64 (12.2–66.3) | 1 (4) |
| 3 | Kernel length (mm) | 5.3 | 5.5 (4.9) | 5.0–5.8 (4.9–5.8) | 37 (44) |
| 4 | Kernel width (mm) | 2.2 | 2.4 (2.2) | 1.9–2.3 (1.9–2.3) | 16 (17) |
| 5 | Length and width ratio | 2.5 | 2.2 (2.2) | 2.2–2.6 (2.2–2.6) | 9 (12) |
| 6 | Volume expansion ratio | 5.3 | — | 4–5.6 (4–5.6) | 5 (14) |
| 7 | Water uptake (ml) | 120.0 | — | 105–265 (105–265) | 117 (139) |
| 8 | Kernel length after cooking (mm) | 8.1 | 10.3 (9.8) | 7.6–11.1 (7.6–11.1) | 92 (115) |
| 9 | Elongation ratio | 1.5 | 1.9 (2.3) | 1.42–2.11 (1.4–2.11) | 94 (116) |
| 10 | Alkali spreading value | 7.0 | 5.0 (5.0) | 3.0–5.0 (3.0–5.0) | — |
| 11 | Amylose content (%) | 26.1 | 23.8 (23.5) | 18.5–29.0 (17.7–29.0) | 13 (12) |
| 12 | Gel consistency (mm) | 65.0 | 56 (58) | 23–80 (23–80) | 3 (5) |
| Sl No. | Trait | Swarna mean | O. nivara mean | Range in BC2F2 families | No. of families showing >5% increase over Swarna |
| 1 | Milling (%) | 66.8 | — | 41.5–76.9 (41.5–76.9) | 31 (32) |
| 2 | Head rice recovery (%) | 60.5 | — | 12.2–64 (12.2–66.3) | 1 (4) |
| 3 | Kernel length (mm) | 5.3 | 5.5 (4.9) | 5.0–5.8 (4.9–5.8) | 37 (44) |
| 4 | Kernel width (mm) | 2.2 | 2.4 (2.2) | 1.9–2.3 (1.9–2.3) | 16 (17) |
| 5 | Length and width ratio | 2.5 | 2.2 (2.2) | 2.2–2.6 (2.2–2.6) | 9 (12) |
| 6 | Volume expansion ratio | 5.3 | — | 4–5.6 (4–5.6) | 5 (14) |
| 7 | Water uptake (ml) | 120.0 | — | 105–265 (105–265) | 117 (139) |
| 8 | Kernel length after cooking (mm) | 8.1 | 10.3 (9.8) | 7.6–11.1 (7.6–11.1) | 92 (115) |
| 9 | Elongation ratio | 1.5 | 1.9 (2.3) | 1.42–2.11 (1.4–2.11) | 94 (116) |
| 10 | Alkali spreading value | 7.0 | 5.0 (5.0) | 3.0–5.0 (3.0–5.0) | — |
| 11 | Amylose content (%) | 26.1 | 23.8 (23.5) | 18.5–29.0 (17.7–29.0) | 13 (12) |
| 12 | Gel consistency (mm) | 65.0 | 56 (58) | 23–80 (23–80) | 3 (5) |
Values for population 2 (BC2F2 from Swarna × IRGC81832) are shown in parentheses corresponding to values for population 1 (BC2F2 from Swarna × IRGC818).
Variation for 12 quality traits in 3 parents and 2 populations derived from Swarna × Oryza nivara (IRGC81848 and IRGC81832) crosses
| Sl No. | Trait | Swarna mean | O. nivara mean | Range in BC2F2 families | No. of families showing >5% increase over Swarna |
| 1 | Milling (%) | 66.8 | — | 41.5–76.9 (41.5–76.9) | 31 (32) |
| 2 | Head rice recovery (%) | 60.5 | — | 12.2–64 (12.2–66.3) | 1 (4) |
| 3 | Kernel length (mm) | 5.3 | 5.5 (4.9) | 5.0–5.8 (4.9–5.8) | 37 (44) |
| 4 | Kernel width (mm) | 2.2 | 2.4 (2.2) | 1.9–2.3 (1.9–2.3) | 16 (17) |
| 5 | Length and width ratio | 2.5 | 2.2 (2.2) | 2.2–2.6 (2.2–2.6) | 9 (12) |
| 6 | Volume expansion ratio | 5.3 | — | 4–5.6 (4–5.6) | 5 (14) |
| 7 | Water uptake (ml) | 120.0 | — | 105–265 (105–265) | 117 (139) |
| 8 | Kernel length after cooking (mm) | 8.1 | 10.3 (9.8) | 7.6–11.1 (7.6–11.1) | 92 (115) |
| 9 | Elongation ratio | 1.5 | 1.9 (2.3) | 1.42–2.11 (1.4–2.11) | 94 (116) |
| 10 | Alkali spreading value | 7.0 | 5.0 (5.0) | 3.0–5.0 (3.0–5.0) | — |
| 11 | Amylose content (%) | 26.1 | 23.8 (23.5) | 18.5–29.0 (17.7–29.0) | 13 (12) |
| 12 | Gel consistency (mm) | 65.0 | 56 (58) | 23–80 (23–80) | 3 (5) |
| Sl No. | Trait | Swarna mean | O. nivara mean | Range in BC2F2 families | No. of families showing >5% increase over Swarna |
| 1 | Milling (%) | 66.8 | — | 41.5–76.9 (41.5–76.9) | 31 (32) |
| 2 | Head rice recovery (%) | 60.5 | — | 12.2–64 (12.2–66.3) | 1 (4) |
| 3 | Kernel length (mm) | 5.3 | 5.5 (4.9) | 5.0–5.8 (4.9–5.8) | 37 (44) |
| 4 | Kernel width (mm) | 2.2 | 2.4 (2.2) | 1.9–2.3 (1.9–2.3) | 16 (17) |
| 5 | Length and width ratio | 2.5 | 2.2 (2.2) | 2.2–2.6 (2.2–2.6) | 9 (12) |
| 6 | Volume expansion ratio | 5.3 | — | 4–5.6 (4–5.6) | 5 (14) |
| 7 | Water uptake (ml) | 120.0 | — | 105–265 (105–265) | 117 (139) |
| 8 | Kernel length after cooking (mm) | 8.1 | 10.3 (9.8) | 7.6–11.1 (7.6–11.1) | 92 (115) |
| 9 | Elongation ratio | 1.5 | 1.9 (2.3) | 1.42–2.11 (1.4–2.11) | 94 (116) |
| 10 | Alkali spreading value | 7.0 | 5.0 (5.0) | 3.0–5.0 (3.0–5.0) | — |
| 11 | Amylose content (%) | 26.1 | 23.8 (23.5) | 18.5–29.0 (17.7–29.0) | 13 (12) |
| 12 | Gel consistency (mm) | 65.0 | 56 (58) | 23–80 (23–80) | 3 (5) |
Values for population 2 (BC2F2 from Swarna × IRGC81832) are shown in parentheses corresponding to values for population 1 (BC2F2 from Swarna × IRGC818).
There were 14 significant correlations among the 12 traits in population 1 and 13 significant correlations in population 2. The trait correlations were almost similar in both the populations. In both the populations, highly significant correlations were observed between kl on one hand and kw, lbr, er, ac, and gc on the other. The trait klac showed the highest positive correlation with er in both populations (0.936, 0.933, P < 0.01), the next was lbr with kl (0.603, 0.612, P < 0.01). A highly negative correlation was found between kw and lbr in both populations (−0.659, −0.643, P < 0.01). Other significant positive correlations were between kw and ver and between wup and asv in both the populations (Supplementary Table 1). Even though 2 different O. nivara accessions from 2 different environments were used in the development of the mapping population, the range of variability for all traits and the correlations were almost similar in the 2 populations. This could be because the 2 O. nivara accessions used in the development of the mapping populations originated from 2 adjoining states of India—Uttar Pradesh and Bihar. Second, the 2 accessions were similar in quality traits and may have produced similar kind of transgressive segregants. Third, even if there was a difference in the 2 populations, it was not obvious as Swarna type of plants were selected in BC1 and BC2. Wild species accessions from different ecogeographical locations are likely to generate different variability.
QTL Analysis
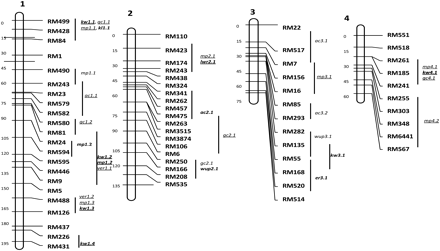
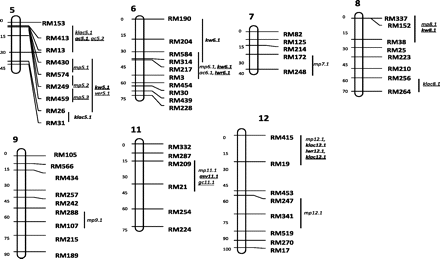
QTLs were identified by IM or CIM or both. Most of the QTLs were derived from Swarna. However, many new and trait-improving QTLs were mapped from O. nivara. In all, 21 QTLs were detected for 9 traits in population 1 and 37 QTLs for 8 traits in population 2 (Table 2). QTLs showing PV of less than 5% are not shown. All the QTLs are depicted in Figure 1.
QTLs identified for 12 grain quality traits by IM and CIM in population 1 (Swarna × Oryza nivara [IRGC81848]) and population 2 (Swarna × Oryza nivara [IRGC81832])
| Trait | Chromosome | Marker interval | Allelic effect | IM | CIM | Trait | Chromosome | Marker interval | Allelic effect | IM | CIM | ||||||||
| LOD | R2 | Additive effect | LOD | R2 | Additive effect | LOD | R2 | Additive effect | LOD | R2 | Additive effect | ||||||||
| Population 1 | Population 2 | ||||||||||||||||||
| Milling percent (%) | Milling percent (%) | ||||||||||||||||||
| mp1.1 | 1 | RM490–RM243 | Swarna | 6.1 | 6 | 5.34 | 4.54 | 8 | 4.02 | mp1.1 | 1 | RM499–RM428 | Swarna | 10.7 | 25 | 14.80 | 5.9 | 35 | 13.48 |
| mp1.2 | 1 | RM24–RM594 | nivara | 4.57 | 5 | −0.69 | mp1.2 | 1 | RM81–RM595 | nivara | 8.4 | 4 | −0.83 | ||||||
| mp6.1 | 6 | RM314–RM3 | Swarna | 5.4 | 21 | 0.79 | mp1.3 | 1 | RM488–RM126 | Swarna | 11.8 | 24 | 0.41 | ||||||
| mp7.1 | 7 | RM172–RM248 | Swarna | 7.02 | 6 | 2.05 | mp2.1 | 2 | RM423–RM438 | Swarna | 9.6 | 5 | 0.23 | ||||||
| mp9.1 | 9 | RM288–RM107 | Swarna | 2.77 | 5 | 3.78 | mp3.1 | 3 | RM7–RM16 | Swarna | 12.1 | 8 | −0.58 | ||||||
| mp11.1 | 11 | RM209–RM21 | Swarna | 8 | 7 | 2.82 | mp4.1 | 4 | RM261–RM241 | Swarna | 12.5 | 22 | 3.48 | ||||||
| mp12.1 | 12 | RM415–RM19 | Swarna | 9.3 | 10 | 0.41 | mp4.2 | 4 | RM241–RM567 | Swarna | 8.0 | 5 | 10.45 | ||||||
| mp5.1 | 5 | RM430–RM574 | Swarna | 9.5 | 17 | 0.98 | 5.68 | 6 | 2.47 | ||||||||||
| mp5.2 | 5 | RM574–RM249 | Swarna | 3.5 | 5 | 0.85 | |||||||||||||
| mp5.3 | 5 | RM249–RM26 | Swarna | 7.4 | 12 | 2.30 | |||||||||||||
| mp8.1 | 8 | RM337–RM38 | Swarna | 5.6 | 6 | 2.34 | |||||||||||||
| mp12.1 | 12 | RM247–RM519 | Swarna | 8.7 | 10 | 13.15 | |||||||||||||
| Kernel length (mm) | |||||||||||||||||||
| kl1.1 | 1 | RM499–RM84 | Swarna | 3.3 | 6 | 0.1 | |||||||||||||
| Kernel width (mm) | Kernel width (mm) | ||||||||||||||||||
| kw3.1 | 3 | RM135–RM168 | nivara | 2.8 | 17 | −0.06 | 2.86 | 16 | −0.06 | kw1.1 | 1 | RM499–RM428 | nivara | 3.3 | 30 | −0.36 | |||
| kw6.1 | 6 | RM190–RM314 | nivara | 2.85 | 9 | −0.04 | kw1.2 | 1 | RM81–RM9 | nivara | 5.2 | 13 | −0.27 | 4.07 | 13 | −0.12 | |||
| kw1.3 | 1 | RM488–RM126 | nivara | 4.0 | 30 | −0.17 | 3.32 | 11 | −0.03 | ||||||||||
| kw1.4 | 1 | RM226–RM431 | nivara | 4.4 | 12 | −0.14 | |||||||||||||
| kw4.1 | 4 | RM185–RM241 | nivara | 2.9 | 12 | −0.25 | |||||||||||||
| kw5.1 | 5 | RM249–RM26 | nivara | 4.2 | 9 | −0.09 | |||||||||||||
| kw6.1 | 6 | RM204–RM3 | nivara | 3.2 | 12 | −0.10 | |||||||||||||
| kw8.1 | 8 | RM152–RM38 | nivara | 4.2 | 13 | −0.10 | |||||||||||||
| L\B ratio | L\B ratio | ||||||||||||||||||
| lwr12.1 | 12 | RM415–RM19 | nivara | 2.56 | 9 | −0.05 | lwr2.1 | 2 | RM174–M324 | nivara | 3.0 | 10 | −0.03 | 2.7 | 7 | −0.03 | |||
| Water uptake | lwr6.1 | 6 | RM584–RM217 | nivara | 3.1 | 8 | −0.03 | 4.23 | 6 | −0.02 | |||||||||
| wup2.1 | 2 | RM250–RM535 | nivara | 3.3 | 24 | −28.15 | Volume expansion ratio | ||||||||||||
| wup3.1 | 3 | RM282–RM55 | Swarna | 2.6 | 15 | 38.79 | ver1.1 | 1 | RM488–RM128 | Swarna | 3.9 | 8 | 0.02 | ||||||
| ver1.2 | 1 | RM226–RM431 | Swarna | 2.8 | 9 | 0.04 | |||||||||||||
| ver5.1 | 5 | RM430–RM26 | Swarna | 4.2 | 5 | 0.05 | |||||||||||||
| Kernel length after cooking (mm) | Kernel length after cooking (mm) | ||||||||||||||||||
| klac5.1 | 5 | RM26–RM31 | nivara | 2.5 | 12 | −1.06 | klac5.1 | 5 | RM153–RM13 | Swarna | 4 | 12 | 20.96 | 5.41 | 15 | 13.71 | |||
| klac12.1 | 12 | RM19–RM453 | nivara | 3.4 | 15 | −21.32 | klac8.1 | 8 | RM256–RM264 | Swarna | 2.49 | 7 | 13.09 | ||||||
| klac12.1 | 12 | RM415–RM19 | nivara | 2.5 | 12 | −7.4 | |||||||||||||
| Elongation ratio | Alkali spreading value | ||||||||||||||||||
| er3.1 | 3 | RM55–RM520 | nivara | 2.7 | 21 | −0.68 | asv11.1 | 11 | RM209–RM21 | nivara | 2.6 | 30 | −0.3 | ||||||
| Amylose content (%) | Amylose content (%) | ||||||||||||||||||
| ac2.1 | 2 | RM262–RM3515 | nivara | 3.4 | 11 | −2.19 | ac1.1 | 1 | RM243–RM582 | Swarna | 2.63 | 5 | 0.27 | ||||||
| ac3.1 | 3 | RM22–RM7 | Swarna | 2.51 | 5 | 0.87 | ac5.1 | 5 | RM413–RM13 | nivara | 2.6 | 9 | −0.34 | ||||||
| ac3.2 | 3 | RM85–RM293 | Swarna | 2.6 | 15 | 2.05 | ac5.2 | 5 | RM153–RM413 | Swarna | 3.54 | 9 | 0.26 | ||||||
| ac6.1 | 6 | RM314–RM3 | Swarna | 2.6 | 23 | 0.86 | |||||||||||||
| Gel consistency (mm) | Gel consistency (mm) | ||||||||||||||||||
| gc2.1 | 2 | RM250–RM535 | Swarna | 3.18 | 14 | 10.1 | gc1.1 | 1 | RM499–RM428 | Swarna | 3.9 | 30 | 5.67 | 4.21 | 29 | 5.49 | |||
| gc1.2 | 1 | RM580–RM81 | Swarna | 3.2 | 21 | 6.45 | |||||||||||||
| gc2.1 | 2 | RM475–RM250 | Swarna | 3.7 | 4 | 0.61 | |||||||||||||
| gc4.1 | 4 | RM185–RM241 | Swarna | 4.2 | 12 | 0.89 | 3.08 | 13 | 4.67 | ||||||||||
| gc11.1 | 11 | RM209–RM21 | Swarna | 2.9 | 30 | 4.25 | 3.36 | 8 | 0.53 | ||||||||||
| Trait | Chromosome | Marker interval | Allelic effect | IM | CIM | Trait | Chromosome | Marker interval | Allelic effect | IM | CIM | ||||||||
| LOD | R2 | Additive effect | LOD | R2 | Additive effect | LOD | R2 | Additive effect | LOD | R2 | Additive effect | ||||||||
| Population 1 | Population 2 | ||||||||||||||||||
| Milling percent (%) | Milling percent (%) | ||||||||||||||||||
| mp1.1 | 1 | RM490–RM243 | Swarna | 6.1 | 6 | 5.34 | 4.54 | 8 | 4.02 | mp1.1 | 1 | RM499–RM428 | Swarna | 10.7 | 25 | 14.80 | 5.9 | 35 | 13.48 |
| mp1.2 | 1 | RM24–RM594 | nivara | 4.57 | 5 | −0.69 | mp1.2 | 1 | RM81–RM595 | nivara | 8.4 | 4 | −0.83 | ||||||
| mp6.1 | 6 | RM314–RM3 | Swarna | 5.4 | 21 | 0.79 | mp1.3 | 1 | RM488–RM126 | Swarna | 11.8 | 24 | 0.41 | ||||||
| mp7.1 | 7 | RM172–RM248 | Swarna | 7.02 | 6 | 2.05 | mp2.1 | 2 | RM423–RM438 | Swarna | 9.6 | 5 | 0.23 | ||||||
| mp9.1 | 9 | RM288–RM107 | Swarna | 2.77 | 5 | 3.78 | mp3.1 | 3 | RM7–RM16 | Swarna | 12.1 | 8 | −0.58 | ||||||
| mp11.1 | 11 | RM209–RM21 | Swarna | 8 | 7 | 2.82 | mp4.1 | 4 | RM261–RM241 | Swarna | 12.5 | 22 | 3.48 | ||||||
| mp12.1 | 12 | RM415–RM19 | Swarna | 9.3 | 10 | 0.41 | mp4.2 | 4 | RM241–RM567 | Swarna | 8.0 | 5 | 10.45 | ||||||
| mp5.1 | 5 | RM430–RM574 | Swarna | 9.5 | 17 | 0.98 | 5.68 | 6 | 2.47 | ||||||||||
| mp5.2 | 5 | RM574–RM249 | Swarna | 3.5 | 5 | 0.85 | |||||||||||||
| mp5.3 | 5 | RM249–RM26 | Swarna | 7.4 | 12 | 2.30 | |||||||||||||
| mp8.1 | 8 | RM337–RM38 | Swarna | 5.6 | 6 | 2.34 | |||||||||||||
| mp12.1 | 12 | RM247–RM519 | Swarna | 8.7 | 10 | 13.15 | |||||||||||||
| Kernel length (mm) | |||||||||||||||||||
| kl1.1 | 1 | RM499–RM84 | Swarna | 3.3 | 6 | 0.1 | |||||||||||||
| Kernel width (mm) | Kernel width (mm) | ||||||||||||||||||
| kw3.1 | 3 | RM135–RM168 | nivara | 2.8 | 17 | −0.06 | 2.86 | 16 | −0.06 | kw1.1 | 1 | RM499–RM428 | nivara | 3.3 | 30 | −0.36 | |||
| kw6.1 | 6 | RM190–RM314 | nivara | 2.85 | 9 | −0.04 | kw1.2 | 1 | RM81–RM9 | nivara | 5.2 | 13 | −0.27 | 4.07 | 13 | −0.12 | |||
| kw1.3 | 1 | RM488–RM126 | nivara | 4.0 | 30 | −0.17 | 3.32 | 11 | −0.03 | ||||||||||
| kw1.4 | 1 | RM226–RM431 | nivara | 4.4 | 12 | −0.14 | |||||||||||||
| kw4.1 | 4 | RM185–RM241 | nivara | 2.9 | 12 | −0.25 | |||||||||||||
| kw5.1 | 5 | RM249–RM26 | nivara | 4.2 | 9 | −0.09 | |||||||||||||
| kw6.1 | 6 | RM204–RM3 | nivara | 3.2 | 12 | −0.10 | |||||||||||||
| kw8.1 | 8 | RM152–RM38 | nivara | 4.2 | 13 | −0.10 | |||||||||||||
| L\B ratio | L\B ratio | ||||||||||||||||||
| lwr12.1 | 12 | RM415–RM19 | nivara | 2.56 | 9 | −0.05 | lwr2.1 | 2 | RM174–M324 | nivara | 3.0 | 10 | −0.03 | 2.7 | 7 | −0.03 | |||
| Water uptake | lwr6.1 | 6 | RM584–RM217 | nivara | 3.1 | 8 | −0.03 | 4.23 | 6 | −0.02 | |||||||||
| wup2.1 | 2 | RM250–RM535 | nivara | 3.3 | 24 | −28.15 | Volume expansion ratio | ||||||||||||
| wup3.1 | 3 | RM282–RM55 | Swarna | 2.6 | 15 | 38.79 | ver1.1 | 1 | RM488–RM128 | Swarna | 3.9 | 8 | 0.02 | ||||||
| ver1.2 | 1 | RM226–RM431 | Swarna | 2.8 | 9 | 0.04 | |||||||||||||
| ver5.1 | 5 | RM430–RM26 | Swarna | 4.2 | 5 | 0.05 | |||||||||||||
| Kernel length after cooking (mm) | Kernel length after cooking (mm) | ||||||||||||||||||
| klac5.1 | 5 | RM26–RM31 | nivara | 2.5 | 12 | −1.06 | klac5.1 | 5 | RM153–RM13 | Swarna | 4 | 12 | 20.96 | 5.41 | 15 | 13.71 | |||
| klac12.1 | 12 | RM19–RM453 | nivara | 3.4 | 15 | −21.32 | klac8.1 | 8 | RM256–RM264 | Swarna | 2.49 | 7 | 13.09 | ||||||
| klac12.1 | 12 | RM415–RM19 | nivara | 2.5 | 12 | −7.4 | |||||||||||||
| Elongation ratio | Alkali spreading value | ||||||||||||||||||
| er3.1 | 3 | RM55–RM520 | nivara | 2.7 | 21 | −0.68 | asv11.1 | 11 | RM209–RM21 | nivara | 2.6 | 30 | −0.3 | ||||||
| Amylose content (%) | Amylose content (%) | ||||||||||||||||||
| ac2.1 | 2 | RM262–RM3515 | nivara | 3.4 | 11 | −2.19 | ac1.1 | 1 | RM243–RM582 | Swarna | 2.63 | 5 | 0.27 | ||||||
| ac3.1 | 3 | RM22–RM7 | Swarna | 2.51 | 5 | 0.87 | ac5.1 | 5 | RM413–RM13 | nivara | 2.6 | 9 | −0.34 | ||||||
| ac3.2 | 3 | RM85–RM293 | Swarna | 2.6 | 15 | 2.05 | ac5.2 | 5 | RM153–RM413 | Swarna | 3.54 | 9 | 0.26 | ||||||
| ac6.1 | 6 | RM314–RM3 | Swarna | 2.6 | 23 | 0.86 | |||||||||||||
| Gel consistency (mm) | Gel consistency (mm) | ||||||||||||||||||
| gc2.1 | 2 | RM250–RM535 | Swarna | 3.18 | 14 | 10.1 | gc1.1 | 1 | RM499–RM428 | Swarna | 3.9 | 30 | 5.67 | 4.21 | 29 | 5.49 | |||
| gc1.2 | 1 | RM580–RM81 | Swarna | 3.2 | 21 | 6.45 | |||||||||||||
| gc2.1 | 2 | RM475–RM250 | Swarna | 3.7 | 4 | 0.61 | |||||||||||||
| gc4.1 | 4 | RM185–RM241 | Swarna | 4.2 | 12 | 0.89 | 3.08 | 13 | 4.67 | ||||||||||
| gc11.1 | 11 | RM209–RM21 | Swarna | 2.9 | 30 | 4.25 | 3.36 | 8 | 0.53 | ||||||||||
QTLs identified for 12 grain quality traits by IM and CIM in population 1 (Swarna × Oryza nivara [IRGC81848]) and population 2 (Swarna × Oryza nivara [IRGC81832])
| Trait | Chromosome | Marker interval | Allelic effect | IM | CIM | Trait | Chromosome | Marker interval | Allelic effect | IM | CIM | ||||||||
| LOD | R2 | Additive effect | LOD | R2 | Additive effect | LOD | R2 | Additive effect | LOD | R2 | Additive effect | ||||||||
| Population 1 | Population 2 | ||||||||||||||||||
| Milling percent (%) | Milling percent (%) | ||||||||||||||||||
| mp1.1 | 1 | RM490–RM243 | Swarna | 6.1 | 6 | 5.34 | 4.54 | 8 | 4.02 | mp1.1 | 1 | RM499–RM428 | Swarna | 10.7 | 25 | 14.80 | 5.9 | 35 | 13.48 |
| mp1.2 | 1 | RM24–RM594 | nivara | 4.57 | 5 | −0.69 | mp1.2 | 1 | RM81–RM595 | nivara | 8.4 | 4 | −0.83 | ||||||
| mp6.1 | 6 | RM314–RM3 | Swarna | 5.4 | 21 | 0.79 | mp1.3 | 1 | RM488–RM126 | Swarna | 11.8 | 24 | 0.41 | ||||||
| mp7.1 | 7 | RM172–RM248 | Swarna | 7.02 | 6 | 2.05 | mp2.1 | 2 | RM423–RM438 | Swarna | 9.6 | 5 | 0.23 | ||||||
| mp9.1 | 9 | RM288–RM107 | Swarna | 2.77 | 5 | 3.78 | mp3.1 | 3 | RM7–RM16 | Swarna | 12.1 | 8 | −0.58 | ||||||
| mp11.1 | 11 | RM209–RM21 | Swarna | 8 | 7 | 2.82 | mp4.1 | 4 | RM261–RM241 | Swarna | 12.5 | 22 | 3.48 | ||||||
| mp12.1 | 12 | RM415–RM19 | Swarna | 9.3 | 10 | 0.41 | mp4.2 | 4 | RM241–RM567 | Swarna | 8.0 | 5 | 10.45 | ||||||
| mp5.1 | 5 | RM430–RM574 | Swarna | 9.5 | 17 | 0.98 | 5.68 | 6 | 2.47 | ||||||||||
| mp5.2 | 5 | RM574–RM249 | Swarna | 3.5 | 5 | 0.85 | |||||||||||||
| mp5.3 | 5 | RM249–RM26 | Swarna | 7.4 | 12 | 2.30 | |||||||||||||
| mp8.1 | 8 | RM337–RM38 | Swarna | 5.6 | 6 | 2.34 | |||||||||||||
| mp12.1 | 12 | RM247–RM519 | Swarna | 8.7 | 10 | 13.15 | |||||||||||||
| Kernel length (mm) | |||||||||||||||||||
| kl1.1 | 1 | RM499–RM84 | Swarna | 3.3 | 6 | 0.1 | |||||||||||||
| Kernel width (mm) | Kernel width (mm) | ||||||||||||||||||
| kw3.1 | 3 | RM135–RM168 | nivara | 2.8 | 17 | −0.06 | 2.86 | 16 | −0.06 | kw1.1 | 1 | RM499–RM428 | nivara | 3.3 | 30 | −0.36 | |||
| kw6.1 | 6 | RM190–RM314 | nivara | 2.85 | 9 | −0.04 | kw1.2 | 1 | RM81–RM9 | nivara | 5.2 | 13 | −0.27 | 4.07 | 13 | −0.12 | |||
| kw1.3 | 1 | RM488–RM126 | nivara | 4.0 | 30 | −0.17 | 3.32 | 11 | −0.03 | ||||||||||
| kw1.4 | 1 | RM226–RM431 | nivara | 4.4 | 12 | −0.14 | |||||||||||||
| kw4.1 | 4 | RM185–RM241 | nivara | 2.9 | 12 | −0.25 | |||||||||||||
| kw5.1 | 5 | RM249–RM26 | nivara | 4.2 | 9 | −0.09 | |||||||||||||
| kw6.1 | 6 | RM204–RM3 | nivara | 3.2 | 12 | −0.10 | |||||||||||||
| kw8.1 | 8 | RM152–RM38 | nivara | 4.2 | 13 | −0.10 | |||||||||||||
| L\B ratio | L\B ratio | ||||||||||||||||||
| lwr12.1 | 12 | RM415–RM19 | nivara | 2.56 | 9 | −0.05 | lwr2.1 | 2 | RM174–M324 | nivara | 3.0 | 10 | −0.03 | 2.7 | 7 | −0.03 | |||
| Water uptake | lwr6.1 | 6 | RM584–RM217 | nivara | 3.1 | 8 | −0.03 | 4.23 | 6 | −0.02 | |||||||||
| wup2.1 | 2 | RM250–RM535 | nivara | 3.3 | 24 | −28.15 | Volume expansion ratio | ||||||||||||
| wup3.1 | 3 | RM282–RM55 | Swarna | 2.6 | 15 | 38.79 | ver1.1 | 1 | RM488–RM128 | Swarna | 3.9 | 8 | 0.02 | ||||||
| ver1.2 | 1 | RM226–RM431 | Swarna | 2.8 | 9 | 0.04 | |||||||||||||
| ver5.1 | 5 | RM430–RM26 | Swarna | 4.2 | 5 | 0.05 | |||||||||||||
| Kernel length after cooking (mm) | Kernel length after cooking (mm) | ||||||||||||||||||
| klac5.1 | 5 | RM26–RM31 | nivara | 2.5 | 12 | −1.06 | klac5.1 | 5 | RM153–RM13 | Swarna | 4 | 12 | 20.96 | 5.41 | 15 | 13.71 | |||
| klac12.1 | 12 | RM19–RM453 | nivara | 3.4 | 15 | −21.32 | klac8.1 | 8 | RM256–RM264 | Swarna | 2.49 | 7 | 13.09 | ||||||
| klac12.1 | 12 | RM415–RM19 | nivara | 2.5 | 12 | −7.4 | |||||||||||||
| Elongation ratio | Alkali spreading value | ||||||||||||||||||
| er3.1 | 3 | RM55–RM520 | nivara | 2.7 | 21 | −0.68 | asv11.1 | 11 | RM209–RM21 | nivara | 2.6 | 30 | −0.3 | ||||||
| Amylose content (%) | Amylose content (%) | ||||||||||||||||||
| ac2.1 | 2 | RM262–RM3515 | nivara | 3.4 | 11 | −2.19 | ac1.1 | 1 | RM243–RM582 | Swarna | 2.63 | 5 | 0.27 | ||||||
| ac3.1 | 3 | RM22–RM7 | Swarna | 2.51 | 5 | 0.87 | ac5.1 | 5 | RM413–RM13 | nivara | 2.6 | 9 | −0.34 | ||||||
| ac3.2 | 3 | RM85–RM293 | Swarna | 2.6 | 15 | 2.05 | ac5.2 | 5 | RM153–RM413 | Swarna | 3.54 | 9 | 0.26 | ||||||
| ac6.1 | 6 | RM314–RM3 | Swarna | 2.6 | 23 | 0.86 | |||||||||||||
| Gel consistency (mm) | Gel consistency (mm) | ||||||||||||||||||
| gc2.1 | 2 | RM250–RM535 | Swarna | 3.18 | 14 | 10.1 | gc1.1 | 1 | RM499–RM428 | Swarna | 3.9 | 30 | 5.67 | 4.21 | 29 | 5.49 | |||
| gc1.2 | 1 | RM580–RM81 | Swarna | 3.2 | 21 | 6.45 | |||||||||||||
| gc2.1 | 2 | RM475–RM250 | Swarna | 3.7 | 4 | 0.61 | |||||||||||||
| gc4.1 | 4 | RM185–RM241 | Swarna | 4.2 | 12 | 0.89 | 3.08 | 13 | 4.67 | ||||||||||
| gc11.1 | 11 | RM209–RM21 | Swarna | 2.9 | 30 | 4.25 | 3.36 | 8 | 0.53 | ||||||||||
| Trait | Chromosome | Marker interval | Allelic effect | IM | CIM | Trait | Chromosome | Marker interval | Allelic effect | IM | CIM | ||||||||
| LOD | R2 | Additive effect | LOD | R2 | Additive effect | LOD | R2 | Additive effect | LOD | R2 | Additive effect | ||||||||
| Population 1 | Population 2 | ||||||||||||||||||
| Milling percent (%) | Milling percent (%) | ||||||||||||||||||
| mp1.1 | 1 | RM490–RM243 | Swarna | 6.1 | 6 | 5.34 | 4.54 | 8 | 4.02 | mp1.1 | 1 | RM499–RM428 | Swarna | 10.7 | 25 | 14.80 | 5.9 | 35 | 13.48 |
| mp1.2 | 1 | RM24–RM594 | nivara | 4.57 | 5 | −0.69 | mp1.2 | 1 | RM81–RM595 | nivara | 8.4 | 4 | −0.83 | ||||||
| mp6.1 | 6 | RM314–RM3 | Swarna | 5.4 | 21 | 0.79 | mp1.3 | 1 | RM488–RM126 | Swarna | 11.8 | 24 | 0.41 | ||||||
| mp7.1 | 7 | RM172–RM248 | Swarna | 7.02 | 6 | 2.05 | mp2.1 | 2 | RM423–RM438 | Swarna | 9.6 | 5 | 0.23 | ||||||
| mp9.1 | 9 | RM288–RM107 | Swarna | 2.77 | 5 | 3.78 | mp3.1 | 3 | RM7–RM16 | Swarna | 12.1 | 8 | −0.58 | ||||||
| mp11.1 | 11 | RM209–RM21 | Swarna | 8 | 7 | 2.82 | mp4.1 | 4 | RM261–RM241 | Swarna | 12.5 | 22 | 3.48 | ||||||
| mp12.1 | 12 | RM415–RM19 | Swarna | 9.3 | 10 | 0.41 | mp4.2 | 4 | RM241–RM567 | Swarna | 8.0 | 5 | 10.45 | ||||||
| mp5.1 | 5 | RM430–RM574 | Swarna | 9.5 | 17 | 0.98 | 5.68 | 6 | 2.47 | ||||||||||
| mp5.2 | 5 | RM574–RM249 | Swarna | 3.5 | 5 | 0.85 | |||||||||||||
| mp5.3 | 5 | RM249–RM26 | Swarna | 7.4 | 12 | 2.30 | |||||||||||||
| mp8.1 | 8 | RM337–RM38 | Swarna | 5.6 | 6 | 2.34 | |||||||||||||
| mp12.1 | 12 | RM247–RM519 | Swarna | 8.7 | 10 | 13.15 | |||||||||||||
| Kernel length (mm) | |||||||||||||||||||
| kl1.1 | 1 | RM499–RM84 | Swarna | 3.3 | 6 | 0.1 | |||||||||||||
| Kernel width (mm) | Kernel width (mm) | ||||||||||||||||||
| kw3.1 | 3 | RM135–RM168 | nivara | 2.8 | 17 | −0.06 | 2.86 | 16 | −0.06 | kw1.1 | 1 | RM499–RM428 | nivara | 3.3 | 30 | −0.36 | |||
| kw6.1 | 6 | RM190–RM314 | nivara | 2.85 | 9 | −0.04 | kw1.2 | 1 | RM81–RM9 | nivara | 5.2 | 13 | −0.27 | 4.07 | 13 | −0.12 | |||
| kw1.3 | 1 | RM488–RM126 | nivara | 4.0 | 30 | −0.17 | 3.32 | 11 | −0.03 | ||||||||||
| kw1.4 | 1 | RM226–RM431 | nivara | 4.4 | 12 | −0.14 | |||||||||||||
| kw4.1 | 4 | RM185–RM241 | nivara | 2.9 | 12 | −0.25 | |||||||||||||
| kw5.1 | 5 | RM249–RM26 | nivara | 4.2 | 9 | −0.09 | |||||||||||||
| kw6.1 | 6 | RM204–RM3 | nivara | 3.2 | 12 | −0.10 | |||||||||||||
| kw8.1 | 8 | RM152–RM38 | nivara | 4.2 | 13 | −0.10 | |||||||||||||
| L\B ratio | L\B ratio | ||||||||||||||||||
| lwr12.1 | 12 | RM415–RM19 | nivara | 2.56 | 9 | −0.05 | lwr2.1 | 2 | RM174–M324 | nivara | 3.0 | 10 | −0.03 | 2.7 | 7 | −0.03 | |||
| Water uptake | lwr6.1 | 6 | RM584–RM217 | nivara | 3.1 | 8 | −0.03 | 4.23 | 6 | −0.02 | |||||||||
| wup2.1 | 2 | RM250–RM535 | nivara | 3.3 | 24 | −28.15 | Volume expansion ratio | ||||||||||||
| wup3.1 | 3 | RM282–RM55 | Swarna | 2.6 | 15 | 38.79 | ver1.1 | 1 | RM488–RM128 | Swarna | 3.9 | 8 | 0.02 | ||||||
| ver1.2 | 1 | RM226–RM431 | Swarna | 2.8 | 9 | 0.04 | |||||||||||||
| ver5.1 | 5 | RM430–RM26 | Swarna | 4.2 | 5 | 0.05 | |||||||||||||
| Kernel length after cooking (mm) | Kernel length after cooking (mm) | ||||||||||||||||||
| klac5.1 | 5 | RM26–RM31 | nivara | 2.5 | 12 | −1.06 | klac5.1 | 5 | RM153–RM13 | Swarna | 4 | 12 | 20.96 | 5.41 | 15 | 13.71 | |||
| klac12.1 | 12 | RM19–RM453 | nivara | 3.4 | 15 | −21.32 | klac8.1 | 8 | RM256–RM264 | Swarna | 2.49 | 7 | 13.09 | ||||||
| klac12.1 | 12 | RM415–RM19 | nivara | 2.5 | 12 | −7.4 | |||||||||||||
| Elongation ratio | Alkali spreading value | ||||||||||||||||||
| er3.1 | 3 | RM55–RM520 | nivara | 2.7 | 21 | −0.68 | asv11.1 | 11 | RM209–RM21 | nivara | 2.6 | 30 | −0.3 | ||||||
| Amylose content (%) | Amylose content (%) | ||||||||||||||||||
| ac2.1 | 2 | RM262–RM3515 | nivara | 3.4 | 11 | −2.19 | ac1.1 | 1 | RM243–RM582 | Swarna | 2.63 | 5 | 0.27 | ||||||
| ac3.1 | 3 | RM22–RM7 | Swarna | 2.51 | 5 | 0.87 | ac5.1 | 5 | RM413–RM13 | nivara | 2.6 | 9 | −0.34 | ||||||
| ac3.2 | 3 | RM85–RM293 | Swarna | 2.6 | 15 | 2.05 | ac5.2 | 5 | RM153–RM413 | Swarna | 3.54 | 9 | 0.26 | ||||||
| ac6.1 | 6 | RM314–RM3 | Swarna | 2.6 | 23 | 0.86 | |||||||||||||
| Gel consistency (mm) | Gel consistency (mm) | ||||||||||||||||||
| gc2.1 | 2 | RM250–RM535 | Swarna | 3.18 | 14 | 10.1 | gc1.1 | 1 | RM499–RM428 | Swarna | 3.9 | 30 | 5.67 | 4.21 | 29 | 5.49 | |||
| gc1.2 | 1 | RM580–RM81 | Swarna | 3.2 | 21 | 6.45 | |||||||||||||
| gc2.1 | 2 | RM475–RM250 | Swarna | 3.7 | 4 | 0.61 | |||||||||||||
| gc4.1 | 4 | RM185–RM241 | Swarna | 4.2 | 12 | 0.89 | 3.08 | 13 | 4.67 | ||||||||||
| gc11.1 | 11 | RM209–RM21 | Swarna | 2.9 | 30 | 4.25 | 3.36 | 8 | 0.53 | ||||||||||
Location of QTLs for 11 grain quality traits identified in Oryza nivara × Swarna-derived backcross populations. Note: QTLs underlined are from population 2 and QTLs in bold are derived from O. nivara.
Milling Percent
Milling quality is an important trait for rice industry. Milling percent depends on the grain size, shape, and chalkiness. The maximum number QTLs were obtained for milling percent, 7 QTLs in population 1 and 12 QTLs in population 2. The QTL mp1.2 in both the populations mapped to the same region RM24–RM595 and showed the highest additive effect for mp in both populations.
Kernel Length and Width
Kernel length, width, and shape are important for minimizing percentage of broken rice after milling and consumer preference. For kernel length, only 1 QTL was identified on chromosome 1 only in population 1. For kernel width, 2 QTLs were detected on chromosomes 3 and 6 in population 1; both were derived from O. nivara. In population 2, 8 QTLs were detected and all had trait-enhancing allele from O. nivara.
Length and Width Ratio
More than the length and width, it is the ratio between them that appears to be more important specially for milling percent as long and slender grains break more often than short and bold grains during milling. One QTL was identified on chromosome 12; it was derived from O. nivara. Two QTLs were identified in population 2 on chromosomes 2 and 6, and both were from O. nivara.
Volume Expansion Ratio and Water Uptake
Cooking quality of rice is evaluated in terms of characteristics, such as grain elongation, volume expansion, and water absorption during cooking. The genetic analysis of these cooking quality traits is very limited. Generally, less water uptake is preferred. Three QTLs were identified only in population 2, and all were derived from Swarna. For water uptake, 2 QTLs were identified only in population 1 and on chromosomes 2 and 3; one of these QTLs was from O. nivara.
Kernel Length after Cooking
Two QTLs were identified for klac and both were derived from O. nivara, and these were located on chromosomes 5 and 12 in population 1. Three QTLs were identified in population 2, but only 1 of these was derived from O. nivara and was on chromosome 12 (Table 2).
Elongation Ratio
Only 1 QTL was identified for er on chromosome 3 in population 1. It explained 21% phenotypic variance and O. nivara allele at this locus increased the elongation ratio.
Alkali Spreading Value
Alkali spreading value determines the gelatinization temperature (GT), which is a critical temperature range at which the starch granules undergo an irreversible process known as gelatinization on cooking. Only 1 QTL was detected in population 2 and the trait-improving allele from O. nivara explained 30% phenotypic variance.
Amylose Content
Amylose content of rice grain determines the cooking and eating quality of rice to a large extent. Rice with low amylose content becomes sticky after cooking and vise versa. In India, nonsticky rice is generally preferred. Four QTLs were identified on 3 chromosomes; 1 QTL was derived from O. nivara in population 1. Three QTLs were detected in population 2, but only 1 QTL was derived from O. nivara. At the locus ac5.1, the O. nivara allele enhanced ac and at ac5.2, the Swarna allele enhanced ac.
Gel Consistency
Gel consistency is a good index of assessing the texture of cooked rice and also important for a good mouth feel. Soft gel consistency is generally preferred by most rice consumers. One QTL was detected on chromosome 2 in population 1 and 5 QTLs were detected in population 2. All the gc increasing alleles were derived from Swarna.
QTL Interaction
There was only 1 significant digenic interaction detected between 2 QTLs. The interacting loci were RM22–RM517 for ac3.1 on chromosome 3 and RM249–RM459 for mp5.3, kw5.1, and ver5.1 on chromosome 5, but the interaction was detected only in population 1.
Discussion
The advantages of using a wild accession are the appearance of transgressive segregants in the elite genetic backgrounds either due to wild allele introgressions or due to de novo arisen genetic variation. In the present investigation, transgressive segregants were observed for all the grain quality traits in both the O. nivara populations. Transgressive segregants have also been reported previously for quality traits in populations derived from O. glaberrima and O. rufipogon as donor parents (Septiningsih et al. 2003; Aluko et al. 2004; Li et al. 2004; Yuan et al. 2010).
The frequency distribution of all the traits followed a normal distribution except only 2 traits, ver and ac, in our study. A bimodal distribution was reported for ac (He et al. 1999) and normal distribution for ver (Tian et al. 2005). The trait correlations were almost similar in both the populations. The mp was highly positively correlated with hrr, kl, and kw. Significant correlations were observed between kl on one hand and kw, lbr, er, ac, and gc on the other hand in both the populations. A highly negative correlation was found between kw and lbr. In a previous study, mp was highly positively correlated with hrr, grain density, and broken rice in O. rufipogon–derived population (Septiningsih et al. 2003). A highly significant positive correlation between klac and er ratio was obtained in our study and also in a RIL population of Zhenshan 97 and Minghui 63 (Ge et al. 2005). Likewise, the highest negative correlation was reported between kw and lbr in our study and also in a RIL population of Pusa1121 and Pusa1342 (Amarawathi et al. 2008).
The highest number of QTLs was identified for mp in both the populations. As expected for a quality trait, the Swarna-derived QTL alleles explained more phenotypic variance in both populations. QTLs mp6.1 and mp12.1 explained 21% phenotypic variance in population 1. Whereas, mp1.1, mp1.3, and mp4.1, each explained more than 20% variance in population 2. Most of the QTLs reported in our study are new and some of them explain high PV compared with that reported earlier. For kl only 1 QTL was detected on chromosome 1 in population 1, and it was derived from Swarna. For kernel width, trait-enhancing QTLs were identified from O. nivara in both the populations. The QTL kw3.1 explained 17% PV in population 1, kw1.1 and kw1.3 each explained 30% PV in population 2. Amarawathi et al. (2008) mapped QTLs explaining 10.1% and 18.9% PV for grain width on chromosomes 1 and 7. The QTLs on chromosome 1 mapped in our study are also on the long arm but at different location than that reported by Amarawathi et al. (2008). Two major loci lwr12.1 and lwr2.1 were detected for grain length and width. The QTL lwr12.1 located at marker interval RM415–RM19 explained 9% PV in population 1 and lwr2.1 located at marker interval RM174–RM324 explained 10% PV in population 2. The QTL lwr12.1 in our study is close to the lwr12.1 from O. glaberrima reported by Li et al. (2004). It is significant to mention here that many QTLs identified for mp in our study were at the same locations as kl, kw, and lbr. This confirms the observation that mp is largely determined by grain dimensions. A major effect QTL wup2.1 was detected on long arm of chromosome 2 for the trait wup. It explained 24% PV and increased the wup by 28 ml. In comparison, the wup QTL from O. sativa Zhenshan 97 was on short arm of chromosome 2 and explained 6% PV and increased wup by only 0.4 ml (Ge et al. 2005).
Trait-enhancing QTLs from O. nivara were detected for klac at marker intervals RM26–RM31 and RM19–RM453 explaining 12% and 15% PV in population 1 (Figure 1). Three QTLs were detected in population 2 between marker intervals RM153–RM13, RM256–RM264, and RM415–RM19 explaining 15%, 7%, and 12% PV, respectively. In O. glaberrima–derived population, a klac QTL ke3.1 was identified on chromosome 3; it had a phenotypic variance of 6% and an additive effect of 0.24 (Li et al. 2004). In a Zhenshan 97– and Delong 208–derived population, QTLs were identified for klac on chromosomes 3, 6, 7, and 8. He et al. (1999) identified QTLs on chromosomes 3 and 6, each one of these explained 28% PV. In Zhenshan 97 and Minghui 63 population, QTLs were detected for width expansion on chromosomes 1, 2, 3, 6, 9, and 11. The QTLs on chromosomes 3 and 6 showed highest PV of 15% and 12%, respectively (Ge et al. 2005).
A significant QTL er3.1 derived from O. nivara was identified for er on chromosome 3. It explained 21% PV and had an additive effect of 0.68. It could be a candidate for use in MAS to increase er. Earlier, a QTL was reported for er on chromosome 11, it showed a phenotypic variance of 6% and an additive effect of 0.45 (Amarawathi et al. 2008).
For asv, 1 major effect QTL with 30% phenotypic variance was mapped at marker interval RM202–RM209 on chromosome 11. This is a new QTL. Earlier, QTLs for asv were reported on chromosome 6 explaining 87% PV in a population derived from Balilla/Nantehao cross (Yan et al. 2001) and 44% PV in Caiapo cultivar background (Aluko et al. 2004) and to a smaller region within that QTL explaining 7% PV in a RIL population (Amarawathi et al. 2008).
Amylose content of rice grain determines the cooking and eating quality of rice to a large extent (Lin et al. 2011). Amylose content is known to be regulated by waxy gene for granule bound starch synthase on chromosome 6. In our study, the QTL on chromosome 6 was derived from O. nivara and explained 23% of phenotypic variance in population 1. Karim et al. (2011) mapped apparent amylose content to waxy region. Ge et al. (2005) also reported a region near waxy locus controlling grain length, width, weight, elongation, width expansion, and water absorption. In the present study, QTLs for ac, mp, and kw were colocated in the waxy region in population 1. In most of the previous QTL mapping studies on cultivated rice, ac was mapped at the waxy locus. However, QTLs for ac were not detected in wx region in our study in population 2 and also in 2 previous studies using O rufipogon (Yuan et al. 2010) and Iranian traditional cultivar (Sabouri 2009).
In the present investigation, QTL alleles from Swarna enhanced gc. One QTL gc2.1 explained 14% phenotypic variance in the population 1, 3 QTLs, gc1.1, gc1.2, and gc11.1, each explained more than 20% PV in population 2. He et al. (1999) identified 2 QTLs for gc on chromosomes 2 (near centromere) and 7. Li et al. (2003) identified QTLs for gc on chromosomes 1, 2, and 6 from O. sativa cross. Li et al. (2004) identified QTLs for gc from O. glaberrima on chromosomes 2 and 7. Thus, it appears that gc-enhancing alleles are selected for in evolution in the cultivated rice species O. sativa and O. glaberrima and that O. nivara has alleles for lower gc than Swarna.
It is significant to note that colocated QTLs were highly correlated. For example, QTLs for kl and kw; klac, wup, lbr, and asv spreading value were colocated and also these traits were significantly correlated.
A comparison was made between QTLs identified for quality traits in our study with previous report of O. nivara (accession IRGC80470)–derived QTLs for shattering and dormancy, 2 domestication related traits. Many QTLs were coincident. QTL er3.1 was located near QTL sh3 between the marker RM303 and RM293. Likewise, mp4.1 and mp4.2 were found in the region sh4 between RM303 and RM280. The QTL for kernel length, kl1.1 was found within the dormancy QTL flanked by RM495 and RM283 on chromosome 1 and klac12.1 was found within the dormancy QTL sd12 flanked by RM19 and RM453 (Li et al. 2006). This indicates that these new QTLs mapped in our study may have been left behind during domestication as they were linked to the undesirable QTLs for shattering and dormancy. More such novel QTLs may be revealed, if different wild accessions from different ecogeographical regions are used in backcrossing programs to increase yield and quality.
A comparison was also made between the location of QTLs for yield and those for quality traits obtained from the same 2 populations. Two major effect yield QTLs from population 1 and 5 major effect QTLs from population 2 were considered for this comparison (Kaladhar et al. 2008; Swamy et al. 2011). It was interesting to observe that QTLs for 4 quality traits were associated with 5 of the 7 major yield QTLs analyzed. The QTL for water uptake, wup2.1 colocated with yld2.3 in population 1. The mp3.1 was located with yld3.1, mp3.3 was near yld3.2, kw8.1 and ver8.1 near yld8.1, and ver8.1 near yld8.2 in population 2. The 18 QTLs reported in this study, which map to previously reported QTLs for the same trait or for meta-QTLs for yield are presented in Supplementary Table 2 (Swamy and Sarla 2008, 2011). This information is useful in breeding for high yielding varieties with different combination of quality traits.
This study clearly demonstrates that O. nivara has useful alleles for improving quality traits, for example, mp and klac, even in an elite genetic background such as Swarna. QTLs mapped for quality traits coincided in the 2 populations and were colocated with QTLs identified for yield and related traits in the same 2 populations. The 7 major effect yield–enhancing QTLs from O. nivara in the 2 populations can possibly be introgressed individually or pooled selectively without compromising grain quality in Swarna. Introgression lines with improved seed quality include the fine grain lines RPBio 4918-14, RPBio 4918-148, and RPBio 4918-166 and their derivatives 14-3, 166-2, 166-9, 175-1, 175-2, RPBio4918-175, and 166-30.
Funding
The project was supported by funds from Department of Biotechnology, Ministry of Science and Technology, Government of India to N.S. as part of the Network Project on Functional Genomics of Rice (BT/AB/03/FG/DRR-II/01/2004-05).
We thank UGC CSIR for fellowship to B.P.M.S. and Venkaiah for help in grain quality analysis.
References
Author notes
Corresponding Editor: Perry Gustafson