-
PDF
- Split View
-
Views
-
Cite
Cite
Tyler A Brown, Nobuo Tsurusaki, Mercedes Burns, Genomic Determination of Reproductive Mode in Facultatively Parthenogenetic Opiliones, Journal of Heredity, Volume 112, Issue 1, January 2021, Pages 34–44, https://doi.org/10.1093/jhered/esaa045
Close - Share Icon Share
Abstract
Sexual reproduction may pose myriad short-term costs to females. Despite these costs, sexual reproduction is near ubiquitous. Facultative parthenogenesis is theorized to mitigate some of the costs of sex, as individuals can participate in occasional sex to limit costs while obtaining many benefits. However, most theoretical models assume sexual reproduction is fixed following mating, with no possibility of clutches of mixed reproductive ontogeny. Therefore, we asked: if coercive males are present at high frequency in a population of facultative parthenogens, will their clutches be solely sexually produced, or will there be evidence of sexually and asexually-produced offspring? How will their offspring production compare to conspecifics in low-frequency male populations? We addressed our questions by collecting females and egg clutches of the facultatively parthenogenetic Opiliones species Leiobunum manubriatum and L. globosum. In L. manubriatum, females from populations with few males were not significantly more fecund than females from populations with higher male relative frequency, despite the potential release of the former from sexual conflict. We used 3 genotyping methods along with a custom set of DNA capture probes to reveal that offspring of L. manubriatum from these high male populations were primarily produced via asexual reproduction. This is surprising because sex ratios in these southern populations approach equality, increasing the probability for females to encounter mates and produce offspring sexually. We additionally found evidence for reproductive polymorphisms within populations. Rapid and accurate SNP genotyping data will continue to allow us to address broader evolutionary questions regarding the role of facultative reproductive modes in the maintenance of sex.
Facultative parthenogenesis is the ability of a female to produce offspring via either sexual or asexual reproduction. The relative rarity (Booth et al. 2012; Burke and Bonduriansky 2017, 2019a) of this mode fascinates evolutionary biologists, because facultative parthenogens theoretically reap the benefits of the combined reproductive strategy (Burke and Bonduriansky 2017) while avoiding a number of known costs. Costs of sexual reproduction are typically borne by the female through her reproductive life, and include the fitness losses associated by undergoing meiosis (Mirzaghaderi and Hörandl 2016), producing sons (Gibson et al. 2017), and the direct harm that may be caused to her via mating (Parker 2006; Boulton and Shuker 2015). Costs of asexual reproduction, such as parthenogenesis, are typically assigned to the species or lineage, and include inbreeding depression (Cáceres et al. 2009) and associated susceptibility to disease or parasitism (Jokela et al. 2009).
Facultatively parthenogenetic species may avoid many of these costs by utilizing both reproductive modes during their lives (Burke and Bonduriansky 2019a). However, the rarity of species that display facultative parthenogenesis has likely led to the paucity of research on their life history, fecundity, and behaviors (Burke and Bonduriansky 2017; Engelstädter 2017). This is distinguished from the comparatively large number and variety of organisms that show partial clonality (i.e., temporally-, environmentally- or life stage-partitioned sexual and vegetative reproduction; Vallejo-Marín et al 2010; Neiman et al 2014; Avise 2015; Van der Kooi et al. 2017). In particular, we do not know whether these species may have single clutches that include offspring produced via parthenogenetic and sexual modes, or the mechanisms that govern reproductive mode switching. We have begun to pursue these open questions by focusing on a group of opilionids (also called “harvestmen” or “daddy-longlegs”) from class Arachnida, which include obligate sexual species and 2 that reproduce through facultative parthenogenesis. Importantly, these species are uniquely distributed in such a way that their populations form a natural laboratory to investigate reproductive mode and behavior in facultatively parthenogenetic species.
Opiliones of Japan
Japan is host to a number of endemic opilionids, including several members of the curvipalpe group of Eupnoi (or “daddy longlegs”-type) Opiliones (Tsurusaki 1985; Hedin et al. 2012). Two species of this group were characterized as facultative parthenogens via penultimate collection and egg rearing, despite the presence of males and gynandromorphs in many populations (Tsurusaki 1986). These species, Leiobunum manubriatum (Figure 1) and L. globosum, are annual species found throughout Gifu, Toyama, and Nagano Prefectures of Honshu Island on north to Hokkaido islands. Although past genomic efforts have found no population substructure consistent with obligatory reproductive modes (Burns et al. 2017), L. manubriatum females are known to have both 2n and 4n cytotypes, while L. globosum appears to be entirely tetraploid (Tsurusaki 1986; Burns et al. 2017). Populations of both species are particularly female-biased at higher latitudes (above 1400 meters) and in northerly locales (Burns et al. 2017), a distributional pattern termed geographic parthenogenesis (Vandel 1928).
Mating pair of Leiobunum manubriatum (female lower left with dark opisthosomal stripe, male upper right with pointed opisthosoma) on mossy rock in Toyama Prefecture. Photograph courtesy of Dr. Sarah Stellwagen.
Recently, a number of works have sought to identify the mechanisms that drive geographical parthenogenesis, alternatively pointing to local adaptation of parthenogens (Kearney 2005), the lack of requirement for mates to achieve reproductive assurance (Tilquin and Kokko 2016), and sexual conflict between males and facultatively parthenogenetic females (Gerber and Kokko 2016: Burke and Bonduriansky 2019b) as causal forces. The latter mechanism is particularly unique to facultative systems, as it assumes males of a species could operate as an “invading” cytotype to female-biased populations and achieve successful matings if equipped for coercive/conflict-based mating or occurring in densities to ensure reproductive interactions will proceed (Kawatsu 2013). Theoretical models specific to the development of geographic parthenogenesis in facultatively parthenogenetic systems typically assume that once females are mated, all offspring will be produced sexually for the rest of the life of the dam (Gerber and Kokko 2016; Burke and Bonduriansky 2019b). However, this assumption may not be realistic given the model system’s functional reproductive morphology and cell biology. Many identified facultatively parthenogenetic animal systems are arthropods (van der Kooi et al. 2017), which often have conflict-induced armaments (Haley and Gray 2011; Perry and Rowe 2012; Burns and Shultz 2015; Hollis et al. 2019) or cryptic female choice responses (Córdoba-Aguilar 2006; Lüpold et al. 2013; Firman et al. 2017), suggesting females of these facultatively parthenogenetic species may be able to exert post-copulatory choice over sperm fate.
To begin to test mechanisms for the production of geographic parthenogenesis in facultatively parthenogenetic species, we must first ask 2 simple questions: do facultatively parthenogenetic females mate? And if so, where? To answer these questions, we collected females of the facultatively parthenogenetic opilionids during their highest activity months (Tsurusaki 1986) from populations that vary in male density. Females and all eggs oviposited by them were genotyped via single nucleotide polymorphisms, or SNPs, using 3 methods to determine rate of sexual versus asexual reproduction: 1) clone probability, 2) homozygous to heterozygous locus transitions (T), and 3) the number of sires inferred via maternal and offspring genotypes.
Without data on mating rate for the curvipalpe group, we recognized early the importance of collecting and genotyping embryos of not only the asexual species, but their closely related obligate sexuals as well. Data gathered from clutches of 2 sexual species, L. tohokuense and L. curvipalpe, was used to formulate comparisons to the facultatively parthenogenetic species to identify differences in fecundity and genotypic evidence of sexual reproduction typical to closely related species. We posited that sexual reproduction would be marked by a high rate of T, low or zero probability of clone identity (decreasing with number of loci typed), and an increased number of sires contributing to clutches as compared with parthenogens. Because southern populations of L. manubriatum have less female-biased sex ratios compared to northern populations, we additionally expected to see genotypic evidence for a higher rate of sexual reproduction in clutches of southern females versus their northern counterparts.
After developing 3 methods for genotyping our samples (MassArray, Genotyping-by-Sequencing, and 3RAD-Capture), we found that although inferred sire number was not significantly different between obligately sexual and facultatively parthenogenetic taxa using higher throughput sequencing methods, mean clone probability was higher for parthenogens than for obligate sexuals, and southern parthenogens had higher numbers of inferred sires than northern clutches. Following these results, we found that northern L. manubriatum clutches had significantly higher T and clone probability than southern L. manubriatum clutches. Together, these results indicate that while parthenogens from populations with higher male frequency do reproduce sexually, the outcrossing rate is still significantly lower than is expected from obligately sexual taxa. Ongoing use of these genotyping methods will be critical for understanding the fitness consequences associated with facultative asexuality, and the potential of sexual conflict to maintain sexual reproduction in these systems.
Methods
Specimen Collection
A total of 200 females of the facultatively parthenogenetic species L. manubriatum (N = 115) and L. globosum (N = 53) and the obligately sexual species L. curvipalpe (N = 18) and L. tohokuense (N = 14) were collected during late June and July in 2015 and 2016. Females were maintained indoors in single terraria with food and water provided ad libitum, and terraria were moistened with sprayed deionized water approximately twice a week. Oviposition substrates, including commercial cricket rearing soil, vermiculite, Kanuma pumice, and bark, were maintained in each terrarium and monitored weekly for appearance of egg clutches. No eggs were ever found in vermiculite or Kanuma pumice substrates. Following senescence, females were preserved in 99% ethanol and stored at −20 °C until DNA extraction.
The presence of eggs was assessed following maternal senescence. A total of 75 females (57 L. manubriatum, including 36 southern and 21 northern females, 9 L. globosum, 8, L. tohokuense, and 1 L. curvipalpe) produced egg clutches. When eggs were found, they were backfilled in soil and maintained at room temperature and twice weekly sprayed with deionized water until December of the collection year, when they were expected to reach their last embryonic stage (Tsurusaki 1986). All eggs were recovered from substrate and fixed in 99% ethanol along with their dam in between December 3rd and 5th of the collection year. Extraction of DNA from embryos was carried out with either the Arcturus PicoPure (Applied Biosystems) or Instagene Chelex Resin Matrix (Bio-Rad; 100–150 uL, with 10 uL of Proteinase K from ThermoFisher Scientific). Extraction of DNA from dams was carried out by excising muscle from 2 to 4 coxae and processing with the Qiagen DNeasy Blood and Tissue Kit.
Array Genotyping Approach
We used one lane of Illumina HiSeq 2500 100bp single-end reads from the Asamushi Forest Park population of L. tohokuense (N = 3) and the southern Mt. Harinoki population of L. manubriatum (N = 6) collected as part of a previous work (Burns et al. 2017) to identify a set of high minor allele frequency (MAF > 0.4) single nucleotide polymorphisms after multiple alignment with PyRAD (Eaton 2014). Because Agena MassArray SNP genotyping requires ~30 base-pairs of flanking sequence, only biallelic SNPs that occurred in the internal 40 bases of this set of Illumina 100bp SE reads could be used. Thirty such sequences for L. manubriatum (8 dams and offspring) and L. tohokuense (8 dams and offspring) collected in 2015 were provided to the University of Arizona Genomics Core (Tucson, AZ).
Genotyping-By-Sequencing Approach
In order to confirm the results of the MassArray genotyping, we also genotyped samples collected in 2015 via 3-enzyme restriction associated DNA sequencing (3RAD; Bayona-Vásquez et al. 2019), using oligonucleotides adapted from Graham et al. (2015). Prior to genome digestion, DNA extractions were quantified using a Qubit 3.0 fluorometer (Thermo Fisher Scientific) and adjusted to ~100ng of genomic DNA in 40 μl of nuclease-free water. Standardized genomic DNA from all L. tohokuense and L. manubriatum dams and embryonic offspring were digested with 100 units each of the enzymes EcoRI-HF (5′-GAATTC-3′), ClaI (5′-ATCGAT-3′), and MspI (5′-CCGG-3′) (New England Biolabs) and pre-annealed, double-stranded EcoRI and ClaI adapters were ligated to the digested genomic DNA using 100 units of T4 Ligase. Ligation was achieved by incubating samples for 2 cycles at 22 °C for 20 min and 37 °C for 10 min, followed by 80 °C for 20 min to stop enzyme activity. Following filtration with Agencourt Ampure XP magnetic beads (Beckman Coulter) to remove deactivated enzymes and unincorporated adaptors, customized row-specific i5 primers and 96 well plate and column-specific i7 primers were added to 6 μL of genomic fragments in a short run PCR (20 μl reaction volume, 95 °C for 2 min, 14 cycles of 98 °C for 20 s, 60 °C for 15 s, 72 °C for 30 s, and a final 72° C for 5 min) with Phusion PCR Kit Master Mix (New England BioLabs). After a second Ampure XP bead clean-up to remove unincorporated primers, all samples were quantified via Qubit 3.0 using the high-sensitivity DNA protocol (Thermo-Fisher). Any samples with concentrations less than 1 ng/uL of DNA detected were amplified with an additional 8 cycles of PCR.
Sample libraries were standardized and combined by 96-well plate row and fragments of 400–600 bp were separated using the BluePippin automated size selection instrument (Sage Sciences). Following a second quantification by Qubit 3.0, nanomolarity of each pooled row was estimated by dividing concentration by average fragment length (500 bp) multiplied by the approximate average molecular weight per base pair of DNA (660 Daltons), and rows were combined into one equimolar genomic library. The library was sequenced by the North Carolina State University Genomics Core (Raleigh, NC) using the Illumina NextSeq platform, producing 75bp paired-end fragments.
3RApture Approach
Egg clutches from 49 female L. manubriatum (28 southern, 21 northern) and 9 female L. globosum, collected from 5 localities during late June and July of 2016, were extracted and sequenced in 2018. The L. globosum specimens were sequenced and genotyped using the GBS methods described previously.
To genotype the 2016 L. manubriatum, we used a customized “3RApture” protocol consisting of 3RADseq adapted from Bayona-Vásquez et al. (2019) followed by a bait capture protocol (Ali et al. 2016; Arbor Biosciences). This procedure utilized all steps described in the previous section on GBS, with the exception of using MultiScreen PCRμl96 Filter Plates (Millipore Sigma) to filter PCR products combined with 80 μl of 1X TE buffer. These filtered products were subsequently re-suspended in 25 μl of nuclease-free water. PCR products were then quantified and combined by rows in equimolar concentrations for size-selection, as for GBS.
Using consensus sequences from the L. manubriatum samples collected in 2015, we developed a custom set of 20,000 DNA probes (Arbor Biosciences). Blocking sequences, used to prevent binding of non-target library fragments, were developed using the mitochondrial genome of the opilionid Phalangium opilio (NCBI Reference Sequence: NC_010766.1; Masta and Boore 2008). Currently, there are no publicly available nuclear reference genomes for Opiliones, so nuclear blocking oligonucleotides were derived from the scorpion species Mesobuthus martensii (GenBank BioProject PRJNA171479; Cao et al. 2013) and Centruroides sculpturatus (GenBank BioProject PRJNA422877), the most closely related (Lozano-Fernandez et al. 2019) arachnid genomes available.
Probes were hybridized with 7 μL of the combined rows of 2016 L. manubriatum 3RAD libraries following Arbor Biosciences protocols and washed with streptavidin-coated magnetic beads to remove off-target and poorly hybridized fragments. Fifteen microliter of washed hybridization supernatant were PCR-amplified using Phusion Master Mix and an equimolar combination of primers after a 5-min incubation of the bead mixture at 95 °C to release target fragments into suspension. Because our libraries averaged approximately 500bp, we used the minimal cycle number of 10 to amplify captured fragments, with the thermal program recommended by Arbor Biosciences. Libraries were lastly bead cleaned, quantified, and pooled using the methods described for the GBS approach, and sequenced as 150 bp paired-end fragments on the Illumina NovaSeq platform at the NC State Genomics Core facility.
Data Analysis
We collected data on embryo development and total eggs produced by all dams, as well as the number of days of captivity for each female in order to verify our husbandry conditions were appropriate. These data were inputted into GraphPad Prism in order to identify any fecundity differences between species.
Sequence data was demultiplexed using the Stacks v. 2.1 (Catchen et al. 2013) de novo pipeline after trimming primers and adapters, with a required stack depth of 3, and maximum possible heterozygosity of 3 sites per fragment. We followed default settings for all additional parameters with the exception that for L. manubriatum genotyped with GBS and 3RApture, we included only dams in the locus catalog due to the large size of the clutches. Output was collected as variant call formatted (*.vcf) files, where all loci needed to be shared by at least 25% of clutches (L. globosum, L. manubriatum) or 75% of clutches (L. tohokuense).
Using regular expressions, we identified all homozygous loci in dams, and calculated the rate of genotypic transition T for each clutch as:
where x is the number of s sites that are heterozygous in o offspring but homozygous in the dam. Because these transitions are not possible when offspring are produced asexually, we calculated T for both obligately sexual and facultatively parthenogenetic dams to compare estimates under obligate and facultatively sexual reproductive modes.
To estimate the number of sires contributing to each clutch and the clone probability of all offspring from facultatively parthenogenetic species, we used Colony v. 2.0.6.5 (Jones and Wang 2010), a Bayesian Likelihood-based application that inputs dominant or codominant variants to identify full and half-sibships. To convert our files to the appropriate format, we used VCF File Converter v. 2.2 (DeWaters 2019; github.com/dandewaters/VCF-File-Converter). In our study, because all dams are known, half-sibships within clutches are indicative of multiple sires. We estimated clone probability and sire number under 3 paternal sibship priors: 1) paternal sibship = maternal sibship (calculated as the average clutch size for the species), 2) paternal = ½ maternal sibship, and 3) paternal sibships = 5. Rate of mistyping was set at 0.5% (Wall et al. 2014).
We additionally performed a discriminant analysis of principal components (DAPC) using the genotypes of the obligate sexual and facultatively parthenogenetic species, following the procedures in R package adegenet (Jombart 2008; Jombart and Ahmed 2011). Genotypes for each dam and associated offspring were applied to the “find clusters” function, allowing for an upper limit of clusters equal to ½ the total number of individuals in each family. The best-fitting cluster model was selected via choice of the lowest Bayesian Information Criterion (BIC), and the number of PCAs retained for each family-level discriminatory analysis was selected via inflection point of PCA scree plot. We retained all discriminant functions and measured the total number of offspring clusters (corresponding to clutch sires) and the proportion of offspring clusters outside of the maternal genotype cluster (corresponding to offspring likely produced sexually). Using this method, we expected to see no offspring outside of the maternal cluster if these offspring were asexually produced, and a number of clusters correlated with the number of sires inferred using the Colony approach if the offspring were produced sexually.
Results
Clutch Comparisons
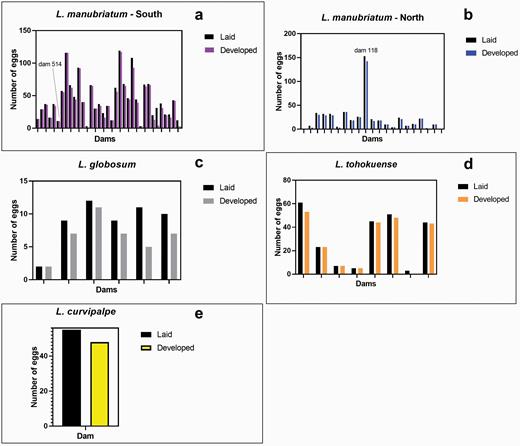
Between the years of 2015 and 2016, we collected a total of 128 L. manubriatum (Southern populations: 60, Northern populations: 68), 53 L. globosum, 14 L. tohokuense, and 18 L. curvipalpe females. Of these females, 70 L. manubriatum (Southern: 36, Northern: 34), 9 L. globosum, 8 L. tohokuense, and 1 L. curvipalpe oviposited. Collectively this amounted to 1567 L. manubriatum (Southern: 1015, Northern: 552), 66 L. globosum, 255 L. tohokuense, and 62 L. curvipalpe eggs attained (Figure 2). All species produced some eggs that did not develop (Figure 2, black bars), though we did not find species-level differences in proportion of developed embryos per clutch (F(3, 53) = 0.8005, P = 0.4991). On average, about 12.5% had no visible embryos and these eggs were subsequently not included in genotyping (Figure 2). If dams produced no eggs with visible embryos, they were also not included in genotyping. Because only one L. curvipalpe specimen produced offspring, we chose not to pursue genotyping of this clutch, and here-on present only data related to husbandry of L. curvipalpe specimens.
Average number of eggs laid (colored) and developed (black) embryos per dam, per species. Leiobunum manubriatum (a: southern N = 36; b: northern N = 21), and (c) L. globosum (N = 6) are facultatively parthenogenetic, while (d) L. tohokuense (N = 8) and (e) L. curvipalpe (N = 1) are obligate sexuals. A one-way ANOVA of species groups reveals a P-value of 0.0130. While L. globosum had significantly lower fecundity than southern L. manubriatum (P = 0.01), we did not find a significant difference in fecundity or fertility between parthenogens and obligate sexual taxa (P = 0.8576). Females of all 4 species produced either no eggs or solely eggs that did not develop embryos (L. manubriatum South N = 24; L. manubriatum North N = 34; L. globosum N = 47; L. tohokuense N = 6; L. curvipalpe N = 17). Females that did not produce offspring did not differ significantly between (F = 2.398; P = 0.054) or within species (F = 0.5409, P = 0.7060) in the days maintained in the laboratory at Tottori University.
Genotyping Methods
Arrays of 25 SNP-containing sequences for L. tohokuense and 27 sequences for L. manubriatum (all southern populations) were designed based on short reads in Burns et al. (2017). All 2015 specimens were subsequently genotyped with their respective panels during the summer of 2017. We acquired unambiguous genotypes (i.e., biallelic and with measurable fluorescence) from 5 of the 8 L. tohokuense dams and 149 offspring, and 8 L. manubriatum dams with 102 offspring.
Genotyping-by-sequencing for 2015 specimens resulted in an average of ~1.4 × 106 paired-end reads, for all 7 L. tohokuense dams and 202 offspring, and all 8 L. manubriatum dams with 122 offspring. This sequencing approach was also used for L. globosum (collected in 2016) in a separate Illumina NextSeq run. This produced an average of ~4.5 × 106 paired-end reads for 5 dams and 32 eggs after quality checking. We outputted 233 SNP loci for L. manubriatum, 614 loci for L. tohokuense, and 518 for L. globosum.
The 3RApture technique was used to sequence SNPs only from L. manubriatum collected from 3 northern and an additional 2 southern populations. These females (N = 35) produced a collective 1071 eggs, and we sequenced an average of 3.2 × 106 paired-end reads for each individual from the data set. Due to the size of this data set, we processed families separately, resulting in an average of 670 loci for subsequent analysis. In comparison to a subset of specimens sequenced with and without bait capture, we found that the 3RApture approach led to a 3.6-fold average increase in sequencing efficiency for genotyping loci. After assessing observed heterozygosity of each species and high-throughput sequencing approach, we found significantly higher heterozygosity in the facultatively parthenogenetic species L. manubriatum as compared to the obligate sexual L. tohokuense (P = 0.0383). This effect was not seen in the MassArray data (P = 0.21), nor between L. manubriatum from northern versus southern populations (P = 0.71).
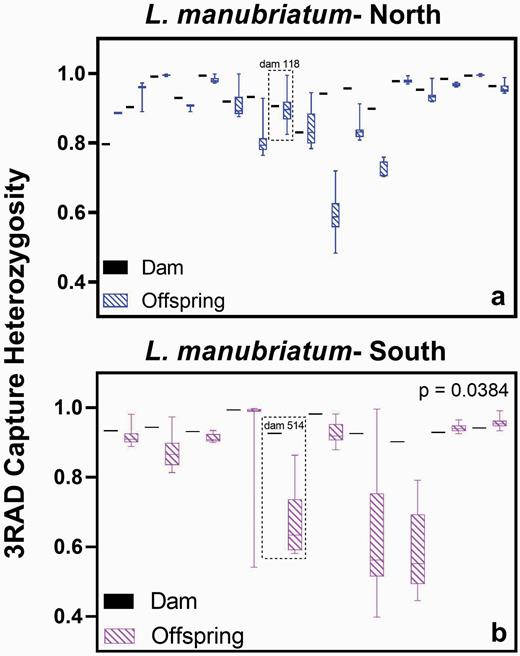
In comparing the heterozygosity of dams in their offspring using a paired t-test, we found only southern L. manubriatum dams and their associated offspring had significantly different values of heterozygosity (Figure 3). Offspring heterozygosity was significantly decreased compared to dams (Figure 3b; 95% CI = −0.1955 to −0.006738).
Heterozygosity of facultatively parthenogenetic L. manubriatum dams and offspring from (a) northern and (b) southern populations. A paired t-test between dam and average offspring heterozygosity was statistically significant only for southern L. manubriatum. However, a one-way ANOVA of combined species groups reveals a P-value of 0.0006, with a post hoc Tukey test on L. tohokuense (GBS) and L. manubriatum-South (GBS) yielding P < 0.05. Dams #118 (North) and #514 (South) are highlighted due to their divergent clone probabilities (Figure 5).
Homozygous Dam to Heterozygous Offspring Locus Transitions
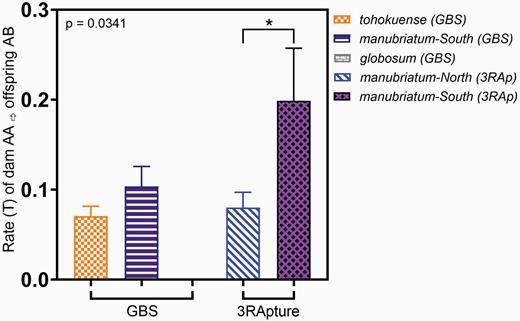
Using the VCF outputs from Stacks, we calculated the number of homozygous loci (s) that were typed as heterozygous in offspring for each dam’s clutch. Using MassArray typing, we found a significantly higher rate of T for the obligate sexual L. tohokuense, and the facultative parthenogen L. manubriatum (t11 = 12.42, P < 0.0001). While there was no significant difference in transition rate T between L. tohokuense and both facultative parthenogens using Genotyping-By-Sequencing (P = 0.9774; Figure 4), we did find southern L. manubriatum had a significantly higher T than their northern conspecifics (P = 0.0341; Figure 4) using the 3RApture method. After adjusting x (the number of heterozygous offspring loci which are homozygous in the dam) to include all missing loci that could potentially be heterozygous, we found that the parthenogen L. globosum additionally had a significantly lower rate of T than the obligate sexual L. tohokuense (P = 0.0055).
Average rate (T) of homozygous dam to heterozygous offspring transitions per loci. Leiobunum tohokuense is an obligate sexual, while L. globosum and L. manubriatum are facultatively parthenogenetic. A one way ANOVA of all GBS groups reveals a P-value of 0.9774, and the asterisk indicates P < 0.05 for an unpaired t-test on L. manubriatum-South (3RAp) and L. manubriatum-North (3RAp).
Clone Probability and Inferred Sires
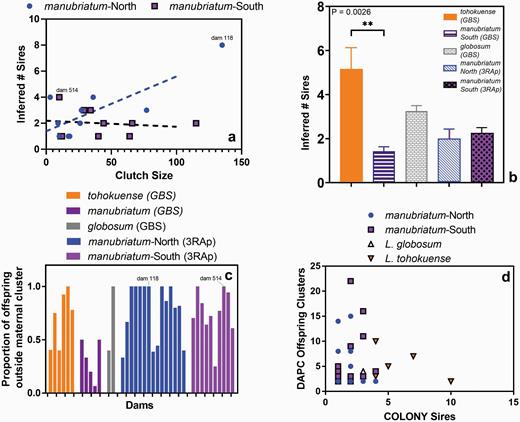
Using the converted VCF files, we ran Colony to determine the average clone probability of all offspring per dam, as well as the number of sires inferred to contribute genetically to each clutch, under 3 different paternal sibship priors. Ultimately, we did not find adjustment of the paternal sibship prior to significantly affect the individual clone probabilities (Figure 6a) of any clutches, so here we display results assuming paternal sibship count is equal to maternal sibship count (average clutch size).
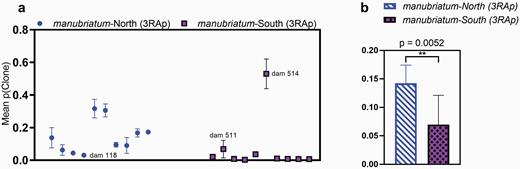
When we allowed for the possibility of obligate sexual L. tohokuense dams to produce cloned offspring in Colony, we found with GBS loci that L. tohokuense offspring had an average clone probability lower than L. manubriatum (P = 0.0025) though this effect was not significant (P = 0.986) with MassArray. Comparing 3RApture typing of northern and southern L. manubriatum also yielded a statistically significant difference in average clone probability (Figure 5b), with southern L. manubriatum showing a lower probability of asexually produced offspring than dams from the north. This trend is particularly clear in Figure 5a, wherein only one northern dam (#118) we typed using 3RApture had a clutch clone probability not significantly different from zero (one-sample t-test; t9 = 4.456, P = 0.0016), while all but 2 of the southern dams’ clutches (#s 511 and 514) had clone probabilities that were not significantly different than zero (t9 = 1.356, P = 0.2082).
Probability that a typed offspring is a clone p(Clone) as inferred by Colony using a 3RAD Capture (3RAp) genotyping method. (a) Mean p(Clone) per dam. Each point represents the mean p(Clone) of all offspring oviposited by a single dam. Blue circles correspond to dams from northern Leiobunum manubriatum population, purple squares correspond to dams from southern L. manubriatum populations. (b) Mean p(Clone) of all dams from a northern vs. southern populations. The double-asterisk indicates P < 0.01 for a Mann-Whitney U test. Dams #118 (North), #511 (South), and #514 (South) are highlighted due to their divergent clone probabilities.
Results from the clone probability assessment did not extend to our examination of the number of inferred sires contributing to each clutch (Figure 6a), as Colony does not remove probable cloned offspring from analyses and will attempt to assign sire genotypes even if asexual reproduction is expected. As a result, we inferred at least one sire for all facultatively parthenogenetic clutches, even those with very high average clone probabilities (Figure 6a). After regression against clutch size, we found that no species’ data yielded a slope significantly different from zero, but L. tohokuense had a larger average number of inferred sires than L. manubriatum (Figure 6b; P = 0.0026).
(a) Number of sires inferred by Colony as a function of clutch size. Each point represents the inferred number of sires for one female’s offspring, assigned a confidence probability of ≥0.05. Blue circles correspond to dams from northern Leiobunum manubriatum populations, purple squares correspond to dams from southern L. manubriatum populations, both genotyped using a 3RAD Capture (3RAp) method. (b) Average number of sires per clutch by species. A one-way ANOVA of species groups reveals a P-value of 0.0036, and the double star indicates P < 0.01 for a post hoc Tukey test between L. manubriatum-South (GBS) and tohokuense (GBS). (c) Proportion of offspring assigned outside of the maternal cluster for dams of obligate sexual and facultatively parthenogenetic species. Dams #118 (North) and #514 (South) are highlighted due to their divergent clone probabilities from Colony analysis (Figure 5a). (d) Scatter plot of numbers of inferred sires from Colony (Figure 6b) and numbers of inferred sires (i.e., non-maternal clusters, Figure 6c) using DAPC clustering.
Many of the dams produced very small numbers of offspring, precluding our ability to infer outcrossing using DAPC. However, we found the majority of dams produced offspring that were assigned to clusters outside of the maternal cluster (Figure 6c). We found a main effect of species in the proportions of offspring not clustering with the dam (P = 0.0207), but this result was driven primarily by the lower proportions of offspring clustering outside of the maternal cluster in southern L. manubriatum genotyped with GBS. Furthermore, sires inferred via DAPC clusters deviated from the number of sires inferred with Colony for all species examined (Figure 6d).
Conclusions
High-Throughput Genotyping Considerations
Opiliones are among a number of non-model organisms that have been adopted for recent focus in studies of reproductive evolution and behavioral ecology (Burns et al. 2016; García-Hernández and Machado 2018; O’Brien et al. 2019; Segovia et al. 2019). Accordingly, we used 3 methods for identifying loci across the genome in order to develop a high-coverage set of SNPs with utility for sibship analyses. SNP genotyping has increased in usage due to the expansion and accessibility of short-read sequencing technologies, and for organisms where such sequencing has already been completed, custom array methods present an additional tier of low-cost genotyping services. We estimated we would need at least 50 high-MAF SNPs to estimate sireship in this dataset (Liu et al. 2006; Hauser et al. 2011; Ellis et al. 2018) but found our results with MassArray, which provided half this number, provided similar conclusions in comparing homozygous to heterozygous locus transitions (T) and sire counts between facultative parthenogens and obligate sexuals. This is likely because we specifically selected these MassArray SNPs from a prior Illumina sequencing project (Burns et al. 2016, 2017) in order to maximize MAF. The downside to using the array approach for SNP genotyping came in the number of ambiguities in typing calls that we were unable to incorporate into our results. This loss of data, coupled with the need to sequencing conspecifics in order to identify loci for the pre-designed arrays leads us to recommend highly-multiplexed GBS for SNP genotyping, followed with a RAD-capture approach if on-going typing is expected.
Comparisons Between Sexual and Facultatively Parthenogenetic Species
We collected and genotyped females and associated offspring of 2 facultatively parthenogenetic opilionid species, Leiobunum manubriatum and L. globosum, as well as a closely related obligate sexual species, L. tohokuense, in order to compare fecundity and fertility of the species, and to identify populations of sexually and asexually reproducing females by estimating the number of contributing sire per clutch. We moreover assessed the potential for the parthenogenetic species to produce mixed clutches of sexually and asexually produced offspring by measuring the heterozygosity, transition rate of homozygous dam to heterozygous offspring loci (T), and clone probability of each clutch. We found that the facultative parthenogens did not have significantly different fecundity or fertility than closely related sexual species, owing to the parthenogen L. manubriatum having similar values to sexual L. tohokuense, while parthenogenetic L. globosum produced significantly fewer eggs than other species (Figure 2). We found evidence for significant differences between L. tohokuense and facultatively parthenogenetic L. manubriatum in the average number of sires contributing to clutches (Figure 6). Moreover, our analysis of clone probability (Figure 5b) and T rate (Figure 4) indicate sexual reproduction is more common in the southern range for parthenogen L. manubriatum. We studied 2 particular dams from the northern (#118) and southern (#514) populations that show clutches with characteristics that suggest reproductive polymorphism. Based on the variance associated with heterozygosity (Figure 3) and clone probability (Figure 5a) in offspring of L. manubriatum, we posit that mixed reproductive strategy for females within single clutches is possible in some cases for this species.
Contrary to studies in a variety of species in which primarily parthenogenetic lineages are associated with low fertility compared to their sexual relatives (Weinzierl et al. 1999; Cosendai et al. 2013), our data indicated that the facultatively parthenogenetic L. manubriatum and L. globosum did not produce a significantly different number of viable offspring than their obligately sexual sister taxa, L. tohokuense. However, we observed that the genotypes of these species differed in several key ways. First, facultatively parthenogenetic taxa tended to have higher heterozygosity than sexual taxa—significantly so, in the case of the comparison of L. tohokuense and L. manubriatum using GBS loci. This could be because we have evidence that L. globosum, and a number of populations of L. manubriatum (including localities we sampled in this study), consist of tetraploid individuals (Tsurusaki 1986; Burns et al. 2017). As this cytotype is consistent with a single genome duplication in the species’ history (Burns et al. 2017), the overall doubling of the genome may produce the increase in heterozygosity seen in facultative parthenogens. However, this hypothesis does not explain why only L. manubriatum and not L. globosum had significantly higher heterozygosity compared with L. tohokuense.
A second hypothesis for the higher heterozygosity of facultatively parthenogenetic species is as a result of Muller’s Ratchet, the theoretical inability of parthenogenetic taxa to remove mutations from their genomes over generations (Vrijenhoek and Lerman 1982; Warren et al. 2018). This maintenance and accumulation of mutations over time causes these species to have elevated levels of genome-wide heterozygosity (Hollister et al. 2015; Engelstädter 2017). However, the consideration that L. manubriatum and L. globosum are facultatively asexual and may at least occasionally mate makes our results consistent with the hypothesis that these facultative parthenogens reproduce sexually at lower rates than closely related obligate sexual taxa. This is in spite of the fact that, for southern L. manubriatum at least, mates are available and often abundant (Tsurusaki 1986; Burns and Tsurusaki 2016; Burns et al. 2017).
Asexual Reproduction in Male-Prevalent Conditions
Our Colony results after inferring the number of sires contributing to clutches of L. tohokuense and L. manubriatum support the hypothesis that facultative parthenogens mate less frequently than sexual species, even if males are present. While both species are observed to mate multiply (Tsurusaki 1985, 1986), clutches of L. tohokuense had an average of 4 additional inferred sires compared to those contributing to L. manubriatum clutches (Figure 6b). Moreover, the high clone probabilities estimated for some clutches L. manubriatum (Figure 5a) suggest that no male actually sired some of the offspring in these clutches, and they were actually produced through asexual reproduction. DAPC analysis found females of most species produced offspring with divergent genotypes (Figure 6c), but L. tohokuense trended towards fewer offspring clusters than the facultatively parthenogenetic species (Figure 6d). This may be because the larger genomes of tetraploid offspring of parthenogenetic species (Burns et al. 2017) also form larger numbers of genotyping clusters using the DAPC method, regardless of whether they were produced sexually or asexually. Though we limited the number of potential DAPC clusters to 50% of the clutch size, automictic thelytoky in the asexual pathway for L. manubriatum means that DAPC clusters could form based on relatively minor genotypic differences in offspring rather than different sires. Future implementation would require a priori knowledge of potential offspring genotypic variance, as could be estimated given some known sire genotypes.
We observed 2 major differences in southern and northern L. manubriatum clutches as genotyped through the 3RApture approach. First, southern L. manubriatum dams were more likely than their northern counterparts to have homozygous loci that were heterozygous in their offspring (Figure 4), a signal of sexual reproduction. Second, southern L. manubriatum had clutches with lower average clone probabilities than their northern counterparts (Figure 5b). These 2 analyses support our hypothesis that northern L. manubriatum likely reproduce asexually more frequently than do southern L. manubriatum.
In agreement with the results of Colony sire estimates for facultatively asexual and sexual taxa, when we compared dam heterozygosity to averaged offspring heterozygosity for each species, significant differences were only found for southern L. manubriatum (Figure 3b). Offspring heterozygosity averages were significantly lower than dam heterozygosity for this group, suggesting asexual reproduction was the most common reproductive mode. This effect was not seen for any other parthenogenetic groups, and was what we would have expected to see in northern L. manubriatum clutches. Therefore we investigated whether results of individual females’ clutches could have driven these conflicting results, and whether our results point to the ability of these facultatively parthenogenetic taxa to produce offspring both sexually and asexually in the same clutch.
Mixed Reproductive Ontogeny: A Matter of Time?
We reviewed individual data for female L. manubriatum, as we have the broadest sampling for the species. L. manubriatum also had the greatest within species variability in fecundity, as exemplified by dam # 118, which produced 4 standard deviations above the mean number of developed eggs for northern populations of the species (Figure 2b). However, females with large clutches did not tend to have significantly different mean offspring heterozygosity (Figure 3), or higher clone probability (Figure 5a). For northern L. manubriatum, large clutches were more likely to have larger numbers of inferred sires using Colony (Figure 6a). Ultimately, although we have evidence supporting the role of asexual reproduction in northern L. manubriatum, dam # 118 likely produced some or all of her offspring sexually, given the presence of some offspring with increased heterozygosity (Figure 3a), low p(clone) (Figure 5a), high numbers of inferred sires (Figure 6a), and a high proportion of offspring with genotypes that clustered outside of her cluster (Figure 6c).
Though the majority of southern L. manubriatum appear to have produced their offspring sexually, the offspring of dam # 514 had the highest clone probability we estimated in this study (Figure 5a), and their heterozygosity was significantly lower than the dam’s (Figure 3b). The variance in heterozygosity and p(clone) for this clutch (as well as dam # 511; Figure 5a) moreover suggest within-clutch variation in reproductive ontogeny. Dam # 514 produced a smaller than average number of offspring (N = 11; Figure 2a), but a comparatively large number of inferred sires (Figure 6a), which additionally suggest sexually produced offspring. As acknowledged, the cytotype variation within populations of L. manubriatum that we sampled could also be responsible for dam #514’s offspring having a large number of inferred sires while p(clone) was also higher than average for the southern region. If dam # 514 was tetraploid, while other specimens from the south were diploid, 514 might have produced a greater variety of genotypes than other dams, even in the absence of sexual reproduction. Additionally, the small number of 514’s offspring could have additionally intensified genotyping errors leading to calculation of our reproductive mode metrics.
Female opilionids lay eggs in distinct clutches multiple times over the summer reproductive season (Machado and Macías-Ordóñez 2007). While we did observe individual L. manubriatum females producing sub-clutches at discrete time points, and observed variation in clone probabilities within individual clutches (Figure 5a), we did not separate eggs by lay time and our methods did not allow us to ascribe offspring to particular mates. Moreover, syngamy in Opiliones appears to occur at the time of oviposition after non-motile sperm are transferred from spermathecal storage (Machado and Macías-Ordóñez 2007), and but we do not currently know whether females actively control sperm fate. Therefore, whether females of L. manubriatum or L. globosum are able to withhold sperm and oviposit unfertilized ova even after mating is unclear. Ongoing research will address whether mating of virgin females of the parthenogenetic species yields oviposition of sexually produced offspring and should additionally assess the period of time between these events.
Sexual Conflict’s Role in Geographic Parthenogenesis
The marked rarity of males in L. globosum and northern populations of L. manubriatum (Tsurusaki 1986; Burns et al. 2017) suggests that density-mediated encounter rate should very simply control the maximum mating rate possible for the species. However, many dense populations of L. manubriatum and L. globosum in Aomori and Hokkaido Prefectures appear to lack males, despite our collecting efforts to prove otherwise and the fact that males in these species (Figure 1) are brightly colored and nearly unmistakable to us in the temperate forests in which they occur. Moreover, our data suggest reduced sexual reproduction in southern L. manubriatum as well, even though males are common. In fact, during a recent collection in the southern range in August 2019, 2 of us (N.T. and M.B.) observed 1–5 male L. manubriatum stationed on the lower trunks of nearly every tree in our sampling area, while females tended to be found above them in foliage. This male posture might be in order to intercept females attempting to reach appropriate oviposition grounds, which include soil and mosses at/near the forest floor (Machado and Macías-Ordóñez 2007).
Given our data, observations, and prior discussion of comparative results between obligately and facultatively sexual species, we suggest sexual conflict plays an additional role in controlling mating rate, which we look to test in ongoing study. Because females in the south are locally adapted to male presence and potential harassment, we suggest females may behaviorally avoid sexual conflict through running away or adopting a head-down posture to preclude intromission (Fowler-Finn et al. 2018) or grasping attempts by males. These behaviors would potentially limit direct reproductive costs to females by minimizing predation risk (Crowley et al. 1991) or harm from mating multiply (Wigby and Chapman 2005; Kokko and Mappes 2013), though we do not yet know whether nor which costs may be incurred by opilionids in this manner.
Theoretically, intersexual conflict would be minimized in a species where females are plentiful, motility is low, and mating interactions have little fitness cost. In this scenario, males should be released from the selective pressure to express traits to improve success in male-male competition (Tilquin and Kokko 2016; Nakano et al 2019) and intersexual interactions (Tilquin and Kokko 2016; Burke and Bonduriansky 2018). However, previous research on males of the Japanese L. curvipalpe group (Burns and Tsurusaki 2016) has shown males of the facultatively parthenogenetic species have larger secondary sex characteristics associated with conflict (Burns and Shultz 2015) than males of other group member species. In Opiliones, male armaments may include modified pedipalps with improved clasping ability and a transition in mechanism of penile protraction from hydraulic to muscular propulsion (Burns and Shultz 2015; Burns and Tsurusaki 2016). The presence of these traits in facultatively parthenogenetic species suggests that there may be active conflict between the sexes that could drive reproductive armaments in males (Kawatsu 2013). Male sexual coercion could be evidenced in genotypic and/or sex-ratio differences between sub-clutches produced by individual females, particularly if female resistance or male coercivity changes throughout the breeding period.
Funding
This work was supported by the National Science Foundation (DBI 1401110 to M.B.) and the Japan Society for the Promotion of Science (JSPS) KAKENHI grants (26440218 and JP17K07534 to N.T.).
Acknowledgments
We would like to thank the AGA 2019 for the invitation to participate in the symposium “Sex and Asex: The genetics of complex life cycles” held in Portland, Oregon, July 2019, and we thank all the participants at the meeting for lively and creative discussions, particularly Dr. Maria Orive, AGA President and Associate Journal Editor. We acknowledge James Barklage for his collections assistance and Mayukha Pakala (UMBC ‘19) for her laboratory support for this project. Dr. Maurine Neiman provided advice on early drafts, and 2 reviewers provided later comments that greatly improved this paper.
Conflict of Interest
The authors declare no conflicts of interest with this manuscript.
Data Availability
We have deposited the primary data underlying these analyses as follows:
Sampling locations, VCF, and Colony files: Dryad Digital Repository: https://doi.org/10.5061/dryad.prr4xgxk3
DNA sequences: NCBI SRA Project number: PRJNA676098