-
PDF
- Split View
-
Views
-
Cite
Cite
Jing Yi, Zhuwei Xu, Ran Zhuang, Jiuping Wang, Yusi Zhang, Ying Ma, Bei Liu, Yun Zhang, Chunmei Zhang, Guolin Yan, Fanglin Zhang, Zhikai Xu, Angang Yang, Boquan Jin, Hantaan Virus RNA Load in Patients Having Hemorrhagic Fever With Renal Syndrome: Correlation With Disease Severity, The Journal of Infectious Diseases, Volume 207, Issue 9, 1 May 2013, Pages 1457–1461, https://doi.org/10.1093/infdis/jis475
Close - Share Icon Share
Abstract
To investigate the role of viral load in the pathogenesis of hemorrhagic fever with renal syndrome, the Hantaan virus RNA load in plasma from 101 patients was quantified, and the relationships between viral load and disease course, severity, and level of specific humoral immunity were analyzed. The viral load, detectable in 79 patients, ranged from 3.43 to 7.33 log10 copies/mL of plasma. In the early stage of disease, patients in severe/critical group were found to have higher viral loads than those in the mild/moderate group (5.90 vs 5.03 log10 copies/mL; P = .001), suggesting an association between Hantaan virus load and disease severity.
Hantaan virus (HTNV), a member of the genus Hantavirus, causes a chronic, asymptomatic infection in its natural host, the striped field mouse Apodemus agrarius [1]. In contrast, HTNV infection in humans manifests as acute hemorrhagic fever with renal syndrome (HFRS). Typically, HFRS occurs in 5 sequential stages: febrile, hypotensive, oliguric, diuretic, and convalescent. Clinical symptoms also include thrombocytopenia and, in severe cases, hemorrhage caused by capillary leak syndrome [2]. As many as 100 000 cases of HFRS have occurred annually worldwide, of which >90% were documented in the mainland of China, with a mortality rate of 0.1%–15% [3].
The pathogenesis of HFRS is considerably far from being completely understood, and aspects of the research are mainly focused on experiments in vitro and limited clinical studies. The basic mechanisms underlying HFRS pathogenesis relate to increased vascular permeability, and the causative agents infect endothelial cells without cytopathic effects, suggesting that viral replication together with the immune response are involved in tissue injury [4]. In recent times, it has been widely recognized that HFRS pathogenesis is largely immune mediated, involving immune complexes, complement activation, B cell response, T cell response, and HTNV-induced cytokine production [3].
HFRS could be caused by other Hantaviruses besides HTNV, such as Dobrava virus, Seoul virus, Amur virus, and Puumala virus. In general, HFRS caused by HTNV, Amur virus, and Dobrava virus is more severe, whereas disease severity caused by Seoul virus and Puumala virus is more moderate or mild [4]. Furthermore, differences in the disease severity among the HFRS cases caused by a certain kind of Hantavirus have been reported. For example, some previous studies have indicated that the disease severity of HFRS correlates with several factors, including serum levels of albumin and inflammatory and regulatory cytokines, cellular immune response to HTNV, HLA phenotype, and level of complement activation.
Despite the limited knowledge on the pathogenesis of HFRS, some researchers have made attempts to investigate the role of antiviral intervention in the therapy of HFRS, at least in vitro. A key question that would help to establish a potential role that antiviral intervention plays in therapy is whether viral load during acute illness could be associated with adverse clinical outcome. Some recent studies have suggested that there might be an association between the Hantavirus RNA load and disease severity. Terajima et al [5] reported a high RNA load of Sin Nombre virus, another member of the genus Hantavirus that causes Hantavirus pulmonary syndrome (HPS), which existed during acute infection in plasma samples from patients with HPS. Furthermore, it has been found by both the Terajima group and the Xiao group [6] that an increased Sin Nombre viral load is likely to be associated with a more severe clinical outcome. Similar results were also obtained on the correlation between viral RNA load and disease severity in the case of Dobrava virus, as demonstrated by Saksida et al [7].
Nevertheless, there has been no report until now about the quantitative HTNV RNA load in patients with HFRS, including the relationship between HTNV load and the pathogenesis and severity of HFRS. Therefore, the HTNV RNA load was measured in plasma samples from patients with HFRS with use of a quantitative 1-step real-time reverse-transcriptase polymerase chain reaction (RT-PCR) assay, and the relationships between viral RNA load and disease course, disease severity, and level of specific humoral immunity were analyzed. Thus, to our knowledge, this study represents the first comprehensive determination of viral load relative to the clinical course and severity of the disease in patients infected with HTNV.
SUBJECTS, MATERIALS, AND METHODS
See Supplementary Data.
RESULTS
To assess the sensitivity of the real-time RT-PCR assay, HTNV RNA preparation was diluted with a 10-fold dilution series, and the data indicated that the detection limit of the assay reached 2700 copies/mL (3.43 log10 copies/mL) in the plasma. In addition, the specificity of the assay was tested using 4 other viruses, namely, hepatitis B virus, hepatitis C virus, influenza, and Seoul viruses. RNA from these virus isolates could not be amplified with the assay. Furthermore, the intra-assay and inter-assay variabilities of the assay were evaluated to be ranging from 0.6% to 4.5% and 3.9% to 7.3%, respectively, which indicates the high reproducibility of the assay.
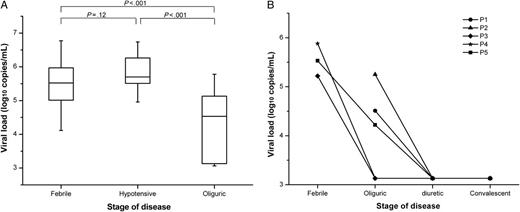
A panel of 198 plasma samples from 101 patients was examined, among which 84 samples from 79 patients could be detected for HTNV RNA load, ranging from 3.43 to 7.33 log10 copies per mL of plasma. The viral RNA could only be detected in plasma samples from patients in febrile, hypotensive, and oliguric stage. Among the 114 samples in which the viral RNA was undetectable, 2 were collected in febrile stage from patients with mild disease, and 14 were collected in oliguric stage; all the samples collected in diuretic (72 samples) and convalescent stage (26 samples) were undetectable for viral load, regardless of whether the patient belonged to the severe/critical group or the mild/moderate group. Viral load of the undetectable samples was recorded as the mean value between the detection limit and zero (1350 copies/mL). The median viral loads for febrile, hypotensive, and oliguric stage were 5.52, 5.90, and 4.53 log10 copies/mL, respectively. Although no statistically significant difference was found between viral load in febrile stage and that, in hypotensive stage (P = .12), the viral loads in these 2 stages were both found to be significantly higher than that in the oliguric stage (P < .001, Figure 1A). It was observed that, in 5 representative patients who had samples for 3 or 4 stages during the course of the disease, viral RNA copy numbers gradually decreased to an undetectable level with the progress of the disease (Figure 1B).
Comparison of HTNV load among patients in different stages of HFRS. Data were obtained from 100 samples collected in the febrile, hypotensive, or oliguric stage. The median, quartile, and standard deviation are shown on the left. Viral load in serial samples from 5 patients who had samples for 3 or 4 stages, P1–P5 represent patient 1 < patient 5, respectively (B). Viral load of the undetectable samples was recorded as the mean value between the detection limit (2700 copies/mL) and zero. The significance of the differences between different groups was determined by the Mann–Whitney U test. P values < .05 were considered to be statistically significant, and Bonferroni correction was used.
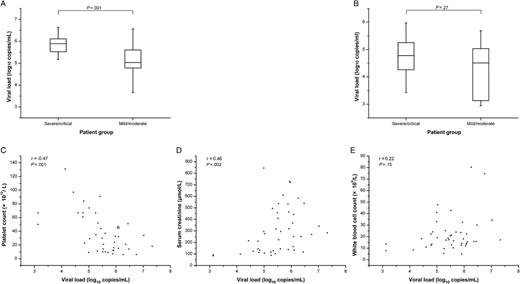
Subsequently, the relationship between disease severity and viral loads in samples collected in febrile/hypotensive stage and oliguric stage was analyzed, respectively. In febrile/hypotensive samples, it was found that the difference between the median viral load in the severe/critical group and in the mild/moderate group was of statistical significance (5.90 vs 5.03 log10 copies/mL; P = .001, Figure 2A). It is noteworthy that 3 patients with high viral load in febrile/hypotensive stage (6.75, 6.26, and 5.91 log10 copies/mL) died later, which also suggests that those patients with high viral loads in the early stage are more likely to have a severe course of disease. However, a comparison of viral loads in oliguric samples from the severe/critical group with those from the mild/moderate group revealed no statistically significant difference (4.78 vs 4.52 log10 copies/mL; P = .27, Figure 2B). On the basis of the data, the relationship between viral load in febrile/hypotensive samples and 3 laboratory parameters that could represent the severity of the disease was further analyzed. The data revealed a significant negative relationship between the viral load and the lowest value of platelet count (P = .001, R = −0.47, Figure 2C) and a significant positive relationship between the viral load and the peak value of serum creatinine (P = .002, R = 0.46, Figure 2D). No such relationship was found with the peak value of white blood cell count (P = .15, Figure 2E).
Relationship between HTNV load and disease severity of HFRS. Comparison of viral load between the severe/critical group and the mild/moderate group in plasma from 45 samples in febrile/hypotensive stage (A) and from 55 samples in oliguric stage (B), Median, quartile, and standard deviation are shown. Relationship between viral load in febrile/hypotensive samples from 44 samples and the lowest platelet count (C), the peak creatinine level (D), and the peak white blood cell count (E). Viral load of the undetectable sample was recorded as the mean value between the detection limit and zero. The significance of the differences between the groups was determined by the Mann–Whitney U test. The relationship between viral load and the laboratory parameters was evaluated using the rank correlation test. P values < .05 were considered to be statistically significant.
DISCUSSION
The data in this study demonstrated that HTNV RNA load could only be detected in plasma of HFRS patients in febrile/hypotensive and oliguric stage (3–7 days and 4–12 days after the onset of disease, respectively), and it gradually decreased to an undetectable level with the progress of the disease, which is similar with the persistence of viremia of Dobrava virus [7] and in accordance with the most important property of HFRS, that is, self-limiting. Some reports have shown that both humoral and cellular immune response might contribute to eliminate the virus and play antiviral roles. On one hand, the protective activity of neutralizing antibodies against HTNV in the clearance of virus has been commonly recognized [8–10]. On the other hand, there are very few reports regarding the immuno-protective roles of specific cellular immune response in the defense against HTNV infection [11] that could be used to draw a conclusion; furthermore, contradictory results exist about the roles of cellular immune response in the pathogenesis of zoonotic disease caused by hantaviruses [12]. Therefore, further experiments must be done to understand virus-host interactions, to clarify the mechanisms of HTNV elimination, and to elucidate the mechanisms of the self-limiting property of HFRS.
The findings also revealed a statistically significant association (P < .05) between HTNV RNA load in plasma and disease severity in patients in the early stage of HFRS. It was demonstrated that patients with high viral loads in febrile/hypotensive stage might be more likely to have a more severe course of disease. In contrast, low viral loads were associated with a relatively mild course of disease. Regarding the oliguric samples, no such significant difference was found between viral loads in the severe/critical group and those in the mild/moderate group. There could be 2 possible reasons: with the progress of the disease, (1) viral loads in both groups decreased considerably and made the difference less obvious and (2) the host's immune responses were induced and the immune mechanisms, rather than viremia, started to play an important role in the pathogenesis of the disease. Moreover, it was reported that viral load in the febrile/hypotensive stage was positively related to the peak value of serum creatinine and negatively related to the lowest value of platelet count, which further certified the relationship between viral load and disease severity. These findings are in accordance with the pantropic property of HTNV and findings from previous studies on Sin Nombre virus RNA load in patients with HPS [5, 6] and on Dobrava virus RNA load in patients with HFRS [7], which revealed that high levels of viremia correlated with disease severity. The results obtained on the relationship between viremia and disease severity in the early stage of HFRS are also in accordance with studies on anti-HTNV–neutralizing mAbs in mice [8], which demonstrates the protective activity of those neutralizing mAbs administrated in the early stage of HTNV infection. Overall, the data in the present study indicate that the initial viral load in patients with HFRS could be used as a prognostic marker for monitoring and/or predicting the severity of clinical manifestations. At present, only clinical status and clinical laboratory parameters are available as guides for HFRS. It is anticipated that prompt determination of HTNV load at hospital admission could eventually guide clinicians toward more- or less-aggressive therapy.
In the present study, the relationship between HTNV load in febrile/hypotensive samples and the peak value of titers of HTNV-NP specific IgM and IgG, which was considered as an index representing humoral immunity against HTNV infection, was also investigated (see Supplementary Data). Different from the previous studies on Puumala virus [13] and Dobrava virus [7], both of which reported an inverse correlation between antibody titers and viral load, no such correlation was found in this study (see Supplementary Figure 1). The reason for the disparity might include differences in virus (HTNV versus Puumala or Dobrava virus). In addition to viral concentration, the level of humoral immune response is determined by several factors, such as genetics (particularly, MHC gene typing), age, sex, and physical status of the host.
In conclusion, this is the first study to describe the plasma HTNV RNA load in patients with HFRS and to discuss its relationship with disease course, disease severity, and level of specific humoral immunity based on a kinetic observation. Using a panel of plasma samples and a real time RT-PCR assay, we showed that the HTNV RNA load gradually decreases to an undetectable level with the progress of the disease and that there is an association between viral load and disease severity in the early stage of the disease, which indicates that the viral load could be used as a prognostic marker and that the viremia might be one of the important factors in the pathogenesis of HFRS. Additional studies are required to determine the mechanisms by which HTNV replication is regulated during acute infection, to define the virus-host interactions, and to gain a greater understanding of how HTNV is recognized by both the innate and adaptive immune systems.
Notes
Acknowledgments. We thank the volunteers who generously participated in this study and Jielai Xia, for assistance in statistical analysis.
Financial support. This work was supported by The National Natural Science Foundation of China (30930087) and the National Basic Research Program of China (2012CB518905).
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
Author notes
J. Y. and Z. X. contributed equally to this study.