-
PDF
- Split View
-
Views
-
Cite
Cite
Sabue Mulangu, Vivian H Alfonso, Nicole A Hoff, Reena H Doshi, Prime Mulembakani, Neville K Kisalu, Emile Okitolonda-Wemakoy, Benoit Ilunga Kebela, Hadar Marcus, Joseph Shiloach, Je-Nie Phue, Linda L Wright, Jean-Jacques Muyembe-Tamfum, Nancy J Sullivan, Anne W Rimoin, Serologic Evidence of Ebolavirus Infection in a Population With No History of Outbreaks in the Democratic Republic of the Congo, The Journal of Infectious Diseases, Volume 217, Issue 4, 15 February 2018, Pages 529–537, https://doi.org/10.1093/infdis/jix619
Close - Share Icon Share
Abstract
Previous studies suggest that cases of Ebola virus disease (EVD) may go unreported because they are asymptomatic or unrecognized, but evidence is limited by study designs and sample size.
A large population-based survey was conducted (n = 3415) to assess animal exposures and behaviors associated with Ebolavirus antibody prevalence in rural Kasai Oriental province of the Democratic Republic of Congo (DRC). Fourteen villages were randomly selected and all healthy individuals ≥1 year of age were eligible.
Overall, 11% of subjects tested positive for Zaire Ebolavirus (EBOV) immunoglobulin G antibodies. Odds of seropositivity were higher for study participants older than 15 years of age and for males. Those residing in Kole (closer to the outbreak site) tested positive at a rate 1.6× higher than Lomela, with seropositivity peaking at a site located between Kole and Lomela. Multivariate analyses of behaviors and animal exposures showed that visits to the forest or hunting and exposure to rodents or duikers predicted a higher likelihood of EBOV seropositivity.
These results provide serologic evidence of Ebolavirus exposure in a population residing in non-EBOV outbreak locations in the DRC and define statistically significant activities and animal exposures that associate with EBOV seropositivity.
(See the editorial commentary by Crozier, on pages 523–5.)
Ebolaviruses, of the family Filoviridae, are highly pathogenic and have been associated with devastating outbreaks of Ebola virus disease (EVD) since 1976. There are 5 known species of Ebolaviruses, named for the geographical location of origin, which include the following: Zaire Ebolavirus (EBOV), Sudan Ebolavirus, Taï Forest Ebolavirus, Reston Ebolavirus (RESTV), and Bundibugyo Ebolavirus [1–4]. Clinical disease in humans arises from infection with all but RESTV, and case fatalities vary from 25% to over 90% depending on the species and location. EBOV is the most pathogenic and is the cause of 7 of the 8 outbreaks in the Democratic Republic of Congo (DRC) [5], the most recent of which occurred from May to July in 2017. The outbreak of EBOV in West Africa that started in 2013 was unprecedented in magnitude and demonstrated how devastating this disease can be in both rural and urban settings, with more than 15000 laboratory-confirmed cases and more than 11000 documented fatalities [5].
Ebola virus disease is a zoonotic disease, and, although still under question, a possible natural reservoir is the fruit bat [6, 7]. The mechanism of EBOV spillover transmission from animals to humans remains unclear; however, once in humans, outbreaks are primarily the result of direct person-to-person or indirect vehicle transmission [8–10]. To date, population-wide estimates of EBOV antibodies are limited, despite documentation of asymptomatic or minimally symptomatic infections of EBOV in both non-outbreak and outbreak locations [11]. A large study of approximately 4500 subjects from randomly selected villages in Gabon (with a history of 4 past outbreaks) found that 15.3% of participants were seropositive for EBOV immunoglobulin (Ig)G, and the prevalence varied according to both age and environment [12]. The sera from this study were collected in 2005 and 2008, 3 and 6 years after the last human and great ape outbreak in Gabon in 2002, respectively [12]. On the other hand, a study in the Central African Republic found that 5.3% of the population tested positive for EBOV, where no outbreak or cases had previously been reported [13]. The sera from this survey were collected in 1985–1987 and 1993. In the DRC, a few weeks after the 1995 Ebola outbreak, EBOV-specific IgG was found in 9.3% of individuals living in unaffected villages near Kikwit with an IgG enzyme-linked immunosorbent assay (ELISA) using antigens prepared by viral culture in Vero E6 cells [14]. In addition, 18.7% of Efé Pygmies in north-eastern DRC were found to be seropositive in a 2002 serosurvey [15]. Serologic evidence of EBOV exposure is limited in DRC because few studies have been conducted, and the available estimates are based on a relatively small number of individuals. It is suggested that asymptomatic Ebolavirus infections is not uncommon: although based on small sample sizes, 1 post-EVD outbreak serosurvey found that 10 of 14 (71%) seropositive individuals had not experienced clinical disease [16], whereas another small study found that 11 of 24 (46%) asymptomatic close contacts of EVD cases were laboratory confirmed as IgM and IgG positive [17]. A recent study in Kono, Sierra Leone has suggested that minimally symptomatic cases are also common in outbreak settings and missed by disease surveillance activities.
The Democratic Republic of Congo has experienced 8 documented outbreaks of Ebolavirus in the last 40 years: Yambuku (1976) [18], Tandala (1977) [19], Kikwit (1995) [20], Mweka/Luebo (2007) [21], Mweka (2008) [6], Isiro (2012) [21, 22], Boende (2014) [22], and Likati (2017) [23]. Currently, the extent of anti-EBOV seropositivity is unknown, particularly in areas where no outbreaks have been recorded. Although the significance of such anti-EBOV positive individuals is unclear, such data may provide a better understanding of the epidemiology of Ebolavirus transmission and may better inform and have implications for outbreak management. Therefore, we conducted a large seroprevalence survey of IgG antibody to EBOV among inhabitants of 2 health zones, Kole and Lomela, within the Sankuru District in the DRC, a site where no outbreaks or cases have been reported.
METHODS
Study Population
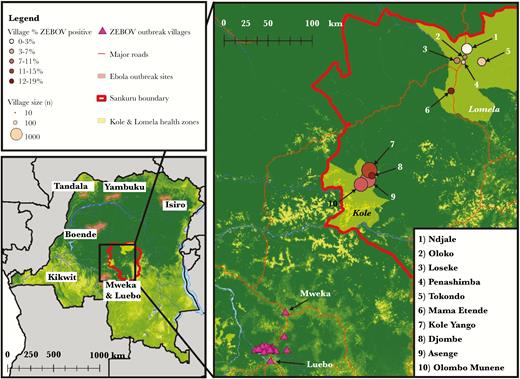
The study population and subject selection have been described elsewhere [24]. In brief, from August to September 2007, we conducted a population-based serosurvey and assessed human exposure to local wild animals and zoonotic pathogens in healthy, rural village populations in the Sankuru District (Kasai Oriental Province) of the DRC (Figure 1). The population comprised persons residing in 14 rural villages from 2 participating health Zones, Kole and Lomela, in the Sankuru District in the Kasai Oriental Province. There are 207 villages within Kole and 167 villages within Lomela. The 14 villages were randomly selected (3 villages—Mamba Etende, Mamba Ewanga and Mamba Edinda—were collapsed into Mamba Etende due to small sample size and close proximity, thus 12 will be referenced in the results) from a list of all villages provided by the DRC Ministry of Health and local health zone offices. Most villages in this region are located in small clearings of tropical forest, surrounded by traditional agricultural fields. The languages spoken in this area are Tetela, Lingala, and French.
Map of the Democratic Republic of Congo and outbreak sites up to 2007, the year of the serosurvey. Shaded areas on map indicate past Ebola outbreak sites with 25-km buffer around the health zones in which the outbreak occurred. Outbreak locations include Yambuku (1976), Tandala (1977), Kikwit (1995), and Mweka/Luebo (2007). The solid thick line indicates the boundary of the Sankuru district, where the Kole and Lomela health zones are located.
Among participants, verbal informed consent was obtained by trained interviewers in local language from all participating adults as well as assent from children 7 to 18 years of age with parental or guardian consent. Parents and guardians of participants under 7 years of age answered the questionnaire on behalf of their children. All responses were double entered into an Access database for quality control purposes.
Serologic Testing: EBOV Nucleoprotein Enzyme-Linked Immunosorbent Assay
We have chosen a serological assay based on recombinant EBOV antigen to avoid the safety issues related to assays using live virus preparation [25–27]. Purified recombinant full-length nucleoprotein (r-NP) from the EBOV was used as antigen in the ELISA to detect anti-EBOV antibodies in serum. Nucleoprotein was chosen because it is one of the most abundant proteins in the virus particle, and the host antibody response is most pronounced for NP during Ebola infection [28–31].
The EBOV r-NP antigen source and preparation are described elsewhere [32, 33]. The EBOV r-NP ELISA protocol was in-house optimized for the amount of the coating antigen, the nature of the blocking agent, the dilution of the test sera, and the species and dilution of the detection antibody for the greatest signal-to-noise ratio. In brief, each well of a 96-well ELISA microplate (Nunc-Immuno Maxisorp plates, Rochester, New York, United States) was coated with 0.25 µg of r-NP at 4°C overnight and blocked with casein-blocking buffer (Sigma, St. Louis, MO) at 37°C. Thereafter, serum samples were incubated at the dilution of 1:200 for 60 minutes at 37°C. The wells were washed 6 times with 0.2% Tween 20 (Sigma-Aldrich, St. Louis, Missouri, United States) in phosphate-buffered saline, followed by incubation for 30 minutes at 37°C with a peroxidase-conjugated F(ab)2 fragment Donkey anti-Human Fc (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA) at a dilution of 1:10000, and by incubation in the dark for 30 minutes at room temperature with o-phenylenediamine dihydrochloride (Sigma), which was used as a chromogenic substrate. The reaction was stopped by adding 100 µL of 1.8 M H2SO4 to each well. Absorbance (490 nm) was measured using an ELISA reader (Victor X3 Plate Reader, PerkinElmer, Waltham, Massachusetts, United States).
The cutoff value for assigning positive test results was based on the average optical density (OD) from 88 Congolese donors. These subjects were considered to be unexposed to Ebolaviruses because they were all born and grew up solely in Kinshasa where an Ebola outbreak has never been observed. The cutoff for positivity was chosen to be highly conservative and set at ≥5 standard deviations from the negative control mean (0.445). The IgG NP ELISA is estimated to have a specificity and sensitivity of 99% and 89%, respectively. The positive predictive value and negative predictive value was 94% and 98%, respectively.
Covariate Assessment
Basic sociodemographic information collected from participants included, but was not limited to, age (years), marital status, education, occupation (primary and secondary), and material possessions. A wealth index, adapted from Malleson et al [34], was created to assess the association of socioeconomic status with serologic response using the following reported assets: domestic animals, fields, radio, bicycle, sewing machine, and motorcycle. Each reported asset was given a value of 1 (maximum value of 6), and each extra point indicated an asset-rich household. Categories for socioeconomic status were created as follows: low (0 reported assets), middle (1–2 reported assets), high (≥3 reported assets).
In addition, information on animal exposures was collected using taxonomic categories derived from accepted local terms from focus group interviews. Participants were shown a representative photo or drawing and asked about the frequency and type of exposure, if any, to the animal in the previous month. We assessed animal exposures by both the type of activity (hunting, butchering skinning, eating, cooking, getting bitten or scratched by, playing with or picking up dead carcasses to eat) or any/no exposure (binary) to animal species and groups.
Statistical Analyses
Among the 3440 subjects for whom serum was tested, 50 were duplicate entries and 92% (23 pairs) had concordant results based upon the specified cutoff value (Supplemental Figure 1). Anti-EBOV NP antibody OD values were averaged for subjects with duplicate entries, resulting in a final sample size of 3415. Basic demographics (age, health zone, and sex) were available for all subjects, whereas 76% of participants (n = 2580) also took part in the questionnaire.
Frequency distributions were run for all covariates included in Table 1. To assess differences in sociodemographic characteristics according to serologic response, χ2 analyses were performed, and then to identify possible independent sociodemographic predictors of seropositivity, univariate logistic regression models were run for variables (see Table 1). Multivariate regression models were initially run with all variables; using backward selection only, significant predictors (alpha ≤0.05) were retained. We then assessed differences in potential exposures of interest, including forest visits and animal exposures (for a full list, see Table 2), using χ2 analyses. For significant exposures, the relationship between the exposure and EBOV seropositivity were further assessed using logistic regression. The final multivariate models for the exposures of interest included significant predictors from the sociodemographic model. All statistical analyses were carried out using SAS software, version 9.4 (SAS Institute, Cary, NC), and maps were generated using ArcGIS software, version 9.3 (ESRI, Redlands, CA).
Frequency Distribution and Regression Analysis of Sociodemographic Characteristics of Serosurvey Study Participants by EBOV Nucleoprotein IgG Results From the Sankuru Province, DRC
| . | All Subjectsn (%) . | EBOV−n (%) . | EBOV+n (%) . | P Valuea . | Regression Analysisb . | |
|---|---|---|---|---|---|---|
| ORcrude (95% CI) . | ORadjc (95% CI) . | |||||
| Aged | ||||||
| 0–14 years | 1407 (42) | 1336 (95) | 71 (5) | <.0001 | ref | ref |
| 15–49 years | 1562 (47) | 1343 (86) | 219 (14) | 3.07 (2.32–4.05) | 3.29 (2.48–4.36) | |
| 50 + years | 362 (11) | 299 (83) | 63 (17) | 3.96 (2.76–5.69) | 4.25 (2.95–6.13) | |
| Sexd | ||||||
| Female | 1930 (58) | 1745 (90) | 185 (10) | .0233 | ref | ref |
| Male | 1404 (42) | 1235 (88) | 169 (12) | 1.29 (1.04–1.61) | 1.51 (1.20–1.89) | |
| Education | ||||||
| None | 256 (10) | 219 (86) | 37 (14) | .0001 | 1.69 (1.13–2.53) | - |
| Some primary education | 1168 (47) | 1062 (91) | 106 (9) | ref | - | |
| Completed primary education | 791 (32) | 678 (86) | 113 (14) | 1.67 (1.26–2.21) | - | |
| Completed secondary education or beyonde | 274 (11) | 228 (83) | 46 (17) | 2.02 (1.39–2.94) | - | |
| Socioeconomic statusf | ||||||
| Low | 1140 (45) | 1053 (92) | 87 (8) | <.0001 | ref | - |
| Middle | 1095 (43) | 930 (85) | 165 (15) | 2.15 (1.63–2.83) | - | |
| High | 312 (12) | 258 (83) | 54 (17) | 2.53 (1.76–3.65) | - | |
| Occupation | ||||||
| Healthcare Workerg,h | ||||||
| No | 2214 (96) | 1944 (88) | 270 (12) | .0047 | ref | - |
| Yes | 84 (4) | 65 (77) | 19 (26) | 2.11 (1.24–3.56) | - | |
| Hunterg,i | ||||||
| No | 1866 (82) | 1660 (88) | 226 (12) | .0472 | ref | - |
| Yes | 411 (18) | 347 (84) | 64 (16) | 1.36 (1.00–1.83) | - | |
| Health Zone | ||||||
| Kole | 2204 (66) | 1939 (88) | 265 (12) | .0003 | 1.58 (1.23–2.03) | 1.58 (1.22–2.04) |
| Lomela | 1140 (34) | 1049 (92) | 91 (8) | ref | ref | |
| . | All Subjectsn (%) . | EBOV−n (%) . | EBOV+n (%) . | P Valuea . | Regression Analysisb . | |
|---|---|---|---|---|---|---|
| ORcrude (95% CI) . | ORadjc (95% CI) . | |||||
| Aged | ||||||
| 0–14 years | 1407 (42) | 1336 (95) | 71 (5) | <.0001 | ref | ref |
| 15–49 years | 1562 (47) | 1343 (86) | 219 (14) | 3.07 (2.32–4.05) | 3.29 (2.48–4.36) | |
| 50 + years | 362 (11) | 299 (83) | 63 (17) | 3.96 (2.76–5.69) | 4.25 (2.95–6.13) | |
| Sexd | ||||||
| Female | 1930 (58) | 1745 (90) | 185 (10) | .0233 | ref | ref |
| Male | 1404 (42) | 1235 (88) | 169 (12) | 1.29 (1.04–1.61) | 1.51 (1.20–1.89) | |
| Education | ||||||
| None | 256 (10) | 219 (86) | 37 (14) | .0001 | 1.69 (1.13–2.53) | - |
| Some primary education | 1168 (47) | 1062 (91) | 106 (9) | ref | - | |
| Completed primary education | 791 (32) | 678 (86) | 113 (14) | 1.67 (1.26–2.21) | - | |
| Completed secondary education or beyonde | 274 (11) | 228 (83) | 46 (17) | 2.02 (1.39–2.94) | - | |
| Socioeconomic statusf | ||||||
| Low | 1140 (45) | 1053 (92) | 87 (8) | <.0001 | ref | - |
| Middle | 1095 (43) | 930 (85) | 165 (15) | 2.15 (1.63–2.83) | - | |
| High | 312 (12) | 258 (83) | 54 (17) | 2.53 (1.76–3.65) | - | |
| Occupation | ||||||
| Healthcare Workerg,h | ||||||
| No | 2214 (96) | 1944 (88) | 270 (12) | .0047 | ref | - |
| Yes | 84 (4) | 65 (77) | 19 (26) | 2.11 (1.24–3.56) | - | |
| Hunterg,i | ||||||
| No | 1866 (82) | 1660 (88) | 226 (12) | .0472 | ref | - |
| Yes | 411 (18) | 347 (84) | 64 (16) | 1.36 (1.00–1.83) | - | |
| Health Zone | ||||||
| Kole | 2204 (66) | 1939 (88) | 265 (12) | .0003 | 1.58 (1.23–2.03) | 1.58 (1.22–2.04) |
| Lomela | 1140 (34) | 1049 (92) | 91 (8) | ref | ref | |
Abbreviations: CI, confidence interval; DRC, Democratic Republic of Congo; EBOV, ebolavirus; IgG, immunoglobulin G; OR, odds ratio; ref, reference; SES, socioeconomic status.
aχ2P value.
bRegression analyses presented as univariate and multivariate analyses based upon backward selection for sociodemographic predictors of seropositivity.
cAdjusted model retains all sociodemographic predictors that meet 0.05 significance level using backward selection; final model includes age, sex, and health zone.
dAge and gender were obtained from all study participants, regardless of participation in questionnaire (n = 3344).
eCategory includes completion of secondary education, apprenticeship, higher education, or university.
fSES categorization is based on calculated wealth index derived from the following reported assets: domestic animals, fields, radio, bicycle, sewing machine, and motorcycle. Each reported asset was given a value of 1 (maximum value of 6), and categories for SES are as follows: low (0 reported assets), middle (1–2 reported assets), high (≥3 reported assets).
gOccupation categories (healthcare worker and hunter) are not mutually exclusive; n = 17 fall within both occupation groups through their response for primary or secondary source of income.
hOccupation of healthcare worker was assigned if participant indicated health worker, midwife, traditional healer, Red Cross, or any other health-related job as either a primary or secondary source of income.
iOccupation of hunter was assigned if participant indicated hunting as either a primary or secondary source of income.
Frequency Distribution and Regression Analysis of Sociodemographic Characteristics of Serosurvey Study Participants by EBOV Nucleoprotein IgG Results From the Sankuru Province, DRC
| . | All Subjectsn (%) . | EBOV−n (%) . | EBOV+n (%) . | P Valuea . | Regression Analysisb . | |
|---|---|---|---|---|---|---|
| ORcrude (95% CI) . | ORadjc (95% CI) . | |||||
| Aged | ||||||
| 0–14 years | 1407 (42) | 1336 (95) | 71 (5) | <.0001 | ref | ref |
| 15–49 years | 1562 (47) | 1343 (86) | 219 (14) | 3.07 (2.32–4.05) | 3.29 (2.48–4.36) | |
| 50 + years | 362 (11) | 299 (83) | 63 (17) | 3.96 (2.76–5.69) | 4.25 (2.95–6.13) | |
| Sexd | ||||||
| Female | 1930 (58) | 1745 (90) | 185 (10) | .0233 | ref | ref |
| Male | 1404 (42) | 1235 (88) | 169 (12) | 1.29 (1.04–1.61) | 1.51 (1.20–1.89) | |
| Education | ||||||
| None | 256 (10) | 219 (86) | 37 (14) | .0001 | 1.69 (1.13–2.53) | - |
| Some primary education | 1168 (47) | 1062 (91) | 106 (9) | ref | - | |
| Completed primary education | 791 (32) | 678 (86) | 113 (14) | 1.67 (1.26–2.21) | - | |
| Completed secondary education or beyonde | 274 (11) | 228 (83) | 46 (17) | 2.02 (1.39–2.94) | - | |
| Socioeconomic statusf | ||||||
| Low | 1140 (45) | 1053 (92) | 87 (8) | <.0001 | ref | - |
| Middle | 1095 (43) | 930 (85) | 165 (15) | 2.15 (1.63–2.83) | - | |
| High | 312 (12) | 258 (83) | 54 (17) | 2.53 (1.76–3.65) | - | |
| Occupation | ||||||
| Healthcare Workerg,h | ||||||
| No | 2214 (96) | 1944 (88) | 270 (12) | .0047 | ref | - |
| Yes | 84 (4) | 65 (77) | 19 (26) | 2.11 (1.24–3.56) | - | |
| Hunterg,i | ||||||
| No | 1866 (82) | 1660 (88) | 226 (12) | .0472 | ref | - |
| Yes | 411 (18) | 347 (84) | 64 (16) | 1.36 (1.00–1.83) | - | |
| Health Zone | ||||||
| Kole | 2204 (66) | 1939 (88) | 265 (12) | .0003 | 1.58 (1.23–2.03) | 1.58 (1.22–2.04) |
| Lomela | 1140 (34) | 1049 (92) | 91 (8) | ref | ref | |
| . | All Subjectsn (%) . | EBOV−n (%) . | EBOV+n (%) . | P Valuea . | Regression Analysisb . | |
|---|---|---|---|---|---|---|
| ORcrude (95% CI) . | ORadjc (95% CI) . | |||||
| Aged | ||||||
| 0–14 years | 1407 (42) | 1336 (95) | 71 (5) | <.0001 | ref | ref |
| 15–49 years | 1562 (47) | 1343 (86) | 219 (14) | 3.07 (2.32–4.05) | 3.29 (2.48–4.36) | |
| 50 + years | 362 (11) | 299 (83) | 63 (17) | 3.96 (2.76–5.69) | 4.25 (2.95–6.13) | |
| Sexd | ||||||
| Female | 1930 (58) | 1745 (90) | 185 (10) | .0233 | ref | ref |
| Male | 1404 (42) | 1235 (88) | 169 (12) | 1.29 (1.04–1.61) | 1.51 (1.20–1.89) | |
| Education | ||||||
| None | 256 (10) | 219 (86) | 37 (14) | .0001 | 1.69 (1.13–2.53) | - |
| Some primary education | 1168 (47) | 1062 (91) | 106 (9) | ref | - | |
| Completed primary education | 791 (32) | 678 (86) | 113 (14) | 1.67 (1.26–2.21) | - | |
| Completed secondary education or beyonde | 274 (11) | 228 (83) | 46 (17) | 2.02 (1.39–2.94) | - | |
| Socioeconomic statusf | ||||||
| Low | 1140 (45) | 1053 (92) | 87 (8) | <.0001 | ref | - |
| Middle | 1095 (43) | 930 (85) | 165 (15) | 2.15 (1.63–2.83) | - | |
| High | 312 (12) | 258 (83) | 54 (17) | 2.53 (1.76–3.65) | - | |
| Occupation | ||||||
| Healthcare Workerg,h | ||||||
| No | 2214 (96) | 1944 (88) | 270 (12) | .0047 | ref | - |
| Yes | 84 (4) | 65 (77) | 19 (26) | 2.11 (1.24–3.56) | - | |
| Hunterg,i | ||||||
| No | 1866 (82) | 1660 (88) | 226 (12) | .0472 | ref | - |
| Yes | 411 (18) | 347 (84) | 64 (16) | 1.36 (1.00–1.83) | - | |
| Health Zone | ||||||
| Kole | 2204 (66) | 1939 (88) | 265 (12) | .0003 | 1.58 (1.23–2.03) | 1.58 (1.22–2.04) |
| Lomela | 1140 (34) | 1049 (92) | 91 (8) | ref | ref | |
Abbreviations: CI, confidence interval; DRC, Democratic Republic of Congo; EBOV, ebolavirus; IgG, immunoglobulin G; OR, odds ratio; ref, reference; SES, socioeconomic status.
aχ2P value.
bRegression analyses presented as univariate and multivariate analyses based upon backward selection for sociodemographic predictors of seropositivity.
cAdjusted model retains all sociodemographic predictors that meet 0.05 significance level using backward selection; final model includes age, sex, and health zone.
dAge and gender were obtained from all study participants, regardless of participation in questionnaire (n = 3344).
eCategory includes completion of secondary education, apprenticeship, higher education, or university.
fSES categorization is based on calculated wealth index derived from the following reported assets: domestic animals, fields, radio, bicycle, sewing machine, and motorcycle. Each reported asset was given a value of 1 (maximum value of 6), and categories for SES are as follows: low (0 reported assets), middle (1–2 reported assets), high (≥3 reported assets).
gOccupation categories (healthcare worker and hunter) are not mutually exclusive; n = 17 fall within both occupation groups through their response for primary or secondary source of income.
hOccupation of healthcare worker was assigned if participant indicated health worker, midwife, traditional healer, Red Cross, or any other health-related job as either a primary or secondary source of income.
iOccupation of hunter was assigned if participant indicated hunting as either a primary or secondary source of income.
Frequency Distribution of Forest Visits, Animal Activities and Exposures Among Serosurvey Study Participants by EBOV Nucleoprotein IgG Results From the Sankuru Province, DRC (n = 2580)
| . | . | Negativen (%) . | Positiven (%) . | P Valuea . |
|---|---|---|---|---|
| Forest visits for regular activitiesb | No | 264 (95) | 15 (5) | .0003 |
| Yes | 1969 (87) | 290 (13) | ||
| Animal Activities | ||||
| Hunt | No | 1859 (89) | 236 (11) | .0206 |
| Yes | 412 (85) | 73 (15) | ||
| Pick up dead | No | 2076 (88) | 277 (12) | .3029 |
| Yes | 195 (86) | 32 (14) | ||
| Butcher/skin | No | 1056 (90) | 121 (10) | .0151 |
| Yes | 1215 (87) | 188 (13) | ||
| Cook | No | 664 (88) | 90 (12) | .9676 |
| Yes | 1607 (88) | 219 (12) | ||
| Eat | No | 108 (92) | 10 (8) | .2304 |
| Yes | 2163 (88) | 299 (12) | ||
| Animal Exposures | ||||
| Rodentsc | No | 207 (93) | 16 (7) | .0209 |
| Yes | 2064 (88) | 293 (12) | ||
| Duiker | No | 253 (92) | 22 (8) | .0316 |
| Yes | 2018 (88) | 287 (12) | ||
| Eutheriad | No | 517 (89) | 61 (11) | .2316 |
| Yes | 1754 (88) | 248 (12) | ||
| Nonhuman Primatee | No | 497 (90) | 57 (10) | .1673 |
| Yes | 1774 (88) | 252 (12) | ||
| Wild Bird | No | 943 (89) | 121 (11) | .4282 |
| Yes | 1328 (88) | 188 (12) | ||
| Wild Boar | No | 932 (89) | 121 (11) | .5280 |
| Yes | 1339 (88) | 188 (12) | ||
| Bat | No | 1545 (88) | 204 (12) | .4776 |
| Yes | 726 (87) | 105 (13) | ||
| Lorisidaef | No | 1588 (88) | 218 (12) | .8220 |
| Yes | 683 (88) | 91 (12) | ||
| Wild Cat | No | 1563 (88) | 212 (12) | .9387 |
| Yes | 708 (88) | 97 (12) | ||
| Reptile | No | 1.719 (88) | 225 (12) | .2708 |
| Yes | 552 (87) | 84 (13) | ||
| Elephant | No | 1815 (88) | 249 (12) | .7850 |
| Yes | 456 (88) | 60 (12) | ||
| . | . | Negativen (%) . | Positiven (%) . | P Valuea . |
|---|---|---|---|---|
| Forest visits for regular activitiesb | No | 264 (95) | 15 (5) | .0003 |
| Yes | 1969 (87) | 290 (13) | ||
| Animal Activities | ||||
| Hunt | No | 1859 (89) | 236 (11) | .0206 |
| Yes | 412 (85) | 73 (15) | ||
| Pick up dead | No | 2076 (88) | 277 (12) | .3029 |
| Yes | 195 (86) | 32 (14) | ||
| Butcher/skin | No | 1056 (90) | 121 (10) | .0151 |
| Yes | 1215 (87) | 188 (13) | ||
| Cook | No | 664 (88) | 90 (12) | .9676 |
| Yes | 1607 (88) | 219 (12) | ||
| Eat | No | 108 (92) | 10 (8) | .2304 |
| Yes | 2163 (88) | 299 (12) | ||
| Animal Exposures | ||||
| Rodentsc | No | 207 (93) | 16 (7) | .0209 |
| Yes | 2064 (88) | 293 (12) | ||
| Duiker | No | 253 (92) | 22 (8) | .0316 |
| Yes | 2018 (88) | 287 (12) | ||
| Eutheriad | No | 517 (89) | 61 (11) | .2316 |
| Yes | 1754 (88) | 248 (12) | ||
| Nonhuman Primatee | No | 497 (90) | 57 (10) | .1673 |
| Yes | 1774 (88) | 252 (12) | ||
| Wild Bird | No | 943 (89) | 121 (11) | .4282 |
| Yes | 1328 (88) | 188 (12) | ||
| Wild Boar | No | 932 (89) | 121 (11) | .5280 |
| Yes | 1339 (88) | 188 (12) | ||
| Bat | No | 1545 (88) | 204 (12) | .4776 |
| Yes | 726 (87) | 105 (13) | ||
| Lorisidaef | No | 1588 (88) | 218 (12) | .8220 |
| Yes | 683 (88) | 91 (12) | ||
| Wild Cat | No | 1563 (88) | 212 (12) | .9387 |
| Yes | 708 (88) | 97 (12) | ||
| Reptile | No | 1.719 (88) | 225 (12) | .2708 |
| Yes | 552 (87) | 84 (13) | ||
| Elephant | No | 1815 (88) | 249 (12) | .7850 |
| Yes | 456 (88) | 60 (12) | ||
Abbreviations: DRC, Democratic Republic of Congo; EBOV, ebolavirus; IgG, immunoglobulin G.
aχ2P value.
bRefers to regular activities completed in the forest in the last month including, but not limited to the following: hunting, cultivating, searching for water or wood, foraging, and fishing.
cRodent category includes reports of exposure to squirrel, African dormouse, porcupine, gambian rat, rat, and mouse.
dEutheria category includes reports of exposure to pangolin and elephant shrew.
eNonhuman primate category includes reports of exposure to Lophocebus aterrimus, Cercopithecus ascanius, Procolobus tholloni, Colobus angolensis, Cercopithecus neglectus, Cercopithecus wolfi, Cercopithecus nicitans, Cercocebus chrysogaster, Pan paniscus, and monkey (not identified).
fLorisidae category includes reports of exposure to potto and galago.
Frequency Distribution of Forest Visits, Animal Activities and Exposures Among Serosurvey Study Participants by EBOV Nucleoprotein IgG Results From the Sankuru Province, DRC (n = 2580)
| . | . | Negativen (%) . | Positiven (%) . | P Valuea . |
|---|---|---|---|---|
| Forest visits for regular activitiesb | No | 264 (95) | 15 (5) | .0003 |
| Yes | 1969 (87) | 290 (13) | ||
| Animal Activities | ||||
| Hunt | No | 1859 (89) | 236 (11) | .0206 |
| Yes | 412 (85) | 73 (15) | ||
| Pick up dead | No | 2076 (88) | 277 (12) | .3029 |
| Yes | 195 (86) | 32 (14) | ||
| Butcher/skin | No | 1056 (90) | 121 (10) | .0151 |
| Yes | 1215 (87) | 188 (13) | ||
| Cook | No | 664 (88) | 90 (12) | .9676 |
| Yes | 1607 (88) | 219 (12) | ||
| Eat | No | 108 (92) | 10 (8) | .2304 |
| Yes | 2163 (88) | 299 (12) | ||
| Animal Exposures | ||||
| Rodentsc | No | 207 (93) | 16 (7) | .0209 |
| Yes | 2064 (88) | 293 (12) | ||
| Duiker | No | 253 (92) | 22 (8) | .0316 |
| Yes | 2018 (88) | 287 (12) | ||
| Eutheriad | No | 517 (89) | 61 (11) | .2316 |
| Yes | 1754 (88) | 248 (12) | ||
| Nonhuman Primatee | No | 497 (90) | 57 (10) | .1673 |
| Yes | 1774 (88) | 252 (12) | ||
| Wild Bird | No | 943 (89) | 121 (11) | .4282 |
| Yes | 1328 (88) | 188 (12) | ||
| Wild Boar | No | 932 (89) | 121 (11) | .5280 |
| Yes | 1339 (88) | 188 (12) | ||
| Bat | No | 1545 (88) | 204 (12) | .4776 |
| Yes | 726 (87) | 105 (13) | ||
| Lorisidaef | No | 1588 (88) | 218 (12) | .8220 |
| Yes | 683 (88) | 91 (12) | ||
| Wild Cat | No | 1563 (88) | 212 (12) | .9387 |
| Yes | 708 (88) | 97 (12) | ||
| Reptile | No | 1.719 (88) | 225 (12) | .2708 |
| Yes | 552 (87) | 84 (13) | ||
| Elephant | No | 1815 (88) | 249 (12) | .7850 |
| Yes | 456 (88) | 60 (12) | ||
| . | . | Negativen (%) . | Positiven (%) . | P Valuea . |
|---|---|---|---|---|
| Forest visits for regular activitiesb | No | 264 (95) | 15 (5) | .0003 |
| Yes | 1969 (87) | 290 (13) | ||
| Animal Activities | ||||
| Hunt | No | 1859 (89) | 236 (11) | .0206 |
| Yes | 412 (85) | 73 (15) | ||
| Pick up dead | No | 2076 (88) | 277 (12) | .3029 |
| Yes | 195 (86) | 32 (14) | ||
| Butcher/skin | No | 1056 (90) | 121 (10) | .0151 |
| Yes | 1215 (87) | 188 (13) | ||
| Cook | No | 664 (88) | 90 (12) | .9676 |
| Yes | 1607 (88) | 219 (12) | ||
| Eat | No | 108 (92) | 10 (8) | .2304 |
| Yes | 2163 (88) | 299 (12) | ||
| Animal Exposures | ||||
| Rodentsc | No | 207 (93) | 16 (7) | .0209 |
| Yes | 2064 (88) | 293 (12) | ||
| Duiker | No | 253 (92) | 22 (8) | .0316 |
| Yes | 2018 (88) | 287 (12) | ||
| Eutheriad | No | 517 (89) | 61 (11) | .2316 |
| Yes | 1754 (88) | 248 (12) | ||
| Nonhuman Primatee | No | 497 (90) | 57 (10) | .1673 |
| Yes | 1774 (88) | 252 (12) | ||
| Wild Bird | No | 943 (89) | 121 (11) | .4282 |
| Yes | 1328 (88) | 188 (12) | ||
| Wild Boar | No | 932 (89) | 121 (11) | .5280 |
| Yes | 1339 (88) | 188 (12) | ||
| Bat | No | 1545 (88) | 204 (12) | .4776 |
| Yes | 726 (87) | 105 (13) | ||
| Lorisidaef | No | 1588 (88) | 218 (12) | .8220 |
| Yes | 683 (88) | 91 (12) | ||
| Wild Cat | No | 1563 (88) | 212 (12) | .9387 |
| Yes | 708 (88) | 97 (12) | ||
| Reptile | No | 1.719 (88) | 225 (12) | .2708 |
| Yes | 552 (87) | 84 (13) | ||
| Elephant | No | 1815 (88) | 249 (12) | .7850 |
| Yes | 456 (88) | 60 (12) | ||
Abbreviations: DRC, Democratic Republic of Congo; EBOV, ebolavirus; IgG, immunoglobulin G.
aχ2P value.
bRefers to regular activities completed in the forest in the last month including, but not limited to the following: hunting, cultivating, searching for water or wood, foraging, and fishing.
cRodent category includes reports of exposure to squirrel, African dormouse, porcupine, gambian rat, rat, and mouse.
dEutheria category includes reports of exposure to pangolin and elephant shrew.
eNonhuman primate category includes reports of exposure to Lophocebus aterrimus, Cercopithecus ascanius, Procolobus tholloni, Colobus angolensis, Cercopithecus neglectus, Cercopithecus wolfi, Cercopithecus nicitans, Cercocebus chrysogaster, Pan paniscus, and monkey (not identified).
fLorisidae category includes reports of exposure to potto and galago.
Ethical Approval
Ethical approval was obtained from UCLA Fielding School of Public Health and Kinshasa School of Public Health.
RESULTS
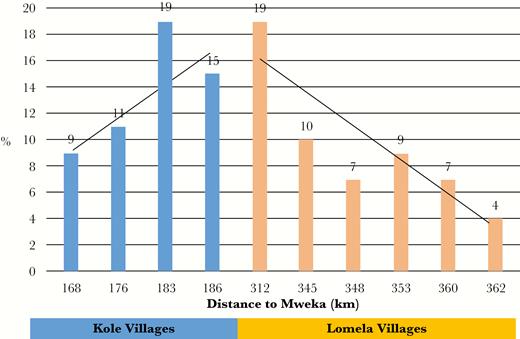
At the time of the survey, 3 outbreaks of EBOV had already occurred in the DRC (in 1976, 1977, and 1995 [Figure 1]), and there was an ongoing outbreak in neighboring Mweka and Luebo health zones (from May to November 2007) [6]. Overall, seroprevalence in the study population was 11% and was significantly associated with health zone (Table 1). In Kole, 12% of residents ≥1 year of age tested positive compared with 8% in Lomela (P = .0003). Because villages in Kole health Zone are located in closest proximity to the outbreak site (Mweka), further analysis across all villages failed to display a linear relationship between the distance from outbreak site (measured from the center of Mweka health zone to the center of the study villages) and seropositivity (P = .1352; data not shown). Indeed, seropositivity peaked at a site located between Kole and Lomela (Figure 2), suggesting that other factors may have contributed to exposure risk.
Ebolavirus (EBOV) seropositivity (%) of study site villages by distance in kilometers from Mweka, the location of the EBOV outbreak during the time of this study, Sankuru Province, Democratic Republic of Congo.
Therefore, we next assessed associations between participant sociodemographic characteristics and serology to evaluate other potential risk factors for EBOV seropositivity. To control for possible confounders, we performed multivariate regression using backward selection to identify significant sociodemographic predictors of serologic response and the final model consisted of age, sex, and health zone. We observed that increasing age, male sex, the extremes of education, increasing socioeconomic status (as measured by reported assets), and occupation (both healthcare workers and hunters compared with other primary and secondary reported occupations) were associated with a positive test result (Table 1). Among survey participants, those 15 years of age or older had over 3-times the odds of seropositivity compared with those <15 years of age, males had 1.5 times the odds of testing positive compared with females (95% confidence interval [CI], 1.2–1.9), and residents of Kole had 1.6 times the odds of seropositivity compared with those living in Lomela (95% CI, 1.2–2.0).
With the identification of significant sociodemographic predictors, we then assessed the following reported animal exposures of interest as drivers of seropositivity: visits to the forest, reported activities resulting in contact with wild animals (hunting, butchering/skinning, picking up dead, cooking and eating), and animals to which participants reported contact (for a full list, see Table 2). In univariate analysis of activities, forest visits, hunting, butchering/skinning, as well as exposure to rodents and duikers were significantly associated with seropositivity in χ2 analysis (P < .05; Table 2). The relationship between these significant independent predictors were further explored in multivariate logistic regression analysis. Controlling for age, sex, and health zone, the odds of seropositivity were significantly higher for those who entered the forest to engage in their regular activities (odds ratio [OR] = 1.75; 95% CI, 1.00–3.05) or had any exposure to rodents (OR = 1.77; 95% CI, 1.02–3.04) (Table 3). In addition, trends were observed among those that hunted (OR = 1.27; 95% CI, 0.95–1.70) and those reporting any exposure to duikers (OR = 1.59; 95% CI, 0.99–2.54).
Regression Analysis of Forest Visits and Animal Exposures for EBOV Seropositivity Among Serosurvey Respondents, Sankuru Province, DRC (n = 2580)
| . | ORcrude (95% CI) . | ORadjusteda (95% CI) . |
|---|---|---|
| Forest visits for regular activitiesa | 2.59 (1.52–4.42) | 1.75 (1.00–3.05) |
| Animal Activitiesb | ||
| Hunting | 1.40 (1.05–1.85) | 1.27 (0.95–1.70) |
| Butcher/skin | 1.35 (1.06–1.72) | 1.00 (0.77–1.29) |
| Animal Exposuresc | ||
| Rodent | 1.84 (1.09–3.10) | 1.77 (1.02–3.04) |
| Duiker | 1.64 (1.04–2.57) | 1.59 (0.99–2.54) |
| . | ORcrude (95% CI) . | ORadjusteda (95% CI) . |
|---|---|---|
| Forest visits for regular activitiesa | 2.59 (1.52–4.42) | 1.75 (1.00–3.05) |
| Animal Activitiesb | ||
| Hunting | 1.40 (1.05–1.85) | 1.27 (0.95–1.70) |
| Butcher/skin | 1.35 (1.06–1.72) | 1.00 (0.77–1.29) |
| Animal Exposuresc | ||
| Rodent | 1.84 (1.09–3.10) | 1.77 (1.02–3.04) |
| Duiker | 1.64 (1.04–2.57) | 1.59 (0.99–2.54) |
Abbreviations: CI, confidence interval; DRC, Democratic Republic of Congo; EBOV, ebolavirus; OR, odds ratio.
aAdjusted models for forest visits, butcher/skin, rodent, and duiker include the following covariates: age, sex, and health zone. Due to collinearity between hunting and sex, adjusted model for hunting includes the following covariates: age and health zone.
bThe reference group are those that did not report the activity-specific exposure of interest for any animal.
cThe reference group are those that did not report any exposure to the animals of interest.
Regression Analysis of Forest Visits and Animal Exposures for EBOV Seropositivity Among Serosurvey Respondents, Sankuru Province, DRC (n = 2580)
| . | ORcrude (95% CI) . | ORadjusteda (95% CI) . |
|---|---|---|
| Forest visits for regular activitiesa | 2.59 (1.52–4.42) | 1.75 (1.00–3.05) |
| Animal Activitiesb | ||
| Hunting | 1.40 (1.05–1.85) | 1.27 (0.95–1.70) |
| Butcher/skin | 1.35 (1.06–1.72) | 1.00 (0.77–1.29) |
| Animal Exposuresc | ||
| Rodent | 1.84 (1.09–3.10) | 1.77 (1.02–3.04) |
| Duiker | 1.64 (1.04–2.57) | 1.59 (0.99–2.54) |
| . | ORcrude (95% CI) . | ORadjusteda (95% CI) . |
|---|---|---|
| Forest visits for regular activitiesa | 2.59 (1.52–4.42) | 1.75 (1.00–3.05) |
| Animal Activitiesb | ||
| Hunting | 1.40 (1.05–1.85) | 1.27 (0.95–1.70) |
| Butcher/skin | 1.35 (1.06–1.72) | 1.00 (0.77–1.29) |
| Animal Exposuresc | ||
| Rodent | 1.84 (1.09–3.10) | 1.77 (1.02–3.04) |
| Duiker | 1.64 (1.04–2.57) | 1.59 (0.99–2.54) |
Abbreviations: CI, confidence interval; DRC, Democratic Republic of Congo; EBOV, ebolavirus; OR, odds ratio.
aAdjusted models for forest visits, butcher/skin, rodent, and duiker include the following covariates: age, sex, and health zone. Due to collinearity between hunting and sex, adjusted model for hunting includes the following covariates: age and health zone.
bThe reference group are those that did not report the activity-specific exposure of interest for any animal.
cThe reference group are those that did not report any exposure to the animals of interest.
DISCUSSION
Most Ebolavirus outbreaks have occurred in remote rural and forest or near-forest villages in central Africa where transportation and communication is difficult, and, as a result, disease surveillance is suboptimal. Because the symptoms can be nonspecific and range from flu-like symptoms to acute hemorrhagic fever, it is probable that there are many cases that go unrecognized, unreported, and attributed to other common illnesses such as malaria, typhoid, or influenza. Our analyses suggest that individuals outside known outbreak zones in central DRC may be exposed to EBOV. In this study of non-EBOV outbreak villages in central DRC, we that observed 11% of subjects tested positive for EBOV antibodies. We identified demographic factors (participants >15 years old age and male sex) that were significant, independent predictors of anti-EBOV IgG seropositivity and observed a relationship with geographic location: those residing in Kole, the health zone in closest proximity to the outbreak in Mweka and Luebo at the time of the survey in 2007, had 1.6 times the odds of a positive test result compared with subjects from Lomela. In multivariate analyses, entering the forest and reported exposure to rodents were significantly associated with EBOV seropositivity, and trends were identified for hunting as well as exposure to duikers.
Although we did not observe a linear relationship between distance from the outbreak site and antibody response, assessment of distance revealed a peak of seropositivity between Kole and Lomela health zones. Two possible explanations for this result were considered. First, most documented transmission during outbreaks occurs through human-to-human contact, so it may be that the peak area represents a shared location for activities that increase human contact. Alternatively, the peak area could harbor a natural reservoir for the virus that is circulating at low levels, which might cause subclinical infection after multiple zoonotic transmissions [35]. It also remains possible that the lack of an observed linear relationship results from the small number of villages. The closest village from the 2 health zones is approximately 150 km away from the outbreak site, thus it may be difficult to identify a possible source of Ebolavirus exposure. It is important to note that villages in these health zones lay along a major roadway, one that is also connected to the active outbreak sites. Given this information, we may have captured a more accurate representation of viral spread and outbreak burden.
In addition, in multivariate analyses of possible drivers of serologic response, reported contact to rodents was significantly associated with seropositivity, with an additional trend observed for duikers. It remains unclear whether exposure to these specific animals is truly associated with exposure to Ebolavirus or whether activities associated with contacting these animals is a proxy for some other exposure. There is some speculation in the literature as to whether rodents and/or duikers play a role in EBOV transmission either directly or as intermediate hosts [36–38]. Despite evidence pointing to fruit bats as the likely reservoir species for Ebola [6, 39], we found no association with any activities involving bats, or bat exposure itself, and serology.
A trend was also observed between hunting and serology. This activity involving wild animals, which may consist of tracking, capturing, killing, handling, and transporting, provides ample opportunities for transmission of EBOV (and other agents) via exposure to tissue and bodily fluids [40–42]. Visiting the forest was significantly associated with seropositivity as well as the following characteristics: hunting as primary or secondary occupation, residing in Kole health zone, any exposure to rodents, and hunting in general. In a previous study within the same population, extensive overlap was observed in the reporting of animal contact, both animal type and exposure activity, further complicating interpretation of these associations [43]. The associations between animal contact, hunting, and other forest activities with seropositivity suggest that intermediate hosts, possibly rodents or duikers, should be evaluated for possible evidence of EBOV infection. Although during an active outbreak human-to-human transmission represents the major mode of exposure, it remains possible that low-level exposures from animal contact could result in subclinical or mild infections that go unreported, and further study of the ecology of Ebolavirus transmission is needed.
Our study expands on previously underpowered studies that found limited to no differences between sociodemographic characteristics and EBOV seroprevalence in the DRC [15, 16]. Furthermore, another large EBOV serological survey of rural populations in Gabon described a similar EBOV-specific IgG seroprevalence of 15.3%, but no associations were found between EBOV IgG prevalence and behavioral and/or sociodemographic characteristics such as gender, age, hunting activity, or contact with specific forest animals in the study area [12]. In the Gabon study, elevated seroprevalence appeared to be associated with proximity to a forest region (19.4%). In contrast to the Gabon study, which surveyed different ecological areas, we only sampled within villages of Kole and Lomela in the DRC. These villages share similar environmental characteristics; therefore, we were unable to assess whether differences in environmental topography or forest cover are associated with variations in seroprevalence to EBOV.
The ELISA test used in the Gabon study had whole EBOV preparation as a source of antigen, and the specificity of the assay was confirmed by Western blot. This ELISA is more specific and sensitive for the detection of Ebolavirus IgG antibodies than the old indirect fluorescent antibody test [27, 44] and can detect cross-species Ebolavirus antibodies [25]. In our study, we used r-NP from EBOV as the antigen to circumvent the safety issue related to whole virus preparation and because NP is abundant in viral particles and it has a robust antigenicity [45, 46]. In addition, we used a very conservative cutoff for positivity (≥5 standard deviations from the negative control mean), which increased the specificity of the assay. Several IgG ELISA using r-NPs have been reported and showed high sensitivity and specificity in detecting Ebolavirus IgG antibodies [45, 47–50]. Therefore, the EBOV seroprevalence observed in Kole and Lomela villages represents a relatively elevated level of exposure to EBOV or to other cross-reactive Ebolavirus species in the study health zones. Given the high case-fatality proportion associated with EBOV, this seroprevalence might have assessed only the survivors and therefore underestimated the level of exposure to EBOV. Alternately, the seropositivity observed in our study could have been the result of exposures to other known or unknown Ebolavirus species of lesser pathogenicity, which may cause cross-reaction with the assay and contribute to the high seroprevalence [50].
CONCLUSIONS
Because our study took place in the forested villages of the northern Sankuru province, an area of high biologic diversity, these findings contribute to the understanding of the role animal contact and associated activities may play in EBOV transmission. Some of the nonsignificant differences observed may be real, but potential recall bias may have reduced the power to detect them. The survey took place during an active EBOV outbreak in Mweka, thus we captured the prevalence of EBOV IgG and a constellation of exposure risks in neighboring, non-EVD-affected villages during this critical time.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Author contributions. A. W. R. designed the study, supervised all aspects of study implementation, collected the data, and directed data analysis with V. H. A., who also conducted data analysis, developed figures and tables, and lead manuscript editing efforts; N. A. H. and R. H. D. assisted with data cleaning and editing; P. M., B. I. K., E. O.-W., and J.-J. M.-T. participated in study design and implementation; L. L. W. and N. K. K. participated in study design, implementation, and editing the manuscript. S. M., H. M., J. S., J. N. P., and N. J. S. conducted the laboratory assays. A. W. R., V. H. A., S. M., and N. J. S. developed figures and cowrote the manuscript.
Acknowledgments. Many individuals contributed to the design of this manuscript and data collection in the field. We thank the healthcare workers in the Sankuru Province and the Democratic Republic of Congo (DRC) Ministry of Health for their surveillance efforts and assistance with the implementation of this study. We also thank Cyrus Sinai for assisting with Figure 1 (DRC map) and Matthew Bramble for providing advice on analyses.
Financial support. Funding support for A. W. R. includes the following: the Eunice Kennedy Shriver National Institute of Child Health and Human Development Global Network for Women’s and Children’s Health Research; National Institute of Allergy and Infectious Diseases, Division of Infectious Diseases and Microbiology (Grant 5K01AI074810-05); Fogarty International Center, Research and Policy for Infectious Disease Dynamics program of the Science and Technology Directorate, Department of Homeland Security; and the Faucett Catalyst Fund. Funding support for N. J. S. includes the following: Intramural Research Program of the Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
Author notes
S. M. and V. H. A. are co-first authors and contributed equally to this work.