-
PDF
- Split View
-
Views
-
Cite
Cite
Wei-Dong Chen, Z. James Han, Joel Skoletsky, Jeff Olson, Jerome Sah, Lois Myeroff, Petra Platzer, Shilong Lu, Dawn Dawson, Joseph Willis, Theresa P. Pretlow, James Lutterbaugh, Lakshmi Kasturi, James K. V. Willson, J. Sunil Rao, Anthony Shuber, Sanford D. Markowitz, Detection in Fecal DNA of Colon Cancer–Specific Methylation of the Nonexpressed Vimentin Gene, JNCI: Journal of the National Cancer Institute, Volume 97, Issue 15, 3 August 2005, Pages 1124–1132, https://doi.org/10.1093/jnci/dji204
Close - Share Icon Share
Abstract
Background: Increased DNA methylation is an epigenetic alteration that is common in human cancers and is often associated with transcriptional silencing. Aberrantly methylated DNA has also been proposed as a potential tumor marker. However, genes such as vimentin, which are transcriptionally silent in normal epithelium, have not until now been considered as targets for cancer-associated aberrant methylation and for use as cancer markers. Methods: We applied methylation-specific polymerase chain reaction to the vimentin gene, which is transcriptionally silent in normal colonocytes, and compared methylation of vimentin exon 1 in cancer tissues and in fecal DNA from colon cancer patients versus control samples from healthy subjects. Results: Vimentin exon-1 sequences were unmethylated in 45 of 46 normal colon tissues. In contrast, vimentin exon-1 sequences were methylated in 83% (38 of 46) and 53% (57 of 107) of tumors from two independently collected groups of colon cancer patients. When evaluated as a marker for colon cancer detection in fecal DNA from another set of colon cancer patients, aberrant vimentin methylation was detected in fecal DNA from 43 of 94 patients, for a sensitivity of 46% (95% confidence interval [CI] = 35% to 56%). The sensitivity for detecting stage I and II cancers was 43% (26 of 60 case patients) (95% CI = 31% to 57%). Only 10% (20 of 198 case patients) of control fecal DNA samples from cancer-free individuals tested positive for vimentin methylation, for a specificity of 90% (95% CI = 85% to 94%). Conclusions : Aberrant methylation of exon-1 sequences within the nontranscribed vimentin gene is a novel molecular biomarker of colon cancer and can be successfully detected in fecal DNA to identify nearly half of individuals with colon cancer.
Aberrant (i.e., increased) methylation of CpG-rich sequences (CpG islands) is an epigenetic change that is common in human cancers ( 1 – 4 ) . Such CpG islands are most frequently located in the promoter regions or in untranslated first exons of human genes ( 1 – 4 ) . Most commonly, increased CpG methylation of gene promoters or first exons is associated with loss of gene transcription ( 1 – 4 ) . In human colon cancers, several genes have been identified that are commonly unmethylated and expressed in normal colon mucosa but are methylated and silenced in colon cancer ( 1 – 7 ) . There has been substantial interest in attempting to adapt such cancer-associated aberrant gene methylation for use as a marker for potential early detection of colon and other cancers ( 3 , 8 – 12 ) .
Colon cancer is the second-leading cause of cancer death in adults in the United States ( 13 ) . When these cancers are detected in early clinical stages, i.e., stages I and II, when the tumors are still confined to the bowel wall, surgical cure rates are 90% and 75%, respectively ( 14 ) . In contrast, chances for cure drop rapidly once colon tumors have spread beyond the confines of the bowel. Initial reports have confirmed the potential for early detection of colon cancer–derived aberrantly methylated DNA in both patient blood and feces, but the sensitivity and specificity of currently identified markers are not optimal ( 8 , 10 ) .
To expand the population of genomic DNA sequences that might potentially be useful as methylated DNA markers of colon cancer, we have investigated whether cancer-associated aberrant DNA methylation might target CpG-rich sequences within a gene that is not expressed by normal colonic epithelium and for which gene silencing would therefore not result from an aberrant methylation event. We chose for this approach the vimentin gene, which encodes a protein constituent of intermediate filaments and whose expression is considered a classic marker of mesenchymal cells, such as fibroblasts ( 15 ) , and which hence should not be expressed by normal colonic epithelium. We describe here the analysis of aberrant methylation of the human vimentin gene and then the assay of vimentin gene methylation as a potential marker of colon cancer in patient tumors and in fecal DNA.
M ATERIALS AND M ETHODS
Tissues, Cell Lines, and Nucleic Acid Isolation
Normal and malignant colon tissue samples were obtained from discarded tissue specimens from the department of surgical pathology at University Hospitals of Cleveland using a tissue procurement protocol approved by the University Hospitals of Cleveland internal review board. These samples included 12 samples of histologically normal colonic mucosa from individuals having resections for noncancer diagnoses (designated normal group 1) and 46 samples of histologically normal colonic mucosa from colon cancer resections (designated normal group 2), along with matching colon cancer tissue from these 46 patients (designated group A). An additional and independent set of 107 colon cancer tumor tissues (designated group B) were collected from consenting patients at the Lahey Clinic (Burlington, MA) and sent to Exact Sciences, which provided these samples for study at Case Western Reserve University. Colon cancer tumors included those arising in the proximal colon (cecum, ascending, and transverse colon), distal colon (descending and sigmoid colon), and rectum. VACO series colon cancer cell lines were established and maintained as described previously ( 16 ) . For initial screening of vimentin gene methylation the 11 cell lines studied were Vaco5, Vaco6, Vaco9m, Vaco10m, Vaco206, Vaco241, Vaco364, Vaco394, Vaco400, Vaco425, Vaco441, and Vaco576. Additional studies also employed Vaco6. RNA and DNA were prepared from colon tissues and cell lines after lysis in guanidine isothiocyanate and fractionation through cesium chloride as previously described ( 17 ) .
Immunohistochemistry
Vimentin protein expression in paraffin-embedded normal colon tissue and colon tumors were evaluated using a mouse anti-vimentin monoclonal antibody, V9 (DAKO Cytomation, Carpinteria, CA). Briefly, 5-μm sections of formalin-fixed, paraffin embedded-tissues were deparaffinized and rehydrated through graded alcohols to water. Antigen unmasking was performed by heat treatment (10 m M citrate, pH 6.0, in an 800-W microwave oven for two 5-minute cycles). Slides were incubated with the V9 anti-vimentin primary antibody at 1 : 100 dilution for 10 minutes and developed using the LSAB2 visualization system (DAKO) with 3,3′ diaminobenzidine tetrahydrochloride substrate, followed by hematoxylin counterstaining. In every analysis, longitudinally cut sections of peripheral nerve were included as a positive control and staining with preimmune mouse serum was performed as a negative control.
Preparation of Colonic Mucosa and Colonic Crypts
Colonic mucosa was prepared by blunt dissection from normal portions of colectomy resections, with tissue maintained at 4 °C throughout. To further prepare colonic crypts, which are epithelial cell-enriched, mucosal samples were cut into 2- to 3-mm strips, incubated with approximately 5 mL of Cell Recovery Solution reagent (Becton Dickinson, Franklin Lakes, NJ) per square centimeter of tissue at 4 °C with gentle rocking for 1 hour, and then passed through a large-bore pipette. Released colonic crypts were collected by low-speed centrifugation at 350 g for 5 minutes at 4 °C.
Real-time Reverse Transcription–Polymerase Chain Reaction
The vimentin transcript was amplified from the isolated RNA of normal colon and colon cancer tissues and colon cancer–derived cell lines in an iCycler instrument (BioRad Laboratories, Hercules, CA) using 400 n M of forward primer, 5′-CACGAAGAGGAAATCCGGAGC-3′, and reverse primer, 5′-CAGGGCGTCATTGTTCCG-3′, to yield a 215-bp product. Each PCR was carried out in a 25-μL volume using SybrGreen Mastermix (BioRad) for 8 minutes, 30 seconds at 95 °C, followed by 50 cycles of 95 °C for 20 seconds, 60 °C for 20 seconds, and 72 °C for 20 seconds. To directly compare vimentin expression in crypt cell preparations and in whole-colonic mucosa, vimentin transcript expression was normalized in both crypt and whole-mucosal preparations to the transcript levels of Muc2, a marker of colonocyte epithelial cell mass. Muc2 transcript was amplified using forward primer 5′-TGAAGAAGACAGAGACCCCCT-3′ and reverse primer 5′-CAGGCAGTCCTCATTGTTCTGAC-3′, spanning exons 14 and 15. The RT-PCR conditions were 50 cycles of 94 °C for 20 seconds, 60 °C for 20 seconds, and 72 °C for 20 seconds. The level of vimentin expression was determined as the ratio of vimentin to Muc2 = 2 CT vimentin − CT Muc2 , where CT vimentin is the cycle number for crossing the iCycler detection threshold in real-time PCR amplification of vimentin, and CT Muc2 is the cycle number for crossing the iCycler detection threshold in real-time PCR amplification of Muc2.
Bisulfite Conversion of Genomic DNA and MS-PCR
Bisulfite conversion of DNA was performed as described previously ( 6 , 18 ) to create a template for methylation-specific PCR (MS-PCR). Briefly, 500 ng to 2 μg of genomic DNA from each sample in a volume of 50 μL was denatured by NaOH (freshly made, final concentration, 0.2 M ) at 37 °C for 15 minutes. Next, 30 μL of 10 m M fresh hydroquinone and 520 μL of freshly prepared 3.0 M NaHSO 3 , pH 5.0 (Sigma, St. Louis, MO) were added, and the mixture was incubated at 55 °C for 16 hours. Bisulfite-modified DNA was purified using the Wizard DNA Cleanup kit (Promega, Madison, WI). The DNA was desulfonated by incubation with NaOH at a final concentration of 0.3 M at room temperature for 15 min and neutralized by adding ammonium acetate, pH 7.0, to a final concentration of 3 M . DNA was precipitated with ethanol and resuspended in distilled water to a final concentration of 5 ng/μL.
Bisulfite-treated DNA was then used as the template for MS-PCR, which was performed as described previously ( 6 , 18 ) . Briefly, 5 μL of bisulfite-converted genomic DNA served as the PCR template. The amplification was in a reaction of 25 μL containing 0.19 m M each dNTP, 1.5 m M MgCl2, 400 n M of forward and reverse primers, and 1.25 U of AmpliTaq Gold in the recommended buffer. Amplification primers and reaction conditions are provided (Supplementary Table, available at http://jncicancerspectrum.oxfordjournals.org/jnci/content/vol97issue15 ). Vimentin MS-PCR reaction #29 employed forward amplification primer 5′-TCGTTTCGAGGTTTTCGCGTTAGAGAC-3′ and reverse amplification primer 5′-CGACTAAAACTCGACCGAC TCGCGA-3′. PCR cycling parameters were as follows: hot start at 95 °C for 9 minutes, followed by 45 cycles of 95 °C (45 seconds), 70 °C (45 seconds), and 72 °C (45 seconds), then 72 °C for 10 minutes, and 10 °C to cool. For amplifications from fecal DNA, both forward and reverse MS-PCR primers were additionally extended by addition of a 5′ tag sequence 5′-GCGGTCCC-3′, which is not derived from the vimentin sequence but which provided, on the second and subsequent cycles of PCR, for more robust amplification of templates that had incorporated the PCR primers. For sequencing of bisulfite-converted DNA, products were amplified with methylation-indifferent primers and cloned into pCR2.1 TOPO TA cloning vector (Invitrogen, Carlsbad, CA); 10–15 individual clones per sample were then sequenced using an automated sequencer (Applied Biosystems, Foster City, CA).
Preparation of Fecal DNA
Stools were collected from a population (n = 198) of average-risk individuals with no prior history of colon cancers or polyps and from a population (n = 94) of colorectal cancer patients, all of whom provided written informed consent, and who represented four different medical care organizations, of which one group contributed half of the total samples studied, with the remaining three groups contributing the balance. Stool samples were frozen within 72 hours after collection and stored at −80 °C. For recovery of human DNA, whole samples were thawed at room temperature and homogenized in excess volume (1 : 7) of EXACT buffer A (EXACT Sciences, Marlborough, MA). Homogenized samples were then archived at −80 °C for an average of 12 months (range = 6–18 months). No effect of the time of sample storage on ultimate sensitivity of the MS-PCR assay was found. To reduce the risk of sample degradation, homogenates were thawed only once, at the time of processing and analysis. At that time, a 4-g stool sample equivalent of each homogenate was centrifuged to remove all particulate matter, and the supernatants were treated with 20 μL of RNase A (2.5 mg/mL) (Roche, Indianapolis, IN) and incubated at 37 °C for 1 hour. Total DNA was then precipitated (by adding 1/10 volume of 3 M NaAc and an equal volume of isopropanol), and the DNA was resuspended in 4 mL of 1× TE buffer (0.01 M Tris, pH 7.4, 0.001 M EDTA) (Pierce, Rockford, IL). Target human vimentin DNA fragments were purified from total DNA preparations by acrylamide gel–based affinity capture as previously described ( 19 ) . Total DNA yields from normal patients (median = 936 genome equivalents, range = 33–18 560 genome equivalents) and cancer patients (median = 1014 genome equivalents, range = 32–3700 genome equivalents) were similar. Total captured DNA from each sample was then subjected to bisulfite-modification and MS-PCR, and the results were analyzed in a manner blinded to patient's disease status.
Statistical Analysis
Exact 95% confidence intervals (CIs) were calculated for all estimated proportions. Clinical variables were adjusted using a logistic regression model, and two-sided P values were calculated for the log-odds ratios using a Wald-type test ( 20 ) . Comparisons were determined to be statistically significant if P <.05. MS-PCR reactions were run independently in quadruplicate on all cell line samples and in duplicate on all patient tissue samples. Due to limitations of sample amount, assays on aberrant crypt foci and on fecal DNAs were single determinations.
R ESULTS
Expression and Methylation of the Vimentin Gene in Colon Cancer Cell Lines, Colon Cancers, and Normal Colonocytes
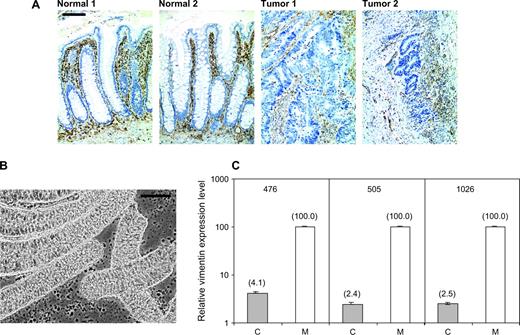
Immunohistochemical assay of vimentin expression in the human colon showed the absence of protein expression in the colonic epithelial cells in both normal colonic crypts and in colon cancers and positive vimentin expression in stromal cells and lymphocytes within both normal colonic crypts and colon cancers ( Fig. 1, A ). To confirm that the vimentin gene is transcriptionally silent in colon epithelial cells, we used real-time RT-PCR to analyze vimentin transcript levels in bluntly dissected normal colonic mucosa, which contains epithelial and stromal cells and in a purified preparation of normal colonic crypts that are highly enriched for colonic epithelial cells ( Fig. 1, B ). On average, colonic crypts retained only 3% (95% CI = 2.9% to 3.1%) of the vimentin transcript level present in the full mucosal tissue ( Fig. 1, C ), strongly suggesting that vimentin transcripts in the normal mucosal tissue are derived essentially completely from the nonepithelial cell population.
Localization of vimentin expression. A ) Immunohistochemical detection of vimentin expression ( brown ) using the monoclonal V9 anti-vimentin antibody (DAKO Cytomation, Carpinteria, CA) in tissue sections from two normal colonic mucosa specimens and two colon cancers. Comparison with a hematoxylin counterstain shows clear vimentin staining in stromal cells and lymphocytes but not in epithelial cells. Bar = 100 μm. B ) Phase-contrast microscopy of colonic crypt preparations showing the enrichment of epithelial cells compared with the colonic mucosa specimens. Bar = 100 μm. C ) Levels of vimentin transcript expression measured by real-time PCR in colonic crypt preparations ( solid bars , labeled C) and in matched colonic mucosal tissue samples ( open bars , labeled M) for three different individuals (case patients 476, 505, and 1026). For each patient, levels of vimentin expression in the mucosal tissue were set to equal 100%. Vimentin expression in mucosa and in crypt preparations was normalized to the epithelial cell mass of the tissue as assessed by the level of expression of the colonic epithelial cell marker Muc2. Graphed values are means and 95% confidence intervals of triplicate determinations. Where 95% confidence intervals are not visible, they represent a range of less than 1%.
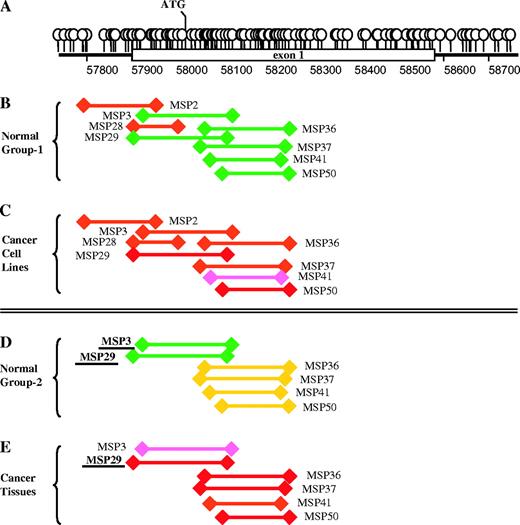
The structure of the vimentin gene demonstrates a dense region of CpG dinucleotides starting upstream of the first exon and continuing across the first two-thirds of this exon ( Fig. 2, A ). To assay this region for potential cytosine methylation, we designed a series of eight MS-PCR primer pairs that defined overlapping fragments spanning the region. These primers were initially used to assay the nonexpressed vimentin gene for DNA methylation in normal colonic mucosal samples from 12 control individuals who did not have colon cancer and in 11 colon cancer cell lines. Although the vimentin gene is transcriptionally silent in colonic mucosal epithelial cells, a 5′ portion of vimentin exon 1, defined by six overlapping MS-PCR primers, was free of detectable DNA methylation ( Fig. 2, B and Table 1 ) in either 11 or 12 of the 12 normal colonic mucosa samples assayed. In contrast, the 11 colon cancer cell lines demonstrated clear acquisition of aberrant methylation across vimentin exon 1, with different MS-PCR primer pairs detecting methylation in from eight to 10 of the 11 cell lines assayed ( Fig. 2, C and Table 1 ).
Distribution of cytosine methylation across the vimentin gene. A ) Balloons designate the distribution of CpG dinucleotides across the promoter and first exon of the vimentin gene. B ) Different vimentin gene domains that were amplified by a panel of methylation specific polymerase chain reactions (MS-PCRs) that were applied to normal colon mucosa samples from 12 control noncancer patients (Normal Group 1). C ) Using the same convention as in panel B, the results of MS-PCR assays for vimentin gene methylation in 11 colon cancer cell lines are shown. D ) The results of MS-PCR testing for vimentin gene methylation in normal colonic mucosa samples from 46 colon cancer patients. E ) The results of MS-PCR testing for vimentin gene methylation in colon tumor samples from the same 46 patients represented in panel D. The color code of the line denoting each MS-PCR indicates the percentage of case patients testing positive for methylation ( green = <10% of samples methylated, yellow = 10%–49%, pink = 50%–80%, and red = >80%.
Vimentin exon-1 methylation in normal and cancer tissues and cancer cell lines *
| Primer set . | Normal group 1 † (n = 12) . | Cancer cell lines (n = 11) . | Normal group 2 ‡ (n = 46) . | Cancer tissues (n = 46) . |
|---|---|---|---|---|
| MSP2 | 83 (52 to 98) | 91 (59 to 99) | — | — |
| MSP3 | 0 (0 to 22) | 82 (48 to 98) | 0 (0 to 6) | 63 (48 to 77) |
| MSP28 | 83 (52 to 98) | 91 (59 to 99) | — | — |
| MSP29 | 0 (0 to 22) | 82 (48 to 98) | 2 (01 to 12) | 83 (69 to 92) |
| MSP36 | 8 (0 to 39) | 82 (48 to 98) | 22 (11 to 36) | 87 (74 to 95) |
| MSP37 | 0 (0 to 22) | 82 (48 to 98) | 24 (13 to 39) | 89 (76 to 96) |
| MSP41 | 0 (0 to 22) | 73 (39 to 94) | 46 (21 to 61) | 89 (76 to 96) |
| MSP50 | 8 (0 to 39) | 82 (48 to 98) | 24 (13 to 39) | 87 (74 to 95) |
| Primer set . | Normal group 1 † (n = 12) . | Cancer cell lines (n = 11) . | Normal group 2 ‡ (n = 46) . | Cancer tissues (n = 46) . |
|---|---|---|---|---|
| MSP2 | 83 (52 to 98) | 91 (59 to 99) | — | — |
| MSP3 | 0 (0 to 22) | 82 (48 to 98) | 0 (0 to 6) | 63 (48 to 77) |
| MSP28 | 83 (52 to 98) | 91 (59 to 99) | — | — |
| MSP29 | 0 (0 to 22) | 82 (48 to 98) | 2 (01 to 12) | 83 (69 to 92) |
| MSP36 | 8 (0 to 39) | 82 (48 to 98) | 22 (11 to 36) | 87 (74 to 95) |
| MSP37 | 0 (0 to 22) | 82 (48 to 98) | 24 (13 to 39) | 89 (76 to 96) |
| MSP41 | 0 (0 to 22) | 73 (39 to 94) | 46 (21 to 61) | 89 (76 to 96) |
| MSP50 | 8 (0 to 39) | 82 (48 to 98) | 24 (13 to 39) | 87 (74 to 95) |
The percentage of subjects demonstrating vimentin gene methylation from methylation-specific polymerase chain reaction assays using the primer sets shown.
Normal group 1 = normal colon mucosal tissues from non–colon cancer patients.
Normal group 2 = matched normal colonic mucosa from colon cancer patients whose cancer tissues were assayed; — = Normal group 2 and cancer tissues groups were not assayed with primer sets 2 and 28.
Vimentin exon-1 methylation in normal and cancer tissues and cancer cell lines *
| Primer set . | Normal group 1 † (n = 12) . | Cancer cell lines (n = 11) . | Normal group 2 ‡ (n = 46) . | Cancer tissues (n = 46) . |
|---|---|---|---|---|
| MSP2 | 83 (52 to 98) | 91 (59 to 99) | — | — |
| MSP3 | 0 (0 to 22) | 82 (48 to 98) | 0 (0 to 6) | 63 (48 to 77) |
| MSP28 | 83 (52 to 98) | 91 (59 to 99) | — | — |
| MSP29 | 0 (0 to 22) | 82 (48 to 98) | 2 (01 to 12) | 83 (69 to 92) |
| MSP36 | 8 (0 to 39) | 82 (48 to 98) | 22 (11 to 36) | 87 (74 to 95) |
| MSP37 | 0 (0 to 22) | 82 (48 to 98) | 24 (13 to 39) | 89 (76 to 96) |
| MSP41 | 0 (0 to 22) | 73 (39 to 94) | 46 (21 to 61) | 89 (76 to 96) |
| MSP50 | 8 (0 to 39) | 82 (48 to 98) | 24 (13 to 39) | 87 (74 to 95) |
| Primer set . | Normal group 1 † (n = 12) . | Cancer cell lines (n = 11) . | Normal group 2 ‡ (n = 46) . | Cancer tissues (n = 46) . |
|---|---|---|---|---|
| MSP2 | 83 (52 to 98) | 91 (59 to 99) | — | — |
| MSP3 | 0 (0 to 22) | 82 (48 to 98) | 0 (0 to 6) | 63 (48 to 77) |
| MSP28 | 83 (52 to 98) | 91 (59 to 99) | — | — |
| MSP29 | 0 (0 to 22) | 82 (48 to 98) | 2 (01 to 12) | 83 (69 to 92) |
| MSP36 | 8 (0 to 39) | 82 (48 to 98) | 22 (11 to 36) | 87 (74 to 95) |
| MSP37 | 0 (0 to 22) | 82 (48 to 98) | 24 (13 to 39) | 89 (76 to 96) |
| MSP41 | 0 (0 to 22) | 73 (39 to 94) | 46 (21 to 61) | 89 (76 to 96) |
| MSP50 | 8 (0 to 39) | 82 (48 to 98) | 24 (13 to 39) | 87 (74 to 95) |
The percentage of subjects demonstrating vimentin gene methylation from methylation-specific polymerase chain reaction assays using the primer sets shown.
Normal group 1 = normal colon mucosal tissues from non–colon cancer patients.
Normal group 2 = matched normal colonic mucosa from colon cancer patients whose cancer tissues were assayed; — = Normal group 2 and cancer tissues groups were not assayed with primer sets 2 and 28.
To quantify the extent of vimentin exon 1 methylation in the 11 cancer cell lines, we prepared bisulfite-converted DNA from three of these cell lines (Vaco5, Vaco6, Vaco400). For each cell line, we sequenced vimentin exon 1 from multiple individual PCR-amplified clones and assessed whether the antecedent cytosine at each CpG site was methylated or unmethylated. In this analysis, every CpG cytosine within the target vimentin exon 1 sequence was methylated in every clone sequenced, demonstrating that in all of these cell lines this region had become essentially 100% methylated (data not shown).
Acquired Increased Vimentin Methylation in Tissues From Primary Colonic Neoplasms
MS-PCR assays for vimentin gene methylation were next used to characterize vimentin gene methylation in matched pairs of normal colonic mucosa and colon cancer tissues obtained from 46 colon cancer patients not mentioned above. In this second set of 46 normal mucosal tissue samples, MS-PCR primer sets 3 and 29 again defined a 216-bp region of vimentin exon 1 that was devoid of any detectable methylation in 45 of 46 samples assayed ( Fig. 2, D ; Table 1 ). In contrast, 83% (38 of 46) of the colon cancers from the same 46 patients had acquired increased methylation in this 216-bp region, particularly when assayed by MS-PCR primer set 29 ( Fig. 2, E , Table 1 ). Among these 46 colon cancers, acquired increased vimentin methylation was detected in 92% of cancers arising in the proximal colon (cecum, ascending and transverse colon), 67% of cancers arising in the distal colon (descending and sigmoid colon), and in 80% of cancers of the rectum ( Table 2 , group A).
Detection of aberrant vimentin exon-1 methylation in colon cancer tissue, according to tumor location and tumor stage *
| . | Group A . | . | . | Group B . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Tumor characteristic . | Sample number . | No. positive . | Positive, % . | Sample number . | No. positive . | Positive, % . | ||||
| Location | ||||||||||
| Proximal colon | 26 | 24 | 92 | 29 | 21 | 72 | ||||
| Distal colon | 15 | 10 | 67 | 29 | 13 | 45 | ||||
| Rectum | 5 | 4 | 80 | 21 | 8 | 38 | ||||
| Unknown | 0 | 0 | 0 | 28 | 15 | 54 | ||||
| Stage | ||||||||||
| I | 0 | 0 | 0 | 2 | 1 | 50 | ||||
| II | 18 | 17 | 94 | 43 | 25 | 58 | ||||
| III | 15 | 11 | 73 | 26 | 15 | 58 | ||||
| IV | 13 | 10 | 77 | 17 | 7 | 41 | ||||
| Unknown | 0 | 0 | 0 | 19 | 9 | 47 | ||||
| Total | 46 | 38 | 83 | 107 | 57 | 53 | ||||
| . | Group A . | . | . | Group B . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Tumor characteristic . | Sample number . | No. positive . | Positive, % . | Sample number . | No. positive . | Positive, % . | ||||
| Location | ||||||||||
| Proximal colon | 26 | 24 | 92 | 29 | 21 | 72 | ||||
| Distal colon | 15 | 10 | 67 | 29 | 13 | 45 | ||||
| Rectum | 5 | 4 | 80 | 21 | 8 | 38 | ||||
| Unknown | 0 | 0 | 0 | 28 | 15 | 54 | ||||
| Stage | ||||||||||
| I | 0 | 0 | 0 | 2 | 1 | 50 | ||||
| II | 18 | 17 | 94 | 43 | 25 | 58 | ||||
| III | 15 | 11 | 73 | 26 | 15 | 58 | ||||
| IV | 13 | 10 | 77 | 17 | 7 | 41 | ||||
| Unknown | 0 | 0 | 0 | 19 | 9 | 47 | ||||
| Total | 46 | 38 | 83 | 107 | 57 | 53 | ||||
Results are shown for two independent sets of colon cancers, group A (with 46 case patients) and group B (with 107 case patients).
Detection of aberrant vimentin exon-1 methylation in colon cancer tissue, according to tumor location and tumor stage *
| . | Group A . | . | . | Group B . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Tumor characteristic . | Sample number . | No. positive . | Positive, % . | Sample number . | No. positive . | Positive, % . | ||||
| Location | ||||||||||
| Proximal colon | 26 | 24 | 92 | 29 | 21 | 72 | ||||
| Distal colon | 15 | 10 | 67 | 29 | 13 | 45 | ||||
| Rectum | 5 | 4 | 80 | 21 | 8 | 38 | ||||
| Unknown | 0 | 0 | 0 | 28 | 15 | 54 | ||||
| Stage | ||||||||||
| I | 0 | 0 | 0 | 2 | 1 | 50 | ||||
| II | 18 | 17 | 94 | 43 | 25 | 58 | ||||
| III | 15 | 11 | 73 | 26 | 15 | 58 | ||||
| IV | 13 | 10 | 77 | 17 | 7 | 41 | ||||
| Unknown | 0 | 0 | 0 | 19 | 9 | 47 | ||||
| Total | 46 | 38 | 83 | 107 | 57 | 53 | ||||
| . | Group A . | . | . | Group B . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Tumor characteristic . | Sample number . | No. positive . | Positive, % . | Sample number . | No. positive . | Positive, % . | ||||
| Location | ||||||||||
| Proximal colon | 26 | 24 | 92 | 29 | 21 | 72 | ||||
| Distal colon | 15 | 10 | 67 | 29 | 13 | 45 | ||||
| Rectum | 5 | 4 | 80 | 21 | 8 | 38 | ||||
| Unknown | 0 | 0 | 0 | 28 | 15 | 54 | ||||
| Stage | ||||||||||
| I | 0 | 0 | 0 | 2 | 1 | 50 | ||||
| II | 18 | 17 | 94 | 43 | 25 | 58 | ||||
| III | 15 | 11 | 73 | 26 | 15 | 58 | ||||
| IV | 13 | 10 | 77 | 17 | 7 | 41 | ||||
| Unknown | 0 | 0 | 0 | 19 | 9 | 47 | ||||
| Total | 46 | 38 | 83 | 107 | 57 | 53 | ||||
Results are shown for two independent sets of colon cancers, group A (with 46 case patients) and group B (with 107 case patients).
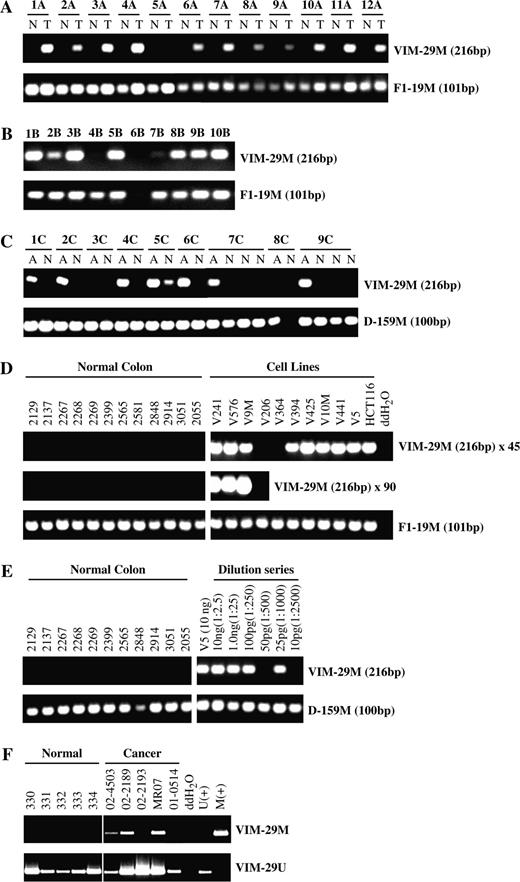
To confirm these results, we used primer set 29 to assay a second independent collection of 107 colon cancer samples. Again, a majority, 53% (57 of 107 case patients), of this second set of colon cancer case patients demonstrated aberrant vimentin methylation. In this second patient series, 72% of cancers of the proximal colon assayed positive for aberrant vimentin gene methylation, and 45% of cancers of the distal colon were methylated ( Table 2 , group B). The smaller proportion of proximal colon cancers in the second patient cohort than the first likely accounts for this series having a somewhat lower overall frequency of vimentin methylation ( Table 2 ). Also, in both series of tumors, early stage I and stage II cancers that have not spread beyond the wall of the colon showed rates of vimentin gene methylation at least equal to those of later stage III and stage IV cancers ( Table 2 ). Detection of vimentin gene methylation was technically robust, and all normal samples that tested negative for vimentin gene methylation tested positive for a constitutively methylated control (MS-PCR assay F1-19M) ( Fig. 3, A ).
Methylation-specific polymerase chain reaction (MS-PCR) detection of vimentin gene methylation in colonic neoplasia. A ) Results of MS-PCR with primer set 29 to detect vimentin gene methylation in 12 colon cancers (T) versus matched normal colon mucosa (N) from the same case patients. The upper panel shows detection of the 216-bp reaction product specific for a methylated vimentin template (VIM-29M). The lower panel shows, as a positive control, MS-PCR detection in each of these samples of a constitutively methylated domain on chromosome 16, as indicated by the 101-bp reaction product, designated F1-19M. B ) Results of MS-PCR with primer set 29 to detect vimentin gene methylation in a cohort of 10 advanced colon adenomas, of size equal or greater than 1 cm. C ) Results of MS-PCR with primer set 29 to detect vimentin gene methylation in a cohort of nine aberrant crypt foci (A) from six different individuals compared with histologically normal mucosa from the same person (N). The lower panel shows detection in these samples of a control constitutively methylated domain on chromosome 10 as indicated by the 100-bp reaction product designated D-159M. D ) Evaluation of the technical performance of vimentin MS-PCR primer set 29. The upper panel shows detection of vimentin methylation by amplification of the 216-bp reaction product specific for methylated vimentin (VIM-29M). Amplifications were performed for 45 cycles. The middle panel shows results of transferring an aliquot of the above reactions to a fresh 45-cycle PCR. The lower panel shows MS-PCR detection in all samples of a constitutively methylated domain F1-19M (positive control). Numbers denote sample numbers of colon cancer cell lines and normal colon tissues. E ) Detection of vimentin methylation by MS-PCR primer set 29 in 10 ng of genomic DNA from colon cancer cell line Vaco5 (V5) and in a series of dilutions of Vaco5 genomic DNA into control, unmethylated normal colon DNA. Reactions contain 25 ng of control unmethylated DNA from normal colon and from 10 ng to 10 pg of methylated Vaco5 DNA. Numbers indicate the amount of methylated Vaco5 DNA and the ratio of Vaco5 DNA relative to the total DNA in each reaction. The lower panel shows detection in all the samples of a constitutively methylated domain D-159M (positive control). F ) Detection of vimentin methylation in fecal DNA analyzed by MS-PCR primer set 29. The upper panel (VIM-29M) shows assay of vimentin gene methylation in fecal DNA samples from five normal and five colon cancer patients. The lower panel (VIM-29U) shows control amplification in all samples of wild-type unmethylated vimentin sequences derived from normal cells. Additional assay controls include a negative water blank and control methylated (M+) and unmethylated (U+) vimentin DNA samples.
To determine the timing during colon carcinogenesis of acquisition of vimentin gene methylation, we next used MS-PCR primer set 29 to test a set of 10 colonic adenomas of 1 cm or greater in size. Of these 10 adenoma lesions, seven were positive for vimentin gene methylation ( Fig. 3, B ). We therefore used MS-PCR primer set 29 to assay DNA extracted from aberrant crypt foci, the microscopic lesions that are recognized as the earliest morphologic abnormality of the colonic mucosa ( 21 ) . Of nine aberrant crypt foci obtained from colons of six different individuals, seven aberrant crypts from five different individuals were positive for vimentin gene methylation ( Fig. 3, C ). In contrast, only one of 14 microdissected regions of histologically normal colon from these individuals tested positive for aberrant vimentin methylation ( Fig. 3, C ).
Sensitivity of Detecting Aberrant Vimentin Methylation
To evaluate the potential use of increased vimentin gene methylation as a cancer biomarker, we tested the technical limits to the sensitivity of detecting DNA methylation by primer set 29. This primer set robustly detected vimentin methylation in colon cancer cell lines but not in normal colonic mucosa obtained from control noncancer colon resections ( Fig. 3, D ). Indeed, DNA from normal colonic mucosa remained negative in this assay, even after subjecting an aliquot of the MS-PCR to a second round of PCR amplification (i.e., 90 cycles total) ( Fig. 3, D ). Moreover, when DNA from a methylated colon cancer cell line was diluted into DNA from normal colon mucosa, the MS-PCR could detect as little as 25–50 pg of input methylated DNA, even in the presence of a 500- to 1000-fold excess of control normal mucosal DNA ( Fig. 3, E ). This amount of DNA corresponds to a detection limit for the assay of approximately 15 methylated cells.
Detection of Vimentin Methylation in Fecal DNA of Colon Cancer Patients
We next evaluated the ability of MS-PCR primer set 29 to perform as a diagnostic marker for detection of colon cancer by testing its ability to detect aberrant vimentin exon 1 methylation in fecal DNA samples prepared from the stools of 94 additional colorectal cancer patients. Fecal DNAs from 43 of these 94 patients tested positive for vimentin methylation in this assay, yielding a 46% clinical sensitivity for detecting the presence of a colon cancer (95% CI = 36% to 56%) ( Table 3 ). To evaluate the clinical specificity of this assay, we next analyzed fecal DNA from stool samples of 198 control individuals, all of whom were negative for colon cancer on colonoscopic exam. Only 20 samples (10%) tested positive for methylation of vimentin exon 1, for a clinical specificity for the assay of 90% (95% CI = 85% to 94%). We also examined sensitivity for earlier and later stage cancers. The assay had a 43% sensitivity among case patients with early ( 14 ) (i.e., stage I and II) colon cancer (26 of 60 samples tested positive) (95% CI = 31% to 57%) and 50% sensitivity among case patients with later ( 14 ) stage III and IV colon cancer (17 of 34 samples tested positive) (95% CI = 32% to 68%) ( Table 3 ).
Detection of vimentin exon-1 methylation in fecal DNA, according to tumor location and tumor stage *
| . | Cancer . | . | . | Normal . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Tumor characteristic . | Total, N . | No. of positive, n . | Percent positive, % . | Total, N . | No. of positive, n . | Percent positive, % . | ||||
| All cases | 94 | 43 | 46 | 198 | 20 | 10 | ||||
| Stage I and II | 60 | 26 | 43 | 0 | 0 | 0 | ||||
| Stage III and IV | 34 | 17 | 50 | 0 | 0 | 0 | ||||
| Proximal colon | 28 | 13 | 46 | 0 | 0 | 0 | ||||
| Distal colon | 60 | 27 | 45 | 0 | 0 | 0 | ||||
| . | Cancer . | . | . | Normal . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Tumor characteristic . | Total, N . | No. of positive, n . | Percent positive, % . | Total, N . | No. of positive, n . | Percent positive, % . | ||||
| All cases | 94 | 43 | 46 | 198 | 20 | 10 | ||||
| Stage I and II | 60 | 26 | 43 | 0 | 0 | 0 | ||||
| Stage III and IV | 34 | 17 | 50 | 0 | 0 | 0 | ||||
| Proximal colon | 28 | 13 | 46 | 0 | 0 | 0 | ||||
| Distal colon | 60 | 27 | 45 | 0 | 0 | 0 | ||||
The percentage of cancer and noncancer control individuals (normal) in whom fecal DNA tested positive for vimentin exon 1 methylation using methylation-specific polymerase chain reaction primer set 29. For six cases, tumor location in the colon was not designated.
Detection of vimentin exon-1 methylation in fecal DNA, according to tumor location and tumor stage *
| . | Cancer . | . | . | Normal . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Tumor characteristic . | Total, N . | No. of positive, n . | Percent positive, % . | Total, N . | No. of positive, n . | Percent positive, % . | ||||
| All cases | 94 | 43 | 46 | 198 | 20 | 10 | ||||
| Stage I and II | 60 | 26 | 43 | 0 | 0 | 0 | ||||
| Stage III and IV | 34 | 17 | 50 | 0 | 0 | 0 | ||||
| Proximal colon | 28 | 13 | 46 | 0 | 0 | 0 | ||||
| Distal colon | 60 | 27 | 45 | 0 | 0 | 0 | ||||
| . | Cancer . | . | . | Normal . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Tumor characteristic . | Total, N . | No. of positive, n . | Percent positive, % . | Total, N . | No. of positive, n . | Percent positive, % . | ||||
| All cases | 94 | 43 | 46 | 198 | 20 | 10 | ||||
| Stage I and II | 60 | 26 | 43 | 0 | 0 | 0 | ||||
| Stage III and IV | 34 | 17 | 50 | 0 | 0 | 0 | ||||
| Proximal colon | 28 | 13 | 46 | 0 | 0 | 0 | ||||
| Distal colon | 60 | 27 | 45 | 0 | 0 | 0 | ||||
The percentage of cancer and noncancer control individuals (normal) in whom fecal DNA tested positive for vimentin exon 1 methylation using methylation-specific polymerase chain reaction primer set 29. For six cases, tumor location in the colon was not designated.
Moreover, detection of vimentin exon 1 methylation in fecal DNA with the primer set 29 assay was equally sensitive for detecting colon cancers arising proximal to the splenic flexure (46% sensitivity) and those arising distal to the splenic flexure (45% sensitivity) ( Table 3 ). Colon cancer patients with positive fecal DNA tests were not statistically significantly different from those with negative tests with respect to age or sex ( Table 4 ). Similarly, among noncancer control subjects, individuals with false-positive fecal DNA tests were not statistically significantly different from those with negative tests with respect to age, sex, or history of colon cancer in a first-degree relative ( Table 4 ). False-positive fecal DNA tests were somewhat more common among control patients with hyperplastic or adenomatous polyps than among those with no evidence of polyps, but these differences were not statistically significant ( Table 4 ).
Study participant characteristics
| Characteristic . | Methylation (+) . | Methylation (−) . | P* . |
|---|---|---|---|
| Normal control (n = 198) | |||
| Mean Age, y (SD) | 68.6 (7.4) | 65.5 (7.3) | .13 |
| Males | 8 | 80 | .69 |
| Females | 12 | 98 | |
| (+) Family History | 4 | 25 | .54 |
| (−) Family History | 16 | 153 | |
| Clean colon | 8 | 99 | (Referent) |
| Hyperplastic polyps | 6 | 23 | .08 |
| Adenomas | 6 | 44 | .55 |
| Other | 0 | 12 | .99 |
| Cancer Patients (n = 94) | |||
| Mean age, y (SD) | 64.3 (12.3) | 66.8 (12.4) | .33 |
| Males | 22 | 25 | .81 |
| Females | 21 | 26 |
| Characteristic . | Methylation (+) . | Methylation (−) . | P* . |
|---|---|---|---|
| Normal control (n = 198) | |||
| Mean Age, y (SD) | 68.6 (7.4) | 65.5 (7.3) | .13 |
| Males | 8 | 80 | .69 |
| Females | 12 | 98 | |
| (+) Family History | 4 | 25 | .54 |
| (−) Family History | 16 | 153 | |
| Clean colon | 8 | 99 | (Referent) |
| Hyperplastic polyps | 6 | 23 | .08 |
| Adenomas | 6 | 44 | .55 |
| Other | 0 | 12 | .99 |
| Cancer Patients (n = 94) | |||
| Mean age, y (SD) | 64.3 (12.3) | 66.8 (12.4) | .33 |
| Males | 22 | 25 | .81 |
| Females | 21 | 26 |
P values (two-sided) were calculated using a Wald-type test. SD = standard deviation.
Study participant characteristics
| Characteristic . | Methylation (+) . | Methylation (−) . | P* . |
|---|---|---|---|
| Normal control (n = 198) | |||
| Mean Age, y (SD) | 68.6 (7.4) | 65.5 (7.3) | .13 |
| Males | 8 | 80 | .69 |
| Females | 12 | 98 | |
| (+) Family History | 4 | 25 | .54 |
| (−) Family History | 16 | 153 | |
| Clean colon | 8 | 99 | (Referent) |
| Hyperplastic polyps | 6 | 23 | .08 |
| Adenomas | 6 | 44 | .55 |
| Other | 0 | 12 | .99 |
| Cancer Patients (n = 94) | |||
| Mean age, y (SD) | 64.3 (12.3) | 66.8 (12.4) | .33 |
| Males | 22 | 25 | .81 |
| Females | 21 | 26 |
| Characteristic . | Methylation (+) . | Methylation (−) . | P* . |
|---|---|---|---|
| Normal control (n = 198) | |||
| Mean Age, y (SD) | 68.6 (7.4) | 65.5 (7.3) | .13 |
| Males | 8 | 80 | .69 |
| Females | 12 | 98 | |
| (+) Family History | 4 | 25 | .54 |
| (−) Family History | 16 | 153 | |
| Clean colon | 8 | 99 | (Referent) |
| Hyperplastic polyps | 6 | 23 | .08 |
| Adenomas | 6 | 44 | .55 |
| Other | 0 | 12 | .99 |
| Cancer Patients (n = 94) | |||
| Mean age, y (SD) | 64.3 (12.3) | 66.8 (12.4) | .33 |
| Males | 22 | 25 | .81 |
| Females | 21 | 26 |
P values (two-sided) were calculated using a Wald-type test. SD = standard deviation.
The fecal DNA samples used in this study were derived from four different medical care organizations, of which one group contributed half of the total samples studied and the remaining three groups contributed the balance of the samples. However, the sensitivity for detection of colon cancer by assay of vimentin gene methylation in fecal DNA was essentially identical for case patients from the largest donor site as for the case patients contributed by the remaining three sites combined (data not shown). Thus, our findings appear to be reproducible in at least two different patient cohorts.
D ISCUSSION
This study has defined a DNA sequence within vimentin exon 1 that is commonly targeted for aberrant DNA methylation by human colon cancers, particularly those arising in the proximal colon. This aberrantly methylated exon was readily detected in fecal DNA as a biomarker that detected the presence of colon cancer in nearly half of colon cancer patients. Moreover, testing for vimentin gene methylation in fecal DNA showed a specificity of 90%.
Although screening for colon cancer is recommended for all average-risk adults age 50 years and older ( 22 , 23 ) , only a minority of the population has demonstrated acceptance of invasive endoscopic-based screening ( 24 ) . Noninvasive colon cancer screening by testing feces for the presence of occult blood has in one recent large study shown only a 14% sensitivity ( 25 ) . Thus, molecular assays of fecal DNA for detection of cancer-specific DNA alterations have been proposed as a new approach for screening and detecting early-stage colon cancer ( 9 , 26 , 27 ) . Our finding of 46% sensitivity with 90% specificity for colon cancer detection by assay of fecal DNA for aberrant vimentin exon 1 methylation compares favorably both with testing for fecal occult blood [14% sensitivity and 95% specificity in one recent study ( 8 , 25 , 28 – 31 ) ] and with previous testing of fecal DNA for other DNA biomarkers tested either individually or in combinations ( 8 , 25 , 28 – 31 ) [52% sensitivity and 94% specificity for a combination panel of such markers ( 8 , 25 , 28 – 31 ) ]. Thus, assay of fecal DNA using vimentin MS-PCR primer pair 29 may have potential clinical utility for colon cancer detection.
Previous investigators have shown that methylated SFRP2 DNA can be detected in the fecal DNA of colon cancer patients with a sensitivity of 77% and a specificity of 77% ( 8 ) . The 90% specificity for the assay of vimentin methylation may make this assay particularly well suited for inclusion in panels that test for multiple colon cancer–associated mutation or DNA methylation events, in which the challenge is to preserve assay specificity. We have found that the sensitivity of hybrid capture of individual DNA targets from feces is fully maintained in such panels (A.S., unpublished data). Examples of other colon cancer–specific DNA changes that have been detected in fecal DNA include mutations in APC, K-Ras, p53, BAT-26, and the presence of long amplifiable DNA ( 26 , 28 – 32 ) . Recent findings of a large prospective trial demonstrated that assessing fecal DNA with panel of these markers achieved 52% sensitivity and 94% specificity for colon cancer detection ( 25 ) . The 46% sensitivity and 90% specificity of the single assay of fecal DNA for vimentin gene methylation suggests that the further incorporation of this assay into such a marker panel may provide one route for the advancement of this approach.
Our findings of 46% sensitivity for detection of vimentin methylation in stool compare favorably with the estimate of 53% occurrence of vimentin exon methylation noted in our 107–tumor sample validation set and is consistent with our observation that the assay for vimentin methylation is technically highly robust. However, the finding of 10% false-positive assays in fecal DNA is somewhat higher than the 2% false-positive rate that we noted in testing normal colon tissues ( Fig. 2, D ; Table 1 ). This may in part reflect the larger sample size of the 198 fecal DNA preparations obtained from normal control individuals compared with the set of 58 normal colon mucosa tissues that we initially examined ( Tables 1 and 2 ). However, it is also possible that the higher rate of false-positive tests in the fecal DNA samples reflects a better representation of the total colonic load of vimentin methylation within aberrant crypt foci by fecal DNA collections than by individual tissue samples.
Because the vimentin gene is not expressed by normal colonic epithelial cells, aberrant vimentin gene methylation at exon 1 would not be expected to alter vimentin gene expression or to confer a selective advantage upon colon cancer cells. Thus, we hypothesize that some structural feature of the vimentin exon 1 region must make this a highly favorable target for aberrant methylation early during colon neoplasia. This finding may thus illustrate the use of this broader class of potentially methylated DNA sequences to be developed as robust biomarkers of different cancers.
The detection of aberrant vimentin exon 1 methylation in human fecal DNA identified nearly half of all colon cancer patients in our study with 90% specificity. Given this initial promise, we will now be using this assay in prospective studies as part of a multicenter trial aimed at defining an optimal biomarker panel for colon cancer detection by molecular testing of fecal DNA.
Supported by PHS grants R01 CA67409 (SDM), R25T CA094186 (PP), R01 CA66725 (TPP), P30CA43703 to the CWRU/Ireland Comprehensive Cancer Center; by gift from the National Colon Cancer Research Alliance (SDM); and by grant from the State of Ohio Biomedical Research and Technology Transfer Commission (BRTT) (SDM). The content reflects the views of the Grantee and does not necessarily reflect the views of the BRTT. SDM is an investigator of the Howard Hughes Medical Institute.
We thank Hui Li for helpful scientific discussion and Kory Thornburg and Pam Shaw for helpful technical assistance. Portions of the technology presented in this article are the subject of commercial discussions between Exact Sciences and Case Western Reserve University. J. Olsen and A. P. Shuber are employees and stockholders of Exact Sciences.
References
Baylin S. Mechanisms underlying epigenetically mediated gene silencing in cancer.
Laird PW. The power and the promise of DNA methylation markers.
Issa JP. The epigenetics of colorectal cancer.
Veigl M, Kasturi L, Olechnowicz J, Ma A, Lutterbaugh J, Periyasamy S, et al. Biallelic inactivation of hMLH1 by epigenetic gene silencing, a novel mechanism causing human MSI cancers.
Moinova HR, Chen WD, Shen L, Smiraglia D, Olechnowicz J, Ravi L, et al. HLTF gene silencing in human colon cancer.
Li H, Myeroff L, Smiraglia D, Romero MF, Pretlow TP, Lakshmi Kasturi L, et al. SLC5A8: A novel sodium transporter, is a tumor suppressor gene silenced by methylation in human colonic aberrant crypt foci and colon cancers.
Muller HM, Oberwalder M, Fiegl H, Morandell M, Goebel G, Zitt M, et al. Methylation changes in faecal DNA: a marker for colorectal cancer screening?
Deenadayalu VP, Rex DK. Fecal-based DNA assays: a new, noninvasive approach to colorectal cancer screening.
Grady WM, Rajput A, Lutterbaugh JD, Markowitz SD. Detection of aberrantly methylated hMLH1 promoter DNA in the serum of patients with microsatellite unstable colon cancer.
Esteller M, Sanchez-Cespedes M, Rosell R, Sidransky D, Baylin SB, Herman JG. Detection of aberrant promoter hypermethylation of tumor suppressor genes in serum DNA from non-small cell lung cancer patients.
Sanchez-Cespedes M, Esteller M, Wu L, Nawroz-Danish H, Yoo GH, Koch WM, et al. Gene promoter hypermethylation in tumors and serum of head and neck cancer patients.
Jemal A, Murray T, Ward E, Samuels A, Tiwari RC, Ghafoor A, et al. Cancer statistics, 2005.
Markowitz S, Dawson DM, Willis J, Willson JK. Focus on colon cancer.
Lodish H, Baltimore D, Berk A, Zipursky SL, Mastudaira P, Darnell J. Molecular cell biology. 3rd ed. New York (NY): Scientific American Books;
Willson J, Bittner G, Oberley T, Meisner G, Weese J. Cell culture of human colon adenomas and carcinomas.
Markowitz S, Wang J, Myeroff L, Parsons R, Sun L, Lutterbaugh J, et al. Inactivation of the type II TGF-b receptor in colon cancer cells with microsatellite instability.
Herman J, Graff J, Myohanen S, Nelkin B, Baylin S. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands.
Whitney D, Skoletsky J, Moore K, Boynton K, Kann L, Brand R, et al. Enhanced retrieval of DNA from human fecal samples results in improved performance of colorectal cancer screening test.
Harrell F. Regression modeling strategies: with applications to linear modeling, logistic regression and survival analysis. New York (NY): Springer;
Pretlow TP, Barrow BJ, Ashton WS, O'Riordan MA, Pretlow TG, Jurcisek JA, et al. Aberrant crypts: putative preneoplastic foci in human colonic mucosa.
Smith RA, Cokkinides V, von Eschenbach AC, Levin B, Cohen C, Runowicz CD, et al. American Cancer Society guidelines for the early detection of cancer.
Winawer S, Fletcher R, Rex D, Bond J, Burt R, Ferrucci J, et al. Colorectal cancer screening and surveillance: clinical guidelines and rationale-Update based on new evidence.
Vernon SW. Participation in colorectal cancer screening: a review.
Imperiale TF, Ransohoff DF, Itzkowitz SH, Turnbull BA, Ross ME. Fecal DNA versus fecal occult blood for colorectal-cancer screening in an average-risk population.
Sidransky D, Tokino T, Hamilton S, Kinzler K, Levin B, Frost P, et al. Identification of ras oncogene mutations in the stool of patients with curable colorectal tumors.
Ahlquist DA, Shuber AP. Stool screening for colorectal cancer: evolution from occult blood to molecular markers.
Traverso G, Shuber A, Levin B, Johnson C, Olsson L, Schoetz DJ Jr, et al. Detection of APC mutations in fecal DNA from patients with colorectal tumors.
Traverso G, Shuber A, Olsson L, Levin B, Johnson C, Hamilton SR, et al. Detection of proximal colorectal cancers through analysis of faecal DNA.
Ahlquist DA, Skoletsky JE, Boynton KA, Harrington JJ, Mahoney DW, Pierceall WE, et al. Colorectal cancer screening by detection of altered human DNA in stool: feasibility of a multitarget assay panel.
Dong SM, Traverso G, Johnson C, Geng L, Favis R, Boynton K, et al. Detecting colorectal cancer in stool with the use of multiple genetic targets.