-
PDF
- Split View
-
Views
-
Cite
Cite
Suzette K. Mouchaty, Anette Gullberg, Axel Janke, Ulfur Arnason, The Phylogenetic Position of the Talpidae Within Eutheria Based on Analysis of Complete Mitochondrial Sequences, Molecular Biology and Evolution, Volume 17, Issue 1, January 2000, Pages 60–067, https://doi.org/10.1093/oxfordjournals.molbev.a026238
Close - Share Icon Share
Abstract
The complete mitochondrial (mt) genome of the mole Talpa europaea was sequenced and included in phylogenetic analyses together with another lipotyphlan (insectivore) species, the hedgehog Erinaceus europaeus, and 22 other eutherian species plus three outgroup taxa (two marsupials and a monotreme). The phylogenetic analyses reconstructed a sister group relationship between the mole and the fruit bat Artibeus jamaicensis (order Chiroptera). The Talpa/Artibeus clade constitutes a sister clade of the cetferungulates, a clade including Cetacea, Artiodactyla, Perissodactyla, and Carnivora. A monophyletic relationship between the hedgehog and the mole was significantly rejected by maximum parsimony and maximum likelihood. Consistent with current systematic schemes, analyses of complete cytochrome b genes including the shrew Sorex araneus (family Soricidae) revealed a close relationship between Talpidae and Soricidae. The analyses of complete mtDNAs, along with the findings of other insectivore studies, challenge the maintenance of the order Lipotyphla as a taxonomic unit and support the elevation of the Soricomorpha (with the families Talpidae and Soricidae and possibly also the Solenodontidae and Tenrecidae) to the level of an order, as previously proposed in some morphological studies.
Introduction
According to most current taxonomic schemes (e.g., MacPhee and Novacek 1993 ), the insectivore order Lipotyphla includes three suborders: Erinaceomorpha, with the family Erinaceidae (hedgehogs, gymnures); Chrysochloromorpha, with the family Chrysochloridae (golden moles); and Soricomorpha, with the families Solenodontidae (solenodons), Tenrecidae (Madagascan tenrecs, African water shrews), Soricidae (shrews), and Talpidae (moles, shrew-moles, desmans). Some taxonomists (e.g., McKenna and Bell 1997 ) have retained the same basic arrangement but ranked the six families as orders, elevating the Lipotyphla to the superordinal level.
Based on detailed morphological analysis, Butler (1988) concluded that monophyly of Lipotyphla was supported by six shared derived characters (synapomorphies), but only two of these characters (hindgut simplification, reduction of the pubic symphysis) were considered synapomorphic by MacPhee and Novacek (1993) . Although relationships among lipotyphlan families have been uncertain because of the morphological diversity of the group (Butler 1972 ), morphological and paleontological evidence is commonly interpreted as supporting two relationships: (1) ancient origin and clear morphological distinction of the Erinaceidae and (2) a close relationship between Talpidae and Soricidae (Butler 1988; Carroll 1988 ; MacPhee and Novacek 1993 ).
The relationship of the Lipotyphla to the other orders of eutherian mammals has remained a major problem in mammalian phylogeny (Butler 1972 ; Novacek 1992 ). The combination of numerous plesiomorphic “primitive” traits and specialized adaptations of the lipotyphlans has confounded attempts to conclusively determine the phylogenetic position of Lipotyphla by morphological comparison, and recent cladistic analyses of morphological data have left Lipotyphla in an unresolved polytomy near the base of the eutherian tree (Novacek, Wyss, and McKenna 1988 ; Novacek 1992 ). Phylogenetic analyses of large mitochondrial data sets support an early divergence of the hedgehog lineage (Erinaceidae) from the rest of Eutheria (Krettek, Gullberg, and Arnason 1995 ; D’Erchia et al. 1996 ; Janke, Xu, and Arnason 1997 ; Penny and Hasegawa 1997 ). However, resolution of the phylogenetic position of Lipotyphla as a whole in these analyses, with the hedgehog as its only representative, depends on the monophyly of Lipotyphla as a taxonomic unit.
Although recent morphological reviews (Butler 1988; MacPhee and Novacek 1993 ) concur on the composition of Lipotyphla, recent molecular studies (Lavergne et al. 1996 ; Springer et al. 1997 ; Stanhope et al. 1998 ) have indicated that Lipotyphla are polyphyletic, with the tenrecs and golden moles being members of a clade composed mainly of taxa endemic to the African continent, to the exclusion of the non-African lipotyphlan families. An important difference between the results of Stanhope et al. (1998) , which were based on nuclear genes and combined trees, and results based on large mitochondrial data sets (Krettek, Gullberg, and Arnason 1995 ; D’Erchia et al. 1996 ; Arnason, Gullberg, and Janke 1997 ; Janke, Xu, and Arnason 1997 ) is the position of this non-African lipotyphlan clade. Stanhope et al. (1988) found that the mole and the hedgehog formed the sister group to Artiodactyla (represented by the cow), while the mitochondrial analyses have consistently placed the hedgehog as the outgroup to all other eutherians.
In this study, we determined the sequence of the complete mtDNA genome of the mole Talpa europaea (family Talpidae) and reconstructed the phylogenetic relationship of the mole to the hedgehog and 22 other eutherian taxa. We also sequenced the complete mitochondrial cytochrome b gene of the shrew Sorex araneus (Soricidae) in order to establish the affinities between the families Talpidae and Soricidae.
Materials and Methods
An enriched mtDNA fraction was isolated from liver and kidney samples of T. europaea following the procedure described in Arnason, Gullberg, and Widegren (1991) . The specimen was collected in Dalby, Sweden. The DNA was digested separately with BclI, EcoRI, HindIII, SpeI, and XhoII and cloned in the phage vectors M13mp18 or M13mp19. Portions of the 12S rRNA and NADH5 genes were amplified by PCR prior to cloning. MtDNA was isolated from a shrew (S. araneus) collected in Madesjö, near Nybro, Sweden, by Maarit Jaarola (specimen 5590). The cytochrome b gene of the shrew was amplified by PCR and then cloned. All sequencing was performed on single-stranded DNA with the dideoxy method using 35S-dATP and both universal and specific oligonucleotide primers. PCR-derived sequences represent the consensus of three different clones. The mitochondrial genome sequence of the mole has been deposited under accession number Y19192, while the cytochrome b sequence of the shrew has accession number AJ245893. Users of these sequences are kindly requested to refer to the present paper in addition to the accession numbers.
The mole was analyzed along with the following species: the platypus Ornithorhynchus anatinus (Janke et al. 1996 ); the opossum Didelphis virginiana (Janke et al. 1994 ); the wallaroo Macropus robustus (Janke, Xu, and Arnason 1997 ); the hedgehog Erinaceus europaeus (Krettek, Gullberg, and Arnason 1995 ); the mouse Mus musculus (Bibb et al. 1981 ); the rat Rattus norvegicus (Gadaleta et al. 1989 ); the guinea pig Cavia porcellus (D’Erchia et al. 1996 ); the rabbit Oryctolagus cuniculus (Gissi, Gullberg, and Arnason 1998 ); the gibbon Hylobates lar (Arnason, Gullberg, and Xu 1996 ); the human, Homo sapiens (Arnason, Xu, and Gullberg 1996 ); the aardvark Orycteropus afer (Arnason, Gullberg, and Janke 1999 ); the armadillo Dasypus novemcinctus (Arnason, Gullberg, and Janke 1997 ); the fruit bat Artibeus jamaicensis (Pumo et al. 1998 ); the harbor seal Phoca vitulina (Arnason and Johnsson 1992 ); the dog Canis familiaris (Kim et al. 1998 ); the domestic cat Felis catus (Lopez et al. 1996 ); the horse Equus caballus (Xu and Arnason 1994 ); the donkey E. asinus (Xu, Gullberg, and Arnason 1996 ); the Indian rhinoceros Rhinoceros unicornis (Xu, Janke, and Arnason 1996 ) the white rhinoceros Ceratotherium simum (Xu and Arnason 1997 ); the pig Sus scrofa (Ursing and Arnason 1998a ); the cow Bos taurus (Anderson et al. 1982 ); the sheep Ovis aries (Hiendleder et al. 1998 ); the hippopotamus, Hippopotamus amphibius (Ursing and Arnason 1998b ); the fin whale, Balaenoptera physalus (Arnason, Gullberg, and Widegren 1991 ); and the blue whale, Balaenoptera musculus (Arnason and Gullberg 1993 ).
The phylogenetic analyses were based on the concatenated sequences of 12 mitochondrial protein-coding genes, excluding the L-strand-encoded NADH6 gene, the composition of which deviates from that of the H-strand-encoded genes. After removing gaps and ambiguous sites adjacent to gaps, the resulting alignment contained 9,870 nt, corresponding to 3,290 amino acids (aa). The analyses were based on both amino acid and nucleotide sequences of first plus second codon positions, with the first positions of leucine codons coded as Y (pyrimidine). Three different methods of phylogenetic reconstruction were used: maximum parsimony (MP; Fitch 1971 ), neighbor joining (NJ; Saitou and Nei 1987 ), and maximum likelihood (ML; Felsenstein 1981 ). The PUZZLE, version 4.01 (Strimmer and von Haeseler 1996 ), and MOLPHY (Adachi and Hasegawa 1996a) programs were used for ML analyses, in which we applied the mtREV-24 rate matrix (Adachi and Hasegawa 1996b ) and the HKY model of sequence evolution (Hasegawa, Kishino, and Yano 1985 ). The same models were used to generate distance matrices for NJ analysis. The ML analysis was performed assuming rate homogeneity and rate heterogeneity with four categories of variable sites and one category of invariable sites. The MP and NJ analyses were carried out using the PHYLIP package (Felsenstein 1991 ). The support for different positions of the mole and the hedgehog within the eutherian tree was investigated both separately and in combination by comparing the log likelihood values obtained from ML analyses of the various topologies.
Phylogenetic analyses of the cytochrome b gene included S. araneus, along with the taxa included in the study of complete mtDNAs. The length of the cytochrome b alignment was 1,122 nt, corresponding to 374 aa, after removal of 18 nt (6 aa) of ambiguous homology at the 3′ end of the gene. The cytochrome b data were analyzed in the same way as the large mitochondrial alignment. Partial sequences of the cytochrome b genes of a number of soricids have been reported by Ohdachi et al. (1997) and Fumagalli et al. (1999) . Analyses of these sequences placed the soricids on a common branch as the sister group of the mole (data not shown).
Congruence with previously published phylogenetic hypotheses based on nuclear data sets, exon 28 of the von Willebrand factor gene (vWF, 318 aa), and the alpha-2 B-adrenergic receptor gene (A2AB, 378 aa), reported by Stanhope et al. (1998) , was tested with the nonparametric test suggested by Templeton (1983) . The test included the seven taxa represented by both vWF and A2AB: the hedgehog, the rat, the rabbit, the human, the mole, the horse, and the cow. The best tree topology for each data set was determined by MP, NJ, and ML. The analyses were performed on all codon positions, as well as on first plus second codon positions. The support of the nuclear sequences for the best mitochondrial tree was also tested.
ResultsCharacteristics of the mtDNA of Talpa europaea
The length of the mole mitochondrial genome described in this paper is 16,884 nt. However, the size of the genome varies in proportion to the number of 16-nt repeats in the control region; here, we report 19 repeats of ACAGGCGTATACACCC in the 1,422-nt- long control region. The two additional clones sequenced had 19 and 24 repeats, respectively. The gene order of the genome conforms with that of other eutherians. The start codon of NADH6 is ATT; all other genes have a methionine (ATG, ATA) start codon. TAA formed the stop codon in eight genes, while incomplete stop codons (TA or T) were found in COIII, NADH2, NADH3, and NADH4; these codons may be converted into complete stop codons by posttranscriptional polyadenylation (Ojala, Montoya, and Attardi 1981 ). The stop codon of cytochrome b is AGA. The nucleotide and amino acid sequence compositions of the mole do not differ significantly from those of other mammals according to a 5% level χ2 test (PUZZLE). The hedgehog fails the same test for nucleotide composition but passes it (P = 0.174) in the corresponding test for amino acid composition.
Phylogenetic Analysis and the Divergence Between Talpa, Chiroptera, and the Cetferungulates
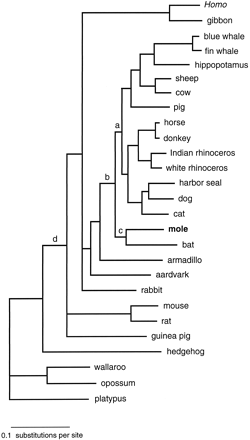
Consistent with a previous study (Krettek, Gullberg, and Arnason 1995 ), the hedgehog represented the most basal eutherian taxon; however, the mole was not found to be the sister group of the hedgehog, as would be expected if Lipotyphla were monophyletic. Instead, the mole was identified as the sister group of the Chiroptera. The position of the Talpa/Chiroptera clade was immediately basal to the cetferungulates (fig. 1 ). This phylogenetic relationship was strongly supported by all three analytical approaches used (MP, NJ, and ML) and was not affected by exclusion of the hedgehog. table 1 summarizes the bootstrap and quartet puzzle (QP) support values for the branches relevant to the positions of the mole and the hedgehog in the eutherian tree.
The topology of the tree shown in figure 1 and different topologies involving the hedgehog and the mole were examined in an exhaustive search with the number of taxa constrained to 10 operational taxonomic units. Due to computational limitations, this required the removal of one taxon from the analysis. Since the guinea pig is not crucial for the definition of the Lipotyphla, this species was not included in the exhaustive search. Under the assumption of rate homogeneity, the ML log likelihood values for the second best positions involving the mole and the hedgehog were all >2 SE worse (table 2 ) than the topology shown in figure 1 . More distant positions became rejectable with progressively higher significance, and, as evident in table 2 , monophyletic grouping of the mole and the hedgehog, either as basal in the eutherian tree (topology 4) or as the sister group of the Chiroptera (topology 6), could be rejected with high statistical significance. It is noteworthy that a transfer of the hedgehog from the base of the eutherian tree to the more apical position as the sister group of the mole is less costly than the reciprocal transfer of the mole to a position as the sister group of the hedgehog. The same observation was made in separate analyses excluding the mole and the hedgehog, respectively (table 2 , trees 7–10 and 11–13). It is probable that the hedgehog lineage, with its early origin among the eutherians, has accumulated more homoplasies than the more recent mole lineage, thereby resulting in different costs for changing the position of the two taxa in the tree. This interpretation would be consistent with the differing number of substitutions in the best trees including only the mole or only the hedgehog.
The cytochrome b sequence of S. araneus is 80.3% and 92.3% identical to the mole sequence at the nucleotide and amino acid levels, respectively. The amino acid difference between the mole and the shrew (7.7%) is less than, for example, the intraperissodactyl difference between Equidae and Rhinocerotidae (8%), the intracarnivoran difference between the cat and the dog (11%), or the intrahominoid difference between Hylobatidae and Hominidae (13%). All three analytical approaches (MP, NJ, and ML) clearly recognized a monophyletic origin of the mole and the shrew with support values between 87% and 99%. The cytochrome b analyses also reconstructed a sister group relationship between the bat and the mole/sorex clade even though the analysis of a single gene did not permit resolution of the relationships among more distantly related taxa.
ML distances calculated using the mtREV-24 model of amino acid sequence evolution (Adachi and Hasegawa 1996b ) were used to estimate the divergence times between the mole and the bat, as well as the origin of the mole/bat clade. The molecular clock was calibrated with the two references A/C-60, the divergence between ruminant Artiodactyla and Cetacea 60 MYA (Arnason and Gullberg 1996 ), and E/R-50, the divergence between Equidae and Rhinocerotidae 50 MYA (Xu, Janke, and Arnason 1996 ; Arnason, Gullberg, and Janke 1998 ). Both references yielded consistent datings, according to which the mole and the bat lineages split ≈74 MYA, while the corresponding dating for the divergence between the mole/bat clade and the cetferungulates was ≈79 MYA. In a previous study, the cetferungulate origin was defined as occurring at the divergence of edentates and cetferungulates (Arnason, Gullberg, and Janke 1997 ). The present study further constrains the cetferungulate origin by placing it at the divergence between Talpa/Chiroptera and the cetferungulate clade.
The branches defining the positions of the guinea pig and the rabbit have been collapsed in figure 1 , as they were not consistently identified by all data sets and analyses. The limited resolution in these parts of the tree have been discussed previously (Arnason, Gullberg, and Janke 1997, 1999 ; Janke, Xu, and Arnason 1997 ) and therefore are not detailed here. The other relationships shown in figure 1 have been described and dated in previous studies (Janke et al. 1994 ; Xu, Janke, and Arnason 1996 ; Janke, Xu, and Arnason 1997 ).
ML analysis of nucleotide sequences using the HKY model of sequence evolution and rate heterogeneity with eight classes of variable sites or four classes of variable sites plus one class of invariable sites did not resolve the phylogenetic position of the hedgehog relative to other eutherian orders, as the lnL differences for the various positions of this taxon differed only marginally. It is probable that this poor resolution is related to the significantly deviating nucleotide composition of the hedgehog. The same analysis using amino acid sequences, however, confirmed a basal position of the hedgehog (table 2 ).
Comparison of Different Data Sets
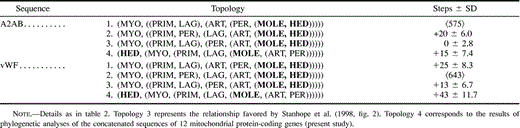
In contrast to the findings presented here, which are based on an alignment with a length of almost 10,000 nt, analyses of nuclear sequences (exon 28 of the vWF gene and the A2AB gene) and of combined nuclear and mitochondrial ribosomal (12S and 16S rDNA) sequences support a mole + hedgehog clade well within the eutherian tree (Stanhope et al. 1998 ). To investigate possible reasons for this inconsistency, we examined the relative phylogenetic signals in the sequences reported by Stanhope et al. (1998) . This examination (table 3 ) revealed significant (2σ) inconsistencies between the phylogenetic signals of the two nuclear sequences and between the vWF and the mitochondrial rDNA data sets, indicating that these genes are subject to differing evolutionary processes or constraints (Bull et al. 1993 ; de Queiroz, Donoghue, and Kim 1995 ).
The best tree topologies for each of the vWF and A2AB data sets were established using MP, NJ, and ML. The support for the best tree from one data set was then tested against the support for the best tree from the other data set using the Wilcoxon signed-ranks test (Templeton 1983 ). As is evident in table 3 , analyses of A2AB support a phylogenetic position of the hedgehog in a clade with the mole, the cow, and the horse, but the branching order is poorly resolved. The vWF data set contains a strong signal supporting the mole + hedgehog clade and would thus appear to be responsible for this topology in analyses of the combined data sets. However, vWF also provides strong support for excluding the horse from a (rabbit, (cow, (mole, hedgehog))) clade, which is the basis of the incongruence between the two nuclear data sets and between the vWF and the mitochondrial rDNA data sets. In cases such as these, combining the data sets may give misleading results (de Queiroz, Donoghue, and Kim 1995 ). Removal of unidentified nucleotides (14%) from the vWF sequence of the hedgehog had no effect on the results.
Discussion
The analyses presented here have identified the hedgehog as the most basal eutherian taxon. This is consistent with the results of previous analyses of concatenated mitochondrial protein-coding sequences, performed using a somewhat different species representation and without the mole (Krettek, Gullberg, and Arnason 1995 ; Arnason, Gullberg, and Janke 1997 ; Penny and Hasegawa 1997 ; Cao et al. 1998 ; Waddell et al. 1999 ). It is noteworthy that one of these studies (Penny and Hasegawa 1997 ) used the LogDet method, an approach which is insensitive to deviations in molecular composition. This consistency among results from different analyses and different taxa samplings makes it unlikely that the basal position of the hedgehog is due to the use of any particular type of analysis or to a random sampling effect.
A nonbasal position of the hedgehog was shown in a tree reported by Sullivan and Swofford (1997) . The findings which were based on a rate heterogeneity model and ML analysis of mitochondrial nucleotide sequences were inconclusive, however, as to the definite position of the hedgehog. The study included no amino acid analysis, and the possible influence of the significantly biased base composition of the hedgehog relative to the other eutherians was not discussed. When rate heterogeneity is taken into account in the ML analysis of amino acid sequences, as in the present study, the hedgehog remains the most basal eutherian taxon. Under the same model of sequence evolution, the monophyly of the mole (Talpidae) and the hedgehog (Erinaceidae) could be excluded with high statistical support. The amino acid composition of the hedgehog does not differ significantly from that of other taxa, even though it remains the most deviating; all analyses of this data set placed the hedgehog basal among the eutherians.
Analyses of two nuclear (vWF and A2AB) and mitochondrial rDNA sequences have supported a sister group relationship between the hedgehog and the mole (Stanhope et al. 1998 ), but trees reconstructed from the analyses showed some inconsistencies, and several ordinal relationships which have received conclusive support in studies of complete mitochondrial genomes remained unresolved. Thus, the analyses of Stanhope et al. (1998) disrupted some phylogenetic relationships, such as the cetferungulate clade, which have been strongly supported in analyses of complete mtDNAs (Arnason et al. 1996 ; D’Erchia et al. 1996 ; Xu, Janke, and Arnason 1996 ; Arnason, Gullberg, and Janke 1997, 1998 ; Janke, Xu, and Arnason 1997 ; Ursing and Arnason 1998a, 1998b ). Our examination of the vWF and A2AB data sets showed that they favored different phylogenies, with the vWF data set containing a strong signal for the mole + hedgehog clade. However, at the same time, vWF did not support other relationships, such as the affinity between Artiodactyla and Perissodactyla, which have been identified in other molecular studies. Our examination of these findings (see table 3 ) indicate that the vWF and A2AB genes are under different evolutionary constraints, which may result in gene phylogenies deviating from the evolutionary history of the species harboring these sequences. Comparable studies of mitochondrial protein-coding genes (Cao et al. 1994, 1998 ) have shown that among the H-strand- encoded genes, only one, NADH1, provides a phylogeny which with respect to one taxon, the rodents, deviates markedly from reconstructions based on the concatenated sequences of all mitochondrial protein-coding genes. Even so, the deviation in this case was still less pronounced than that occurring in the nuclear data sets used by Stanhope et al. (1998) .
The phylogenetic position of Lipotyphla has remained obscure in morphological studies because of the combination of numerous “primitive” traits and specialized adaptations of lipotyphlan families. Cladistic analyses of morphological characters have recognized Lipotyphla as both monophyletic and basal in the eutherian tree (Novacek and Wyss 1986 ; Novacek, Wyss, and McKenna 1988 ; Novacek 1989, 1990 ). The present molecular analyses of 12 mitochondrial protein-coding genes do not support lipotyphlan monophyly. Instead, a phylogenetic position of the hedgehog at a basal position in the eutherian tree and of the mole/bat as the sister group of the cetferungulates was strongly supported by all three methods of phylogeny reconstruction used, MP, NJ, and ML (table 1 ). Alternative placements of the mole were without statistical support (table 2 ). Inclusion or exclusion of the hedgehog did not affect the position of the mole. The results challenge the retention of the Lipotyphla as a monophyletic systematic unit. Furthermore, the findings support phylogenetic reevaluation of several morphological characteristics, as proposed in a recent provocative study of eutherian evolution (Werdelin and Nilsonne 1999 ).
Morphologists have long recognized a clear distinction between moles and hedgehogs, advocating separation of the Lipotyphla into two primary lineages, the Erinaceomorpha, with the family Erinaceidae, and the Soricomorpha, traditionally including the shrews (Soricidae) and the talpid moles (Gregory 1910 ; Butler 1972, 1988 ; McKenna 1975 ). As discussed by MacPhee and Novacek (1993) the morphological evidence in support of lipotyphlan monophyly is weak, and the molecular results indicating lipotyphlan polyphyly are therefore not contradicted by morphological conclusions. The most important result of the present study is the strong statistical support for the phylogenetic position of Talpidae distant from the Erinaceidae in the eutherian tree. Furthermore, our results support the traditional view of a close phylogenetic relationship between Soricidae and Talpidae in the Soricoidea (Gregory 1910 ; Hutchinson 1968 ; Butler 1972, 1988 ). McDowell (1958) and McKenna and Bell (1997) have suggested a sister group relationship between Talpidae and Erinaceidae. The morphological support for this relationship is not strong, however, and the characters listed by McDowell have been interpreted as being plesiomorphic (Butler 1988 ).
The molecular data presented here are consistent with the placement of the families Talpidae and Soricidae within the Soricomorpha (Gregory 1910 ; McKenna 1975 ; Butler 1972, 1988 ; MacPhee and Novacek 1993 ) and the ordinal-level classification of the Erinaceomorpha and Soricomorpha (McKenna 1975 ). The study has placed the Erinaceomorpha and the Soricomorpha at very different positions in the eutherian phylogenetic tree, a finding that is inconsistent with lipotyphlan monophyly.
Pekka Pamilo, Review Editor
Keywords: Phylogenetics, insectivore, Lipotyphla, mole, Talpa europaea, mitochondrial DNA.
Address for correspondence and reprints: Ulfur Arnason, Department of Genetics, Division of Evolutionary Molecular Systematics, University of Lund, Sölvegatan 29, S-223 62 Lund, Sweden. E-mail: ulfur.arnason@gen.lu.se.
Fig. 1.—Phylogenetic position of the mole Talpa europaea in a maximum-likelihood tree from the concatenated amino acid sequences of 12 mitochondrial protein-coding genes. Support values for labeled branches are shown in table 1 . For the scientific names of the taxa included, see Materials and Methods.
Table 1 Bootstrap and Quartet Support Values for Selected Branches of the Tree Shown in figure 1
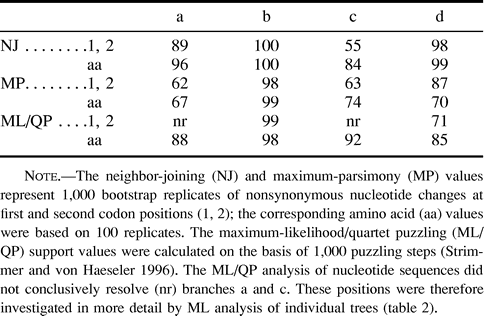
Table 1 Bootstrap and Quartet Support Values for Selected Branches of the Tree Shown in figure 1

Table 2 Maximum-Likelihood Analyses of Different Positions of the Mole (MOLE) and the Hedgehog (HED) in the Mammalian Tree

Table 2 Maximum-Likelihood Analyses of Different Positions of the Mole (MOLE) and the Hedgehog (HED) in the Mammalian Tree

We thank Bengt and Kerstin Olsson for the mole specimen and Karl Fredga and Maarit Jaarola for the shrew specimen. Scott K. Davis, Bengt Olle Bengtsson, and W. Patrick Luckett provided valuable comments on the manuscript. S.K.M. was the recipient of a Fulbright fellowship for the academic year 1996–1997. This work was supported by the Erik Philip-Sörensen Foundation, by the Swedish Natural Sciences Research Council and by contract ERBCHR XCT 930254 from the European Commission. The alignments used will be made available to other researchers on request.
References
Adachi, J., and M. Hasegawa. 1996a. MOLPHY version 2.3: programs for molecular phylogenetics based on maximum likelihood. Comput. Sci. Monogr. 28:1–150.
———. 1996b. Model of amino acid substitution in proteins encoded by mitochondrial DNA. J. Mol. Evol. 42:459–468.
Anderson, S., M. H. L. de Bruijn, A. R. Coulson, I. C. Eperon, F. Sanger, and I. G. Young.
Arnason, U., and A. Gullberg.
———.
Arnason, U., A. Gullberg, and A. Janke.
———.
———.
Arnason, U., A. Gullberg, A. Janke, and X. Xu.
Arnason, U., A. Gullberg, and B. Widegren.
Arnason, U., A. Gullberg, and X. Xu.
Arnason, U., and E. Johnsson.
Arnason, U., X. Xu, and A. Gullberg.
Bibb, M. J., R. A. Van Etten, C. T. Wright, M. W. Walberg, and D. A. Clayton.
Bull, J. J., J. P. Huelsenbeck, C. W. Cunningham, D. L. Swofford, and P. J. Waddell.
Butler, P. M.
———.
Cao, Y., J. Adachi, A. Janke, S. Pääbo, and M. Hasegawa.
Cao, Y., A. Janke, P. J. Waddell, M. Westerman, O. Takenaka, S. Murata, N. Okada, S. Paabo, and M. Hasegawa.
D’Erchia, A. M., C. Gissi, G. Pesole, C. Saccone, and U. Arnason.
de Queiroz, A., M. J. Donoghue, and J. Kim.
Felsenstein, J.
———.
Fitch, W. M.
Fumagalli, L., P. Taberlet, D. T. Stewart, L. Gielly, J. Hausser, and P. Vogel.
Gadaleta, G., G. Pepe, G. de Candia, C. Quagliariello, E. Sbisa, and C. Saccone.
Gissi, C., A. Gullberg, and U. Arnason.
Hasegawa, M., H. Kishino, and T. Yano.
Hiendleder, S., H. Lewalski, R. Wassmuth, and A. Janke.
Hutchinson, J. H.
Janke, A., G. Feldmaier-Fuchs, W. K. Thomas, A. von Haeseler, and S. Pääbo.
Janke, A., N. J. Gemmell, G. Feldmaier-Fuchs, A. von Haeseler, and S. Pääbo.
Janke, A., X. Xu, and U. Arnason.
Kim, K. S., S. E. Lee, H. W. Jeong, and J. H. Ha.
Kishino, H., and M. Hasegawa.
Kishino, H., T. Miyata, and M. Hasegawa.
Krettek, A., A. Gullberg, and U. Arnason.
Lavergne, A., E. Douzery, T. Stichler, F. M. Catzeflis, and M. S. Springer.
Lopez, J. V., M. Culver, S. Cevario, and S. J. O’Brien.
McKenna, M. C.
McKenna, M. C., and S. K. Bell.
MacPhee, R. D. E., and M. J. Novacek.
Novacek, M. J.
———.
Novacek, M. J., and A. R. Wyss.
Novacek, M. J., A. A. Wyss, and M. C. McKenna.
Ohdachi, S., R. Masuda, H. Abe, J. Adachi, N. E. Dokuchaev, V. Haukisalmi, and M. C. Yoshida.
Ojala, D., J. Montoya, and G. Attardi.
Pumo, D. E., P. S. Finamore, W. R. Franek, C. J. Carleton, S. Tarzami, and D. Balzarano.
Saitou, N., and M. Nei.
Springer, M. S., G. C. Cleven, O. Madsen, W. W. De Jong, V. G. Waddell, H. M. Amrine, and M. J. Stanhope.
Stanhope, M. J., V. G. Waddell, O. Madsen, W. W. de Jong, S. B. Hedges, G. C. Cleven, D. Kao, and M. S. Springer.
Strimmer, K., and A. von Haeseler.
Sullivan, J., and D. L. Swofford.
Templeton, A. R.
Ursing, B. M., and U. Arnason. 1998a. The complete mitochondrial DNA sequence of the pig (Sus scrofa). J. Mol. Evol. 47:302–306.
———. 1998b. Analyses of mitochondrial genomes strongly support a hippopotamus-whale clade. Proc. R. Soc. Lond. B Biol. Sci. 265:2251–2255.
Waddell, P. J., Y. Cao, J. Hauf, and M. Hasegawa.
Werdelin, L., and A. Nilsonne.
Xu, X., A. Gullberg, and U. Arnason.
Xu, X., and U. Arnason.
———.