-
PDF
- Split View
-
Views
-
Cite
Cite
Agatha Schlüter, Stéphane Fourcade, Raymond Ripp, Jean Louis Mandel, Olivier Poch, Aurora Pujol, The Evolutionary Origin of Peroxisomes: An ER-Peroxisome Connection, Molecular Biology and Evolution, Volume 23, Issue 4, April 2006, Pages 838–845, https://doi.org/10.1093/molbev/msj103
Close - Share Icon Share
Abstract
The peroxisome is an essential eukaryotic organelle, crucial for lipid metabolism and free radical detoxification, development, differentiation, and morphogenesis from yeasts to humans. Loss of peroxisomes invariably leads to fatal peroxisome biogenesis disorders in man. The evolutionary origin of peroxisomes remains unsolved; proposals for either a symbiogenetic or cellular membrane invagination event are unconclusive. To address this question, we have probed with a peroxisomal proteome, an “ensemble” of 19 representative eukaryotic complete genomes. Molecular phylogenetic and sequence comparison tools allowed us to identify four proteins as peroxisomal markers for unequivocal in silico peroxisome detection. We have then detected the Apicomplexa phylum as the first group of organisms devoid of peroxisomes, in the presence of mitochondria. Finally, we deliver evidence against a prokaryotic ancestor of peroxisomes: (1) the peroxisomal membrane is composed of purely eukaryotic bricks and is thus useful to trace the eukaryotes in their evolutionary paths and (2) the peroxisomal matrix protein import system shares mechanistic similarities with the endoplasmic reticulum/proteasome degradation process, indicating a common evolutionary history.
Introduction
The indispensable role of peroxisomes is stressed by the fatal consequences of the mutations inactivating peroxisomal proteins essential for biogenesis and matrix and membrane protein import: the human diseases known as peroxisome biogenesis disorders (PBD) (Wanders 2004). Zellweger syndrome (ZS), neonatal adrenoleukodystrophy (NALD), infantile Refsum disease, and rhizomelic chondrodysplasia punctata are characterized by a strong reduction in number and size or even complete absence of peroxisomes and are syndromes not compatible with life or normal development. From all organisms studied until present, 35 different peroxins (proteins involved in peroxisome biogenesis) have been identified, of which only 18 are present in human. Most peroxins are peroxisomal membrane proteins or interact through docking sites with the peroxisomal membrane. A complex peroxin interaction network controls biogenesis and division (Pex11, Pex23, Pex25, Pex27, Pex28, Pex29, Pex30, Pex31, and Pex32) and allows for the recognition of peroxisome target proteins through specific receptors (Pex5, Pex5r, Pex7, Pex18, Pex20, and Pex21), for membrane protein assembly (Pex3, Pex15, Pex16, Pex19, and Pex24), for the docking of these receptors (Pex13, Pex14, and Pex17), for receptor recycling and protein import (Pex1, Pex4, Pex6, Pex8, Pex9, Pex22, and Pex26), and for the translocation of proteins to peroxisomal matrix (Pex2, Pex10, and Pex12) (Lazarow 2003; Titorenko and Rachubinski 2004).
Knowledge on organelle biogenesis, metabolic functions and their key players, and evolutionary history is still incomplete. In contrast to mitochondria and chloroplasts, peroxisomes lack DNA and are surrounded by a single membrane, but nevertheless the origin of the organelle has been suggested to be symbiogenetic, derived from an enslaved anaerobic hydrogen-producing prokaryote (de Duve 1996; Cavalier-Smith 1997). Here we have undertaken a comprehensive comparative genomics approach to address these questions. The establishment of phylogenetic distribution, by means of profile comparisons and molecular phylogenetic reconstructions, respectively, enables the comparison of experimental results obtained from different species. The results can be used to identify essential, identity conferring core elements to trace back the origin of the organelle or to infer functional associations.
Materials and Methods
The Peroxisomal Proteome
Using annotated data derived from a variety of online resources (Gene Ontology [GO], Kyoto Encyclopedia of Genes and Genomes, National Center for Biotechnology Information [NCBI]) and the literature, we have retrieved 103 peroxisomal proteins as components of the peroxisomal proteome, including the whole human peroxisomal proteome (80 proteins) and 23 additional fungi and mouse proteins without human orthologue. This proteome contains the 35 known peroxins (including the three different isoforms of Pex11: Pex11α, Pex11β, and Pex11γ and the Pex5-like peroxin [Amery et al. 2001]), 57 enzymes, five carriers, and six proteins of unknown function. It is worth noting that the actual number of human proteins declared as such in GO is 51. We have created a database (www.peroxisomeDB.org; A. Schlüter, S. Fourcade, E. Domènech, G. Berthommier, R. J. A. Wanders, J. L. Mandel, O. Poch, and A. Pujol, in preparation) that compiles the current knowledge focused on peroxisomal genes, their encoded proteins, the metabolic routes they belong to, and the diseases they may cause.
Psi-Blast Analysis
The 103 proteins that are included in the peroxisome database have been analyzed using the Psi-Blast algorithm (Altschul et al. 1997) until five iterations when possible. Psi-Blast process was performed for each protein by our structural genomics platform for annotation and integrative analysis (GSCOPE, R. Ripp and O. Poch, unpublished data). In order to select the proteins lacking regions of homology in bacterial/archaeal sequences, we screened in the last Psi-Blast iteration, with a cutoff of E value < 10−3.
Blast Analysis
The proteins from our peroxisomal proteome that did not show motifs of similarity in Bacteria/Archea were analyzed by Blast algorithm (Altschul et al. 1997) and further, by multiple alignment analysis performed by ClustalW (Thompson, Higgins, and Gibson 1994). In search for orthologues, we complemented the sequence comparison data with the construction and analysis of phylogenetic trees, which helped to discriminate between orthology and paralogy relationships. We identified as homologues, proteins having regions of similarity covering more than 50% of the query sequence and bearing the conserved domains identified in the query sequence by CD search. For those cases where “conspicuous proteins” appeared, we did reciprocal searches by Blast algorithm in order to confirm the homology. When orthologues were not identified, we proceeded to analyze the genomes by TBlastN.
Organisms
We have chosen the following relevant organisms among the complete (or nearly completed) genomes.
Fully sequenced genomes: Thalassiosira pseudonana (http://genome.jgi-psf.org/thaps1/thaps1.download.html), Cyanidioschyzon meroale (http://merolae.biol.s.u-tokyo.ac.jp/download/), Dictyostelium discoideum (http://www.genedb.org/genedb/dicty/index.jsp), and Tetraodon nigroviridis (http://www.genoscope.cns.fr/externe/tetraodon/); Oryza sativa, Arabidopsis thaliana, Plasmodium falciparum, Plasmodium yoelii yoelii, Cryptosporidium parvum, Encephalitozoon cuniculi, Saccharomyces cerevisiae, Schizosaccharomyces pombe, Anopheles gambiae, Drosophila melanogaster, Caenorhabditis elegans, Ratus norvegicus, Mus musculus, and Homo sapiens in NCBI (http://www.ncbi.nlm.nih.gov).
Genomes near to completion and annotation: Giardia Lamblia (http://gmod.mbl.edu/perl/site/giardia?page=download). Other genomes screened but not yet fully sequenced at the time of analysis: Trypanosoma brucei, Leishmania major and Leishmania donovani, Toxoplasma gondii, and Candida albicans (http://www.ncbi.nlm.nih.gov).
Supertree
The programs Phylo_win, version 1.2 (Galtier, Gouy, and Gautier 1996), and MEGA, version 2.1 (Kumar et al. 2001), were used for phylogenetic analysis. The supertree (Daubin, Gouy, and Perriere 2001) allowed for integration in a single phylogenetic tree of the set of 25 peroxisomal proteins from complete eukaryotic genomes and without overall homologies to bacterial/archeal genomes. Briefly, it was performed by the concatenation of each protein matrix into a supermatrix, building a phylogenetic tree using the neighbor-joining method and percent accepted mutations correction as a distance matrix, under complete gap deletion. Matrix protein length was corrected in order to have the same weight in the supertree matrix. For each peroxisomal protein, bootstrap values for each tree were computed by resampling 500 times among the orthologues of a given protein. Each tree obtained from a given peroxisomal protein was coded into a matrix of informative sites repeated proportionally to the approximate bootstrap probability associated to each internal branch. The number of sites repeated was linearly depending on bootstrap values over 60%. This matrix was coded with modifications to the Baum (1992) and Ragan (1992) ones. Instead of binary matrix when two orthologues did not share the same branch, the symbols in the matrix were encoded randomly between 19 different ones. When a given protein was not found in a species, the symbol was encoded as unknown. The Pex11 tree was built taking the three paralogues together prior to splitting into the three respective trees.
Results and Discussion
Finding Markers and Tracking the Tree of Life
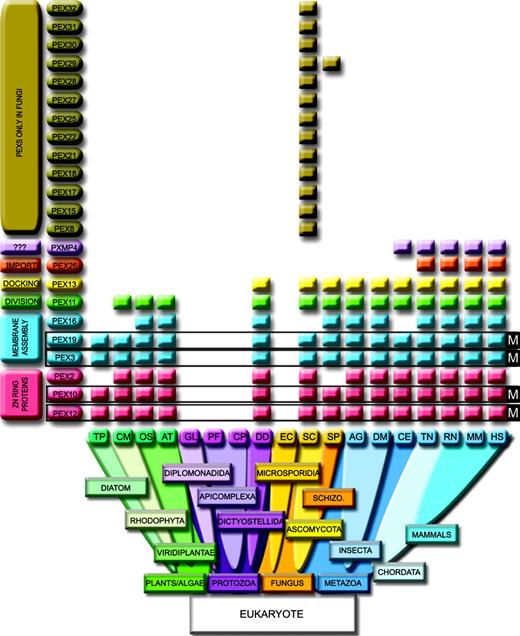
Firstly, we have generated a database that compiles all known human peroxisomal proteins plus a set of well-characterized yeast proteins not present in higher eukaryotes, comprising a proteome containing 103 proteins (http://www.peroxisomedb.org/proteome.php; A. Schlüter, S. Fourcade, E. Domènech, G. Berthommier, R. J. A. Wanders, J. L. Mandel, O. Poch, and A. Pujol, in preparation). By Psi-Blast searches, we have detected 35 peroxisomal proteins that do not show overall homology in Bacteria or Archea (table 1). From this set of 35 proteins, 29 are peroxins, 6 are other peroxisomal membrane proteins, and strikingly, none of the membrane or matrix enzymes have been retrieved following this selection procedure. Peroxins that showed restricted domains with homology to Bacteria/Archea are listed in Supplementary Table 1 (Supplementary Material online). Removing the proteins that had sequence similarities with proteins from organisms which are lacking peroxisomes, E. cuniculi and G. lamblia (Cavalier-Smith 1993), we come up with an ensemble of 25 proteins. We have then combined Blast, pairwise and profile-based searches at the DNA and protein sequence levels, in a representative set of 19 fully sequenced genomes. The result allowed us to generate a protein-profile distribution, derived from the presence/absence of a given gene in the set of genomes under study (fig. 1). Besides the well-known richness of the S. cerevisiae peroxisomal proteome, this analysis reveals the absence of the organelle in the Apicomplexa phylum. Our study has taken place after completion of three Apicomplexa genomes, P. falciparum, P. yoelii, and C. parvum, and including the available sequences of a fourth, T. gondii. In the latter, the peroxisome existence was postulated on the basis of catalase detection by immunohistochemistry (Kaasch and Joiner 2000). We thus provide a new warning against the misleading use of catalase as peroxisomal marker. To date, absence of peroxisomes has been only documented in amitochondriate parasites such as E. cuniculi, G. lamblia, or Entamoeba histolytica. Apicomplexans are so far the first group of organisms devoid of peroxisomes in the presence of mitochondria.
Peroxisomal Proteins Lacking Overall Homology in Bacteria/Archea Genomes
Protein . | Domain . | Function . | Compartment . | Kind of Protein . | Comments . |
|---|---|---|---|---|---|
| Pex2 | Zn-RING | Protein import | Membrane | Peroxin | Thalassiosira pseudonanaA, Schizosaccharomyces pombeA |
| Pex3 | — | Membrane assembly | Membrane | Peroxin | MARKER |
| Pex4 | Ubiquitin-ligase | Protein import | Cytosol and peroxisome docking | Peroxin | EukaryoteR |
| Pex8 | — | Matrix protein import | Membrane | Peroxin | Some SaccharomycetalesR |
| Pex9 | — | Matrix protein import | Membrane | Peroxin | Yarrowia lipolyticaR |
| Pex10 | Zn-RING | Protein import | Membrane | Peroxin | MARKER |
| Pex11, Pex11β, Pex11γ | — | Division Proliferation | Membrane | Peroxin | T. pseudonanaA |
| Pex12 | Zn-RING | Protein import | Membrane | Peroxin | MARKER |
| Pex13 | SH3 | Docking of receptors | Membrane | Peroxin | photosyntheticsA |
| Pex15 | Phosphorylation | Membrane assembly | Membrane | Peroxin | Some SaccharomycetalesR |
| Pex16 | — | Membrane assembly | Membrane | Peroxin | Caenorhabditis elegansA, Saccharomyces cerevisiaeA, Cyanidioschyzon merolaeA, T. pseudonanaA |
| Pex17 | — | Docking of receptors | Membrane | Peroxin | Some SaccharomycetalesR |
| Pex18 | — | PTS targeting | Cytosol and peroxisome docking | Peroxin | Some SaccharomycetalesR |
| Pex19 | Farnesylation | Membrane assembly | Cytosol and peroxisome docking | Peroxin | MARKER |
| Pex20 | — | PTS targeting | Cytosol and peroxisome docking | Peroxin | Neurospora crassar and Y. lipolyticar |
| Pex21 | — | PTS targeting | Cytosol and peroxisome docking | Peroxin | Some Saccharomycetalesr |
| Pex22 | — | Protein import | Membrane | Peroxin | Some Saccharomycetalesr |
| Pex23 | Dysferlin | Proliferation | Membrane | Peroxin | Y. lipolyticaR |
| Pex24 | — | Membrane assembly | Membrane | Peroxin | Y. lipolyticaR |
| Pex25 | — | Proliferation | Membrane | Peroxin | Some SaccharomycetalesR |
| Pex26 | — | Protein import and recruitment | Membrane | Peroxin | VertebrataR |
| Pex27 | — | Proliferation | Membrane | Peroxin | Some SaccharomycetalesR |
| Pex28 | — | Proliferation | Membrane | Peroxin | FungiR |
| Pex29 | — | Proliferation | Membrane | Peroxin | FungiR |
| Pex30 | Dysferlin | Proliferation | Membrane | Peroxin | FungiR |
| Pex31 | Dysferlin | Proliferation | Membrane | Peroxin | FungiR |
| Pex32 | Dysferlin | Proliferation | Membrane | Peroxin | FungiR |
| Mpv17, Pmp2, FLJ12592, MGC12972 | Mpv17 | Unknown | Membrane | PMP | EukaryotesR |
| Pxmp4 | — | Unknown | Membrane | PMP | MammalsR, C. elegansR, FungiR, InsectaA |
| Pmp34 | — | ATP transporter | Membrane | PMP | AmitochondriatesA |
Protein . | Domain . | Function . | Compartment . | Kind of Protein . | Comments . |
|---|---|---|---|---|---|
| Pex2 | Zn-RING | Protein import | Membrane | Peroxin | Thalassiosira pseudonanaA, Schizosaccharomyces pombeA |
| Pex3 | — | Membrane assembly | Membrane | Peroxin | MARKER |
| Pex4 | Ubiquitin-ligase | Protein import | Cytosol and peroxisome docking | Peroxin | EukaryoteR |
| Pex8 | — | Matrix protein import | Membrane | Peroxin | Some SaccharomycetalesR |
| Pex9 | — | Matrix protein import | Membrane | Peroxin | Yarrowia lipolyticaR |
| Pex10 | Zn-RING | Protein import | Membrane | Peroxin | MARKER |
| Pex11, Pex11β, Pex11γ | — | Division Proliferation | Membrane | Peroxin | T. pseudonanaA |
| Pex12 | Zn-RING | Protein import | Membrane | Peroxin | MARKER |
| Pex13 | SH3 | Docking of receptors | Membrane | Peroxin | photosyntheticsA |
| Pex15 | Phosphorylation | Membrane assembly | Membrane | Peroxin | Some SaccharomycetalesR |
| Pex16 | — | Membrane assembly | Membrane | Peroxin | Caenorhabditis elegansA, Saccharomyces cerevisiaeA, Cyanidioschyzon merolaeA, T. pseudonanaA |
| Pex17 | — | Docking of receptors | Membrane | Peroxin | Some SaccharomycetalesR |
| Pex18 | — | PTS targeting | Cytosol and peroxisome docking | Peroxin | Some SaccharomycetalesR |
| Pex19 | Farnesylation | Membrane assembly | Cytosol and peroxisome docking | Peroxin | MARKER |
| Pex20 | — | PTS targeting | Cytosol and peroxisome docking | Peroxin | Neurospora crassar and Y. lipolyticar |
| Pex21 | — | PTS targeting | Cytosol and peroxisome docking | Peroxin | Some Saccharomycetalesr |
| Pex22 | — | Protein import | Membrane | Peroxin | Some Saccharomycetalesr |
| Pex23 | Dysferlin | Proliferation | Membrane | Peroxin | Y. lipolyticaR |
| Pex24 | — | Membrane assembly | Membrane | Peroxin | Y. lipolyticaR |
| Pex25 | — | Proliferation | Membrane | Peroxin | Some SaccharomycetalesR |
| Pex26 | — | Protein import and recruitment | Membrane | Peroxin | VertebrataR |
| Pex27 | — | Proliferation | Membrane | Peroxin | Some SaccharomycetalesR |
| Pex28 | — | Proliferation | Membrane | Peroxin | FungiR |
| Pex29 | — | Proliferation | Membrane | Peroxin | FungiR |
| Pex30 | Dysferlin | Proliferation | Membrane | Peroxin | FungiR |
| Pex31 | Dysferlin | Proliferation | Membrane | Peroxin | FungiR |
| Pex32 | Dysferlin | Proliferation | Membrane | Peroxin | FungiR |
| Mpv17, Pmp2, FLJ12592, MGC12972 | Mpv17 | Unknown | Membrane | PMP | EukaryotesR |
| Pxmp4 | — | Unknown | Membrane | PMP | MammalsR, C. elegansR, FungiR, InsectaA |
| Pmp34 | — | ATP transporter | Membrane | PMP | AmitochondriatesA |
NOTE.—The comments column states from which organisms/lineages proteins are restricted (R) or absent (A). Pex, peroxin; PMP, peroxisomal membrane protein.
Peroxisomal Proteins Lacking Overall Homology in Bacteria/Archea Genomes
Protein . | Domain . | Function . | Compartment . | Kind of Protein . | Comments . |
|---|---|---|---|---|---|
| Pex2 | Zn-RING | Protein import | Membrane | Peroxin | Thalassiosira pseudonanaA, Schizosaccharomyces pombeA |
| Pex3 | — | Membrane assembly | Membrane | Peroxin | MARKER |
| Pex4 | Ubiquitin-ligase | Protein import | Cytosol and peroxisome docking | Peroxin | EukaryoteR |
| Pex8 | — | Matrix protein import | Membrane | Peroxin | Some SaccharomycetalesR |
| Pex9 | — | Matrix protein import | Membrane | Peroxin | Yarrowia lipolyticaR |
| Pex10 | Zn-RING | Protein import | Membrane | Peroxin | MARKER |
| Pex11, Pex11β, Pex11γ | — | Division Proliferation | Membrane | Peroxin | T. pseudonanaA |
| Pex12 | Zn-RING | Protein import | Membrane | Peroxin | MARKER |
| Pex13 | SH3 | Docking of receptors | Membrane | Peroxin | photosyntheticsA |
| Pex15 | Phosphorylation | Membrane assembly | Membrane | Peroxin | Some SaccharomycetalesR |
| Pex16 | — | Membrane assembly | Membrane | Peroxin | Caenorhabditis elegansA, Saccharomyces cerevisiaeA, Cyanidioschyzon merolaeA, T. pseudonanaA |
| Pex17 | — | Docking of receptors | Membrane | Peroxin | Some SaccharomycetalesR |
| Pex18 | — | PTS targeting | Cytosol and peroxisome docking | Peroxin | Some SaccharomycetalesR |
| Pex19 | Farnesylation | Membrane assembly | Cytosol and peroxisome docking | Peroxin | MARKER |
| Pex20 | — | PTS targeting | Cytosol and peroxisome docking | Peroxin | Neurospora crassar and Y. lipolyticar |
| Pex21 | — | PTS targeting | Cytosol and peroxisome docking | Peroxin | Some Saccharomycetalesr |
| Pex22 | — | Protein import | Membrane | Peroxin | Some Saccharomycetalesr |
| Pex23 | Dysferlin | Proliferation | Membrane | Peroxin | Y. lipolyticaR |
| Pex24 | — | Membrane assembly | Membrane | Peroxin | Y. lipolyticaR |
| Pex25 | — | Proliferation | Membrane | Peroxin | Some SaccharomycetalesR |
| Pex26 | — | Protein import and recruitment | Membrane | Peroxin | VertebrataR |
| Pex27 | — | Proliferation | Membrane | Peroxin | Some SaccharomycetalesR |
| Pex28 | — | Proliferation | Membrane | Peroxin | FungiR |
| Pex29 | — | Proliferation | Membrane | Peroxin | FungiR |
| Pex30 | Dysferlin | Proliferation | Membrane | Peroxin | FungiR |
| Pex31 | Dysferlin | Proliferation | Membrane | Peroxin | FungiR |
| Pex32 | Dysferlin | Proliferation | Membrane | Peroxin | FungiR |
| Mpv17, Pmp2, FLJ12592, MGC12972 | Mpv17 | Unknown | Membrane | PMP | EukaryotesR |
| Pxmp4 | — | Unknown | Membrane | PMP | MammalsR, C. elegansR, FungiR, InsectaA |
| Pmp34 | — | ATP transporter | Membrane | PMP | AmitochondriatesA |
Protein . | Domain . | Function . | Compartment . | Kind of Protein . | Comments . |
|---|---|---|---|---|---|
| Pex2 | Zn-RING | Protein import | Membrane | Peroxin | Thalassiosira pseudonanaA, Schizosaccharomyces pombeA |
| Pex3 | — | Membrane assembly | Membrane | Peroxin | MARKER |
| Pex4 | Ubiquitin-ligase | Protein import | Cytosol and peroxisome docking | Peroxin | EukaryoteR |
| Pex8 | — | Matrix protein import | Membrane | Peroxin | Some SaccharomycetalesR |
| Pex9 | — | Matrix protein import | Membrane | Peroxin | Yarrowia lipolyticaR |
| Pex10 | Zn-RING | Protein import | Membrane | Peroxin | MARKER |
| Pex11, Pex11β, Pex11γ | — | Division Proliferation | Membrane | Peroxin | T. pseudonanaA |
| Pex12 | Zn-RING | Protein import | Membrane | Peroxin | MARKER |
| Pex13 | SH3 | Docking of receptors | Membrane | Peroxin | photosyntheticsA |
| Pex15 | Phosphorylation | Membrane assembly | Membrane | Peroxin | Some SaccharomycetalesR |
| Pex16 | — | Membrane assembly | Membrane | Peroxin | Caenorhabditis elegansA, Saccharomyces cerevisiaeA, Cyanidioschyzon merolaeA, T. pseudonanaA |
| Pex17 | — | Docking of receptors | Membrane | Peroxin | Some SaccharomycetalesR |
| Pex18 | — | PTS targeting | Cytosol and peroxisome docking | Peroxin | Some SaccharomycetalesR |
| Pex19 | Farnesylation | Membrane assembly | Cytosol and peroxisome docking | Peroxin | MARKER |
| Pex20 | — | PTS targeting | Cytosol and peroxisome docking | Peroxin | Neurospora crassar and Y. lipolyticar |
| Pex21 | — | PTS targeting | Cytosol and peroxisome docking | Peroxin | Some Saccharomycetalesr |
| Pex22 | — | Protein import | Membrane | Peroxin | Some Saccharomycetalesr |
| Pex23 | Dysferlin | Proliferation | Membrane | Peroxin | Y. lipolyticaR |
| Pex24 | — | Membrane assembly | Membrane | Peroxin | Y. lipolyticaR |
| Pex25 | — | Proliferation | Membrane | Peroxin | Some SaccharomycetalesR |
| Pex26 | — | Protein import and recruitment | Membrane | Peroxin | VertebrataR |
| Pex27 | — | Proliferation | Membrane | Peroxin | Some SaccharomycetalesR |
| Pex28 | — | Proliferation | Membrane | Peroxin | FungiR |
| Pex29 | — | Proliferation | Membrane | Peroxin | FungiR |
| Pex30 | Dysferlin | Proliferation | Membrane | Peroxin | FungiR |
| Pex31 | Dysferlin | Proliferation | Membrane | Peroxin | FungiR |
| Pex32 | Dysferlin | Proliferation | Membrane | Peroxin | FungiR |
| Mpv17, Pmp2, FLJ12592, MGC12972 | Mpv17 | Unknown | Membrane | PMP | EukaryotesR |
| Pxmp4 | — | Unknown | Membrane | PMP | MammalsR, C. elegansR, FungiR, InsectaA |
| Pmp34 | — | ATP transporter | Membrane | PMP | AmitochondriatesA |
NOTE.—The comments column states from which organisms/lineages proteins are restricted (R) or absent (A). Pex, peroxin; PMP, peroxisomal membrane protein.
Distribution among the Eukarya of the selected peroxisomal proteins that lack significant homologies within Bacteria/Archea taxa. Peroxisomal markers (M): Pex3, Pex19, Pex10, and Pex12. Pex26 is restricted to vertebrate and Pxmp4 to Fungi and Metazoa (excluding Insecta). Pex13 is excluded from the photosynthtetic lineage. The amitochondriates Encephalitozoon cuniculi (EC) and Giardia lamblia (GL) have no peroxisomal proteins. The Apicomplexa Plasmodium falciparum (PF) and Cryptosporidium parvum (CP) contain mitochondria but no peroxisomes. HS, Homo sapiens; RN, Ratus norvegicus; MM, Mus musculus; TN, Tetraodon nigroviridis; DM, Drosophila melanogaster; AG, Anopheles gambiae; CE, Caenorhabditis elegans; SC, Saccharomyces cerevisiae; SP, Schizosaccharomyces pombe; AT, Arabidopsis thaliana; OS, Oryza sativa; DD, Dictyostelium discoideum; CM, Cyanidioschyzon merolae; TP, Thalassiosira Pseudonana.
As deduced from figure 1, we have identified four peroxins that can be considered peroxisomal markers because they are ubiquitously present in all peroxisome-containing organisms but absent from the organisms devoid of peroxisomes: Pex3, Pex19, Pex10, and Pex12. The Pex3 and Pex19 peroxisomal markers, together with Pex16, are required to elaborate and maintain the membrane. Their loss of function leads to peroxisome absence in ZS patients (Pex16 and Pex19) and in NALD patients (Pex3) (reviewed in Wanders 2004). In these patients, neither peroxisome nor peroxisomal membranes (ghosts) are detected (Honsho et al. 1998; Shimozawa et al. 2000). The transexpression of Pex3, Pex19, or Pex16 restores the affected peroxisomal membrane, allowing for the import of matrix proteins, thus leading to postulate a “de novo” regeneration of peroxisomes (Matsuzono et al. 1999; South and Gould 1999; Muntau et al. 2000). The markers Pex10 and Pex12 are integral membrane proteins characterized by the presence of a Zn-RING domain. Mutations within the sequence of the Zn-RING proteins affect the import of peroxisomal proteins across the organelle membrane leading to PBD. This domain is involved in mediating protein-protein interactions, and Pex10/Pex12 complex leads to the Pex5-dependent cargo import after recognition of the peroxisome-targeting signal 1. Screening for the presence of the four markers will be particularly useful for unambiguous in silico detection of the organelle in the growing amount of new sequenced genomes, especially on those deep branching at the boundaries of the Eukarya kingdom.
In spite of being ubiquitously distributed in peroxisome-containing organisms, the Pex5 receptor had to be discarded as a specific marker because Psi-Blast analysis detected a limited domain homologous to bacteria. However, the region of bacterial homology in Pex5 is restricted to the tetratricopeptide (TPR) repeat motif and covers only 18% of the protein sequence (Supplementary Table 1, Supplementary Material online). The TPR domain is widely spread in many functionally and genetically unrelated proteins from Bacteria, Archea, and Viridae genomes. Excluding the depicted domains in Supplementary Table 1 (Supplementary Material online), no true orthologues of these Pexs could be detected in noneukaryotic organisms.
The peroxisome of the diatom alga T. pseudonana seems to be reduced to its minimal expression. Compared to the red alga Cyanidioschyzon merolae, the smallest genome among the photosynthetic eukaryotes, T. pseudonana lacks Pex2, Pex14, and Pex11. The Pex11 gene controls peroxisome division and abundance (Schrader et al. 1998), and it has been reported to be missing from C. merolae, in agreement with the presence of a single peroxisome (microbody) (Matsuzaki et al. 2004) in this organism. However, we succeed to identify a well-conserved orthologue of mammalian Pex11 in C. merolae, whose function remains to be tested experimentally. Because diatoms arose from a secondary endosymbiotic event between a red alga and a eukaryotic cell, we can speculate that this protein has been lost in T. pseudonana. Both genomes contain Pex5, Pex1, and Pex6. Other interesting findings are the presence of Pex26 (fig. 1) and the three Pex11 paralogues in the genome of the fish T. nigroviridis, thus expanding to the chordata features believed to be restricted to mammals (Li et al. 2002). Pex13 is absent from all photosynthetic organisms, from T. pseudonana to O. sativa. The peroxisome seems to have evolved under different environmental pressures that led to its diversification and specialization, resulting in a peroxisomal tree distribution paralleling the shape of the tree of life (Supplementary Figure 1, Supplementary Material online). It should be noted that C. elegans is branched with metazoan, although lacking significant bootstrap values, most likely due to annotation errors in the available C. elegans sequences. For instance, the Pex10 sequence is found as a chimeric product with tryptophanyl tRNA synthetase (C34E10.4).
Membrane Composition, Biogenesis, and the Origin of Peroxisomes
Omnis membrana e membrana (Günther Blobel, Nobel Prize 1999). Thus, intracellular membranes and organelles can originate from two main mechanisms: by invagination and fission, as for the formation of the nuclear envelope continuous with the rough and smooth endoplasmic reticulum (ER) or by symbiotic capture of another cell. As relic of their symbiogenetic history, mitochondria and chloroplasts are enwrapped in two, three, or four membranes, from which only the external envelope is of host origin. Although peroxisomes lack DNA and are surrounded by a single membrane, an endosymbiotic origin of the organelle has been suggested on theoretical bases: it would be derived from an anaerobic hydrogen-producing prokaryote that was enslaved before mitochondria and chloroplasts and, therefore, would have lost DNA and membranes long ago (Cavalier-Smith 1997) and would proliferate by growth and division of preexisting organelles (Lazarow and Fujiki 1985). Other authors have suggested a common origin for Golgi, lysosomes, and peroxisomes, directly derived by invagination from the cell membrane or as merely outgrowths of the ER. Direct proof for any of these hypotheses is lacking, and the issue has never been addressed with comparative genomic tools.
Here we show that, unlike the enzymes residing in the peroxisomal matrix, such as the various oxidases, there is no evidence for proteins of prokaryotic origin at the peroxisomal membrane (although we found restricted motifs of homology widespread in proteins playing the most unrelated functions, see Supplementary Table 1, Supplementary Material online). In contrast, the protein import machinery in membranes of mitochondria and chloroplasts contains a large set of translocases from eubacterial or cyanobacterial origin (Dyall, Brown, and Johnson 2004), such as Omp85. The role of Omp85-like proteins (such as the plastidial Toc75 or mitochondrial Sam50) is critical for outer envelope biogenesis (Genevrois et al. 2003) and for converting endosymbiotic bacteria to organelles (Gentle et al. 2004). In peroxisomes, the insertion of protein into the membrane relies on peroxins, such as Pex16 or the peroxisomal markers Pex3 and Pex19, which are truly eukaryotic proteins, in the absence of any Omp85 or mitochondrial/plastidial homologues. Moreover, we were unable to identify secondary sequence homologies, such as beta-barrels typical of the pore-type mitochondrial and bacterial translocons. Altogether this is strong evidence for the eukaryotic nature of the proteins of the peroxisomal membrane.
Concerning biogenesis and proliferation, peroxisomes are rather particular organelles. Peroxisomal membranes normally arise by division of preexisting membranes of the same type, but, unlike mitochondria, chloroplasts, the nuclear envelope, and rough endoplasmic reticulum membranes, they can also be regenerated de novo in a matter of hours upon transfection of the missing gene—this argues against a symbiogenetic origin for peroxisomes (Matsuzono et al. 1999; South and Gould 1999; Muntau et al. 2000). The source of this newly synthetized membrane and organelle remained, however, controversial. Some groups presented evidence (in Yarrowia lipolytica) of the ER membranes as donor of the required components (Titorenko, Ogrydziak, and Rachubinski 1997), while others sustained the hypothesis of elusive peroxisomal remnants or proto-peroxisomal membranes (Hazra et al. 2002; Lazarow 2003). While this article was in preparation, the issue has elegantly and definitely been settled: the peroxisomal markers essential for biogenesis, Pex3 and Pex19, initially localize to the ER before maturing into import-competent peroxisomes, at least in S. cerevisiae (Hoepfner et al. 2005).
Players of a Mechanistic ER-Peroxisome Connection
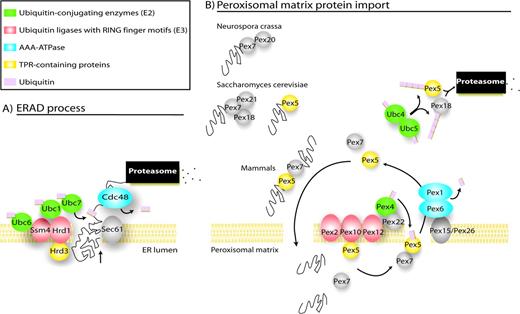
Our results indicate that the other two markers, the components of the import machinery Pex10 and Pex12, also exhibit a relationship to ER, although of different nature. These proteins are E3 ubiquitin ligases and belong to a subclass that harbors a Zn-RING finger domain, able to bind to E2 ubiquitin-conjugating enzymes. This Zn-RING domain is widely represented in Eukarya, absent in Prokarya although strikingly, and also present in a few viruses. Interestingly, two membrane-bound ER resident proteins are E3s ubiquitin ligases with a Zn-RING domain that participate in the ER-associated degradation (ERAD) process for turnover of misfolded and short-lived proteins (Jarosch et al. 2002a) (depicted in fig. 2A). The ERAD process is based on retrograde protein translocation from the ER membrane to the cytosol, mediated by targeting through a TPR-containing protein (Hrd3), association with ubiquitin-conjugating enzymes (E2s) (Bays et al. 2001) which are linked to the membrane by the E3 ubiquitin ligases, subsequent ubiquitination, and finally recruiting of the 26S proteasome to the ER membrane by the AAA-adenosine triphosphatase (ATPase) Cdc48 complex, for substrate degradation (Jarosch et al. 2002b).
ER-peroxisome connection (A) ERAD process, adapted from McCracken and Brodsky (2003). (B) Essential components of the import machinery in peroxisomes. Based on orthology/phylogenetic criteria, we propose that peroxisome and ER would use a similar mechanism to recycle the peroxisomal receptors and export proteins to the cytosol, respectively. First, the proteins directed to the peroxisome would be binding to the peroxisomal Pex5 or Pex7 receptors (Pex7 binds to Pex20 in Neurospora crassa, to Pex18 and Pex21 in Saccharomyces cerevisiae, and to the long form of Pex5 in mammals). Once bound to their cargoes, Pex5 and Pex7 would approach the peroxisomal membrane and get translocated into the peroxisome. In the matrix, receptors will be uncoupled and exported to the cytosol for recycling. The attachment of ubiquitin to Pex5 emerging from the export channel, most likely by Pex4, would drive the receptor dislocation. Pex1 and Pex6 would then recognize this single ubiquitin as signal, then unfold the complex and mediate the export to the cytosol using ATP hydrolysis. Other cytosolic E2, such as Ubc4 and Ubc5, would polyubiquitinate Pex5 and Pex18 and address them to the proteasome for degradation.
Striking analogies exist between the ERAD process components and the components of the peroxisome import system (fig. 2B): (1) Pex2, Pex12, and Pex10 are membrane embedded, RING finger E3 ubiquitin ligases like the ER proteins Hrd1 or Ssm4; (2) Pex5 is a TPR-containing protein like the ER Hrd3p; (3) Pex4-conjugating enzyme family (E2) like the E2 components of the ER, Ubc7, Ubc6, and Ubc1; and (4) finally Pex6 and Pex1 are bona fide AAA-ATPases (Birschmann et al. 2003), belonging a monophyletic cluster with Cdc48 (35% identity the protein level with Cdc48p). Very recently, a proteasome-independent function for Cdc48 has been identified; it mediates the reassembly of Golgi cisternae after mitosis, in an ubiquitin-dependent manner (Wang et al. 2004). In a similar manner, ubiquitin modification could serve as regulatory signal for sorting in the peroxisomal scenario, independent of the proteasome degradation process. Thus, we propose that peroxisome and ER would use a similar mechanism to recycle the peroxisomal receptors (implicating ubiquitination) and to export proteins to the cytosol, respectively. Figure 2 highlights the resemblances between both systems. Noteworthy, two components of this core complex are the essential Pex12 and Pex10 markers, plus the quasi-marker Pex5.
Along these lines, and while this article was under preparation and review, other authors have found experimental evidences linking ERAD and peroxisome receptor recycling processes (Erdmann and Schliebs 2005; Kragt et al. 2005); for instance (1) Pex1 and Pex6 dislocate Pex5 from peroxisome to cytosol in an ATP-dependent manner (Miyata and Fujiki 2005; Platta et al. 2005) and (2) Pex5 is ubiquitinated after receptor docking and RING finger protein intercession (Erdmann and Schliebs 2005; Kiel et al. 2005; Kragt et al. 2005; Platta et al. 2005)
In view of these observations, we propose that a common ancestor of the complex E2/E3/AAA-ATPase existed in the primitive eukaryotic cell membranes, which served to form the first ERs and the first peroxisomes. The core translocation machinery, the mechanistics, and the functional units would be conserved to present day. Although we cannot exclude the possibility that peroxisomal enzymes were donated by an early endosymbiont, the fact that the peroxisomal membrane is purely constituted by eukaryotic proteins provides robust evidence against the symbiogenetic theory.
Geoffrey McFadden, Associate Editor
This study was supported by funds from the Institut National de la Santé et de la Recherche Médicale, the Centre National de la Recherche Scientifique, the European Commission contract no. LSHM-CT2004-502987, the Association Française contre les Myopathies (AFM) (Project no. 9315), the European Leukodystrophy Association, and the Asociación Española contra la Leucodistrofia (ALE-ELA España). A.S. was a fellow of the AFM, Decrypton Program. S.F. was a fellow of the ELA and the European Commission.
References
Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman.
Amery, L., H. Sano, G. P. Mannaerts, J. Snider, J. Van Looy, M. Fransen, and P. P. Van Veldhoven.
Baum, B. R.
Bays, N. W., R. G. Gardner, L. P. Seelig, C. A. Joazeiro, and R. Y. Hampton.
Birschmann, I., A. K. Stroobants, M. van den Berg, A. Schafer, K. Rosenkranz, W. H. Kunau, and H. F. Tabak.
———.
Daubin, V., M. Gouy, and G. Perriere.
Dyall, S. D., M. T. Brown, and P. J. Johnson.
Erdmann, R., and W. Schliebs.
Galtier, N., M. Gouy, and C. Gautier.
Genevrois, S., L. Steeghs, P. Roholl, J. J. Letesson, and L. P. van der.
Gentle, I., K. Gabriel, P. Beech, R. Waller, and T. Lithgow.
Hazra, P. P., I. Suriapranata, W. B. Snyder, and S. Subramani.
Hoepfner, D., D. Schildknegt, I. Braakman, P. Philippsen, and H. F. Tabak.
Honsho, M., S. Tamura, N. Shimozawa, Y. Suzuki, N. Kondo, and Y. Fujiki.
Jarosch, E., R. Geiss-Friedlander, B. Meusser, J. Walter, and T. Sommer.
Jarosch, E., C. Taxis, C. Volkwein, J. Bordallo, D. Finley, D. H. Wolf, and T. Sommer.
Kaasch, A. J., and K. A. Joiner.
Kiel, J. A., K. Emmrich, H. E. Meyer, and W. H. Kunau.
Kragt, A., T. M. Voorn-Brouwer, B. M. Van den, and B. Distel.
Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei.
Lazarow, P. B.
Li, X., E. Baumgart, G. X. Dong, J. C. Morrell, G. Jimenez-Sanchez, D. Valle, K. D. Smith, and S. J. Gould.
Matsuzaki, M., O. Misumi, I. Shin et al. (42 co-authors).
Matsuzono, Y., N. Kinoshita, S. Tamura, N. Shimozawa, M. Hamasaki, K. Ghaedi, R. J. Wanders, Y. Suzuki, N. Kondo, and Y. Fujiki.
McCracken, A. A., and J. L. Brodsky.
Miyata, N., and Y. Fujiki.
Muntau, A. C., P. U. Mayerhofer, B. C. Paton, S. Kammerer, and A. A. Roscher.
Platta, H. W., S. Grunau, K. Rosenkranz, W. Girzalsky, and R. Erdmann.
Ragan, M. A.
Schrader, M., B. E. Reuber, J. C. Morrell, G. Jimenez-Sanchez, C. Obie, T. A. Stroh, D. Valle, T. A. Schroer, and S. J. Gould.
Shimozawa, N., Y. Suzuki, Z. Zhang, A. Imamura, K. Ghaedi, Y. Fujiki, and N. Kondo.
South, S. T., and S. J. Gould.
Thompson, J. D., D. G. Higgins, and T. J. Gibson.
Titorenko, V. I., D. M. Ogrydziak, and R. A. Rachubinski.
Titorenko, V. I., and R. A. Rachubinski.
Wanders, R. J.
Author notes
*Institut de Génétique et de Biologie Moléculaire et Cellulaire, Centre National de la Recherche Scientifique/Institut National de la Sante et de la Recherche Medicale/Université Louis Pasteur/Collège de France, Illkirch, France; †Centre de Genètica Mèdica i Molecular, Institut de Recerca Oncològica/Institut d'Investigació Biomèdica de Bellvitge, Hospital Duran i Reynals, Hospitalet de Llobregat, Barcelona, Spain; and ‡Institució Catalana de Recerca i Estudis Avançats, Barcelona, Spain