-
PDF
- Split View
-
Views
-
Cite
Cite
Haixiang Yu, Weijuan Yang, Obtin Alkhamis, Juan Canoura, Kyung-Ae Yang, Yi Xiao, In vitro isolation of small-molecule-binding aptamers with intrinsic dye-displacement functionality, Nucleic Acids Research, Volume 46, Issue 8, 4 May 2018, Page e43, https://doi.org/10.1093/nar/gky026
Close - Share Icon Share
Abstract
Aptamer-based sensors offer a powerful tool for molecular detection, but the practical implementation of these biosensors is hindered by costly and laborious sequence engineering and chemical modification procedures. We report a simple strategy for directly isolating signal-reporting aptamers in vitro through systematic evolution of ligands by exponential enrichment (SELEX) that transduce binding events into a detectable change of absorbance via target-induced displacement of a small-molecule dye. We first demonstrate that diethylthiatricarbocyanine (Cy7) can stack into DNA three-way junctions (TWJs) in a sequence-independent fashion, greatly altering the dye's absorbance spectrum. We then design a TWJ-containing structured library and isolate an aptamer against 3,4-methylenedioxypyrovalerone (MDPV), a synthetic cathinone that is an emerging drug of abuse. This aptamer intrinsically binds Cy7 within its TWJ domain, but MDPV efficiently displaces the dye, resulting in a change in absorbance within seconds. This assay is label-free, and detects nanomolar concentrations of MDPV. It also recognizes other synthetic cathinones, offering the potential to detect newly-emerging designer drugs, but does not detect structurally-similar non-cathinone compounds or common cutting agents. Moreover, we demonstrate that the Cy7-displacement colorimetric assay is more sensitive than a conventional strand-displacement fluorescence assay. We believe our strategy offers an effective generalized approach for the development of sensitive dye-displacement colorimetric assays for other small-molecule targets.
INTRODUCTION
Aptamers are nucleic acid-based molecules that can be isolated in vitro through systematic evolution of ligands by exponential enrichment (SELEX) techniques to bind various targets with high specificity and affinity (1,2). They are increasingly being used as recognition elements in biosensing platforms due to their low cost of production, ease of modification, chemical stability, and long shelf life (2–4). The generation of a sensor readout typically requires an aptamer to undergo some manner of binding-induced change, and most aptamer-based sensors utilize structure-switching aptamers that perform a major conformational rearrangement upon target binding. However, most aptamers do not innately undergo a measureable binding-induced conformational change, and the development of a structure-switching aptamer typically entails a multi-stage process of sequence analysis and chemical modification (3–5).
SELEX methods have been developed that make it possible to isolate aptamers with inherent structure-switching functionality. For example, Ellington (6) and Li (7) have utilized a ‘strand-displacement’ strategy to directly isolate structure-switching aptamers. Their approach begins with the hybridization of library molecules with an immobilized complementary strand. Library strands that bind to the target undergo a conformational change, dissociate from the complementary strand, and are collected in the supernatant. After several rounds of isolation and enrichment, these aptamers are sequenced, chemically modified with a fluorophore, and can then be directly employed in strand-displacement assays with a quencher-modified complementary strand. This approach eliminates the need for sequence engineering, but there is an inherent conflict between the requirements for aptamer isolation and sensor development. This is because the isolation of high-affinity aptamers requires strong hybridization of the complementary strand with the library molecules in order to create an energetic barrier for stringent selection. However, since the complementary strands have high binding affinity for the aptamer, they inadvertently inhibit target binding and consequently reduce the sensitivity of strand-displacement assays. This is particularly problematic for the detection of small-molecule targets, which usually have micromolar affinities for aptamers. As such, the limit of detection of strand-displacement assays for small molecules is usually comparable to or even higher than the dissociation constant (KD) of the aptamer being employed (7–11). Moreover, such assays require a heating-and-cooling process lasting ≥ 30 min to ensure complete hybridization between the complementary strand and aptamer to achieve low background. This time-consuming step greatly hinders the use of these assays for rapid on-site detection. Nonetheless, the strand-displacement assay is presently the only generally applicable method for developing aptamer-based small-molecule sensors directly from isolated aptamers without further engineering.
Dye-displacement strategies offer an appealing alternative in this regard (12–15). In such assays, a small-molecule dye is initially associated with the aptamer-binding domain. The target displaces the dye from the binding domain, resulting in a change in the absorbance or fluorescence of the dye. These assays do not require any additional labeling or chemical modification of the aptamer. Since the aptamers typically bind to these small-molecule targets and dyes with similar affinities, target-induced dye-displacement is more thermodynamically feasible than the displacement of a tightly-bound complementary strand. As a result, dye-displacement assays can achieve a detection limit that is at least 10-fold lower than the KD of the aptamer being used (12–15). However, this assay strategy has not been generalized, as there are currently no sequence engineering strategies or SELEX techniques that can directly isolate aptamers with dye-displacement functionality.
Herein, we for the first time have developed a new library-immobilized SELEX strategy for the isolation of small-molecule-binding aptamers with inherent dye-displacement functionality. The isolated aptamers can rapidly report binding events through target-induced dye-displacement, which can then be employed in a generalized sensor platform for the detection of small-molecule targets. Previous research has demonstrated that diethylthiatricarbocyanine (Cy7) can bind to a cocaine-binding aptamer via its three-way junction (TWJ) binding domain (12), and we have now demonstrated that such Cy7 binding is a general and sequence-independent feature of DNA TWJ structures. This interaction produces a distinctive change in absorbance at 670 and 760 nm, which allowed us to isolate aptamers that undergo target binding-induced dye-displacement from a TWJ-containing structured library. These aptamers can then be directly employed to optically report the presence of their respective target through displacement of Cy7. As a demonstration, we isolated an aptamer for 3,4-methylenedioxypyrovalerone (MDPV), a synthetic cathinone designer drug. The isolated aptamer retains a TWJ-structured binding domain and has a KD of 6.1 μM. The addition of MDPV displaced Cy7 from the TWJ domain, resulting in a change in the absorbance of the dye. Using this label-free assay, we were able to detect MDPV at a concentration as low as 300 nM—20-fold lower than the KD of the aptamer—within seconds. Our Cy7-displacement assay was more sensitive and rapid than an equivalent strand-displacement fluorescence assay, which used a fluorophore-modified version of the same aptamer along with a quencher-modified complementary strand. Importantly, our isolated aptamer is also cross-reactive to other synthetic cathinones, which represent a diverse emerging class of drugs of abuse, while remaining non-responsive to other structurally similar non-cathinone molecules and commonly-used cutting agents. Our method should be generally applicable as a means to enable the rapid and sensitive detection of a variety of small-molecule targets that bind to three-way junction–derived receptors.
MATERIALS AND METHODS
Materials
All oligonucleotides were ordered from Integrated DNA Technologies (IDT), purified with HPLC and dissolved in PCR quality water (Invitrogen). DNA concentrations were measured using a NanoDrop 2000 spectrophotometer (Thermo Scientific). Diethylthiatricarbocyanine iodide (Cy7), dehydroisoandrosterone-3-sulfate (DIS) sodium salt dihydrate, deoxycorticosterone-21 glucoside (DOG) and all other chemicals were purchased from Sigma-Aldrich unless otherwise noted. Drug standards, including 3,4-methylenedioxypyrovalerone (MDPV), mephedrone, methylone, naphyrone and pentylone were purchased from Cayman Chemicals. Tween 20 and formamide were purchased from Fisher Scientific. Streptavidin-coated agarose resin (binding capacity: 1−3 mg biotinylated BSA/ml resin), One Shot Chemically Competent Escherichia coli, and SYBR Gold were purchased from Thermo Scientific. 500 μl micro-gravity columns were purchased from BioRad.
SELEX procedure
The isolation of MDPV-binding aptamers follows a previously-reported SELEX method (10) with some modifications. Briefly, during each round of SELEX, 100 pmole of either the TWJ-containing library or the enriched pool from a given round was mixed with 500 pmole of biotinylated complementary strand (cDNA-bio) in 250 μl selection buffer (10 mM Tris–HCl, 0.5 mM MgCl2, 20 mM NaCl, pH 7.4). The mixture was heated at 95°C for 5 min, slowly cooled to room temperature over 30 min, and then loaded into a micro-gravity column containing 250 μl of streptavidin-coated agarose resin for library immobilization. Unconjugated library and complementary strands were removed by flowing 250 μl of selection buffer through the library-immobilized column at least 10 times. Target-bound aptamers were then eluted at room temperature with 250 μl of selection buffer containing either 100 μM (Rounds 1–5) or 50 μM (Rounds 6–10) MDPV. Each elution was completed in 2 min. The elution was repeated two more times to increase the yield. The combined eluted aptamers were concentrated using a 3K molecular weight cut-off spin filter (Millipore) and mixed with 1 μM forward primer (Supplementary Table S1, FP) and 1 μM biotinylated reverse primer (Supplementary Table S1, RP-bio) in 1 ml GoTaq Hot Start Master Mix (Promega). Polymerase chain reaction (PCR) was then performed using a BioRad C1000 thermal cycler. Amplification conditions were as follows: 1 cycle of 95°C for 2 min; 13 cycles of 95°C for 15 s, 58°C for 30 s and 72°C for 45 s; and 1 cycle of 72°C for 2 min. The double-stranded PCR product was immobilized on a micro-gravity column containing 200 μl of streptavidin-coated agarose resin. The column was washed five times with 200 μl of 10 mM Tris–HCl buffer (pH 7.4) containing 20 mM NaCl to remove unconjugated strands, and then incubated with 300 μl of 0.2 M NaOH for 10 min. Finally, the eluent was collected and neutralized with 0.2 M HCl in a 1.5 ml microcentrifugation tube and concentrated using a 3K molecular weight cut-off spin filter. The enriched pool was used for the next round of SELEX after measuring its concentration with a NanoDrop 2000.
Counter-SELEX was performed against methamphetamine (Rounds 4–5), amphetamine (Rounds 6–7) and dopamine (Rounds 8–10). Specifically, the library-immobilized column was washed 10 times with 250 μl of 100 μM counter-SELEX target in selection buffer, followed by 20 times with 250 μl of selection buffer prior to target elution.
Cloning and sequencing
After 10 rounds of SELEX, the final enriched pool was PCR amplified with unlabeled forward and reverse primers (Supplementary Table S1, FP and RP) under the conditions described above, with a prolonged 30-min extension step at 72°C to add an A-tail. The PCR product was cloned into E. coli using the TOPO TA cloning kit (Invitrogen). Fifty colonies were randomly picked and sequenced at the Florida International University DNA Core Facility. Multiple sequence alignments were carried out using BioEdit software, and the sequence logo was generated using WebLogo (16).
Cy7 displacement assay for detection of MDPV
5 μl of MDPV-binding aptamer (Supplementary Table S1, MA) (final concentration: 7 μM), 5 μl of Cy7 (final concentration = 3 μM), 5 μl of varying concentrations of MDPV, and 35 μl of reaction buffer (10 mM Tris–HCl, 0.5 mM MgCl2, 20 mM NaCl, 0.01% Tween 20, 1% DMSO, pH 7.4) were mixed in wells of a 384-well plate. UV–vis spectra were immediately recorded from 450–900 nm using a Tecan Infinite M1000 PRO at room temperature. Signal gain was calculated by (R – R0)/R0, where R0 and R are the 670/760 nm absorbance ratio without and with MDPV, respectively. The same assay was performed with 50 and 250 μM of naphyrone, methylone, pentylone and mephedrone to evaluate the cross-reactivity of the assay. Additionally, we tested the specificity of the assay using 50 and 250 μM of interfering agents such as amphetamine, methamphetamine, dopamine, lidocaine, benzocaine and caffeine.
RESULTS
Binding mechanism of Cy7 to TWJs
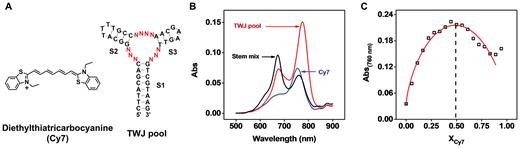
Stojanovic et al. have previously reported that Cy7 (Figure 1A) specifically binds to the TWJ-structured binding domain of a cocaine-binding aptamer via hydrophobic interactions (12). Cocaine displaces Cy7 from the TWJ, resulting in a significant reduction in the absorbance of the dye at 760 nm. We demonstrated the generality of this Cy7-displacement mechanism using another two reported TWJ-structured aptamers which bind to dehydroisoandrosterone-3-sulfate (DIS)- and deoxycorticosterone-21 glucoside (DOG) (10). Cy7 was immediately displaced from both aptamers upon the addition of their respective targets, yielding a significant decrease in the absorbance of the dye at 760 nm (Supplementary Figure S1). Based on these results, we hypothesized that Cy7 binds to our TWJs in a sequence-independent manner, a principle that could guide the SELEX-based isolation of new small-molecule binding aptamers with intrinsic Cy7-displacement functionality.
Cy7 binding to a TWJ-containing structured DNA pool in a sequence-independent manner. (A) Chemical structure of Cy7 and the sequence of the randomized TWJ pool. (B) When combined with the TWJ pool, the absorbance of Cy7 monomer (760 nm) and dimer (670 nm) is enhanced. In contrast, when combined with a 1:1:1 mixture of isolated S1, S2 and S3 stem–loops, only the absorbance of the dimer is enhanced. (C) Binding stoichiometry of Cy7 monomer to the TWJ pool as characterized by a Job plot.
To confirm our hypothesis, we chemically synthesized a 46-nt TWJ-structured DNA library, in which each strand contains three fully complementary stems (S1, S2 and S3) and 10 randomized nucleotides within the junction itself (Figure 1A and Supplementary Table S1, TWJ pool). After mixing 2 μM Cy7 with 10 μM TWJ pool, we observed an increase in the absorbance of the dye at 760 and 670 nm, which respectively corresponds to binding of the Cy7 monomer and dimer (Figure 1B) (17,18). To assess the binding of Cy7 to various domains within the TWJ pool, we synthesized the isolated S1 stem with an added terminal TTT loop (termed the S1 stem–loop), the S2 stem–loop and the S3 stem–loop (Supplementary Table S1 and Supplementary Figure S2A) and characterized the binding of Cy7 to each of them. We observed that none of the stem–loops bound to the monomer (Supplementary Figure S2B), and the dimer only bound to the S1 stem–loop—most likely within its minor groove (18). When we incubated Cy7 with a 1:1:1 mixture of the three isolated stem–loops, only dimer binding was observed (Figure 1B). These results clearly indicated that the Cy7 monomer can only bind to the fully-assembled TWJ domain of the aptamer.
We then obtained the binding affinity of Cy7 monomer for the TWJ pool by titrating different concentrations of the pool into 2 μM Cy7. As the concentration of the TWJ pool increased, we observed an increase in absorbance at 760 nm and a slight decrease in absorbance at 670 nm (Supplementary Figure S3A). This provided further evidence that the TWJ domain preferentially binds to the Cy7 monomer. The decreased absorbance of the Cy7 dimer at higher DNA concentrations implies an equilibrium shift from dimer to monomer that favors Cy7 binding to the TWJ pool. Based on the absorbance at 760 nm, we obtained a KD of 6.4 ± 0.8 μM (Supplementary Figure S3B). We further characterized the binding stoichiometry of the Cy7 monomer to the TWJ pool using a Job plot (19). We measured the absorbance at 760 nm at various Cy7:DNA ratios, with a constant total concentration of 5 μM. The maximum absorbance was observed at a mole fraction (XCy7) of 0.5, indicating that the TWJ pool binds to Cy7 in a 1:1 ratio. Since the Cy7 monomer exclusively binds to the TWJ domain, each randomized TWJ in the pool should bind to a single Cy7 monomer (Figure 1C). These results confirmed our initial hypothesis that the binding of Cy7 to TWJs is sequence-independent, and indicated that it should therefore be feasible to isolate new small-molecule-binding aptamers with intrinsic dye-displacement functionality from TWJ-structured libraries.
Library design and isolation process
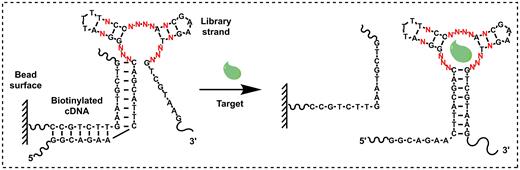
We designed a structured library in which we extended both termini of the TWJ pool by 12 nucleotides, creating partial PCR primer binding sites (Figure 2, Supplementary Table S1, Library). We have previously demonstrated that changes in the sequence adjacent to the TWJ binding domain greatly affected the affinity of a cocaine-binding aptamer (13,20). With this in mind, we randomized one base-pair in stems 2 and 3 to further enhance the potential target-binding affinity of our aptamer. The resulting library is designed to primarily form TWJ-structured target-binding domains. Although there is a small possibility of isolating non-TWJ-structured aptamers in the final enriched pool, these sequences can be identified and removed after sequencing. It should be noted that the randomness of this library (414 sequences) is considerably lower than many reported SELEX libraries (7,11,21). However, this should not be of concern, as small-molecule targets can only interact with a limited number of nucleotides within an aptamer's binding domain. In fact, several small-molecule-binding aptamers have been successfully isolated using a TWJ-structured library representing no more than 48 different sequences (10). Increasing the number of random nucleotides in the TWJ binding domain could also lead to the isolation of aptamers with non-TWJ structures, which lack dye-displacement functionality.
Isolation of TWJ-structured aptamers for a Cy7-displacement small-molecule detection assay. A TWJ-containing structured DNA library is immobilized onto a streptavidin-coated bead surface using a biotinylated cDNA strand (left). Target-binding strands are eluted due to a conformational change in the aptamer (right).
We then used this TWJ-containing library along with a previously reported library-immobilized SELEX strategy (10) to isolate aptamers for MDPV, an emerging drug of abuse in the synthetic cathinone family. Since there is currently no reliable presumptive test for any synthetic cathinone, the development of such an assay would be highly valuable. We performed SELEX in a low ionic strength buffer (10 mM Tris–HCl, 20 mM NaCl, 0.5 mM MgCl2, pH 7.4), as an excessive amount of ions can induce non-specific hydrophobic interactions (22) between the target and the TWJ binding domain, which disfavors the isolation of highly target-specific aptamers. We immobilized our library onto streptavidin-coated agarose beads through hybridization to a biotinylated complementary DNA (cDNA) strand (Supplementary Table S1, cDNA-bio) that disrupts the TWJ structure (Figure 2A, left). In the presence of MDPV, library molecules that recognized this target underwent a conformational change and refolded into a TWJ structure, thereby detaching themselves from the biotinylated cDNA (Figure 2A, right). These aptamers were collected, PCR amplified, and used for the next round of selection. To further improve aptamer specificity, we sequentially performed counter-SELEX at various rounds against three molecules that are structurally similar to MDPV: amphetamine, methamphetamine and dopamine.
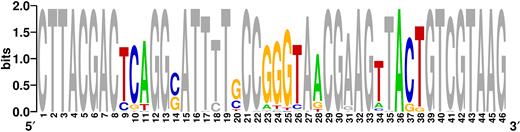
After each round of SELEX, we estimated the target affinity and specificity of the enriched pool with a target elution assay (see Supplementary Data for detailed methods). After 10 rounds of selection, the enriched pool achieved saturated target affinity and low cross-reactivity to the counter-SELEX targets (Supplementary Figure S4). The isolated aptamers were then cloned and sequenced. We observed that all sequences retained the TWJ structure present in the original library. The 10 random nucleotides (Figure 3, positions 9–11, 23–26, 36–38) in the TWJ binding domain exhibited a high level of consensus, with 41 out of 50 clones sharing an identical sequence at these sites. The four random nucleotide positions within the stems were more varied (Figure 3, positions 14, 20, 28 and 34), but predominantly retained standard Watson-Crick base-pairing. We selected the most highly-represented sequence and truncated the 12-nucleotide partial primer-binding sites at both termini to generate the MDPV-binding aptamer termed MA (Supplementary Table S1).
Sequence logo for 50 clones isolated after 10 rounds of SELEX, showing the relative frequency of every nucleotide at each position. Nucleotides at pre-determined positions are marked gray, while positions with randomized nucleotides are color-coded to show the distribution of different nucleotides. Higher frequency nucleotides appear in a larger font-size.
Aptamer characterization and development of the label-free Cy7-displacement assay
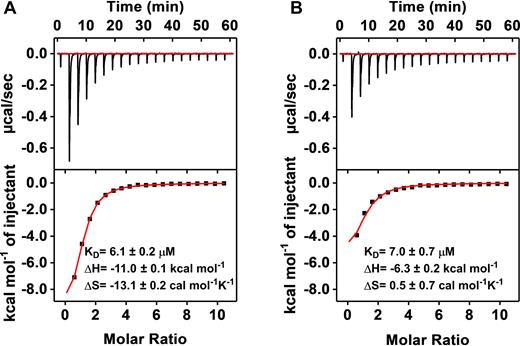
We determined the binding affinity of MA using isothermal titration calorimetry (ITC), titrating 1 mM MDPV into a 20 μM solution of the aptamer. We found that MA binds MDPV with a KD of 6.1 ± 0.2 μM (Figure 4A). Binding is driven by a negative enthalpy change (ΔH = –11.0 ± 0.2 kcal mol−1) but with a moderate entropy cost (ΔS = –13.1 ± 0.2 cal mol−1K−1) due to the fact that MDPV is constrained within the TWJ binding domain. Based on our prior findings, we anticipated that Cy7 would also bind to the TWJ domain of MA but could be displaced by MDPV. This was confirmed by performing an ITC experiment in which we titrated 1 mM MDPV into 20 μM Cy7–MA complex (1:1). The presence of Cy7 did not significantly reduce the MDPV-binding affinity of MA (KD = 7.0 ± 0.7 μM) (Figure 4B). However, the binding heat (ΔH = –6.3 ± 0.2 kcal/mol) was notably lower than in the absence of Cy7 (ΔH = –11.0 ± 0.2 kcal mol−1), indicating that a certain amount of energy was required for target-induced displacement of the dye. Additionally, the higher binding entropy obtained with Cy7 (ΔS = 0.5 ± 0.7 cal mol−1 K−1) suggested positive entropy associated with the MDPV-mediated release of the dye from the aptamer. These results indicate that both Cy7 and MDPV can bind to the TWJ-structured binding domain, and that MDPV can efficiently displace Cy7.
ITC characterization of MA binding to MDPV in the (A) absence or (B) presence of Cy7. The top panels display raw data, showing the heat generated from each titration of MDPV. The bottom panels show the integrated heat of each titration after correcting for dilution heat of the titrant.
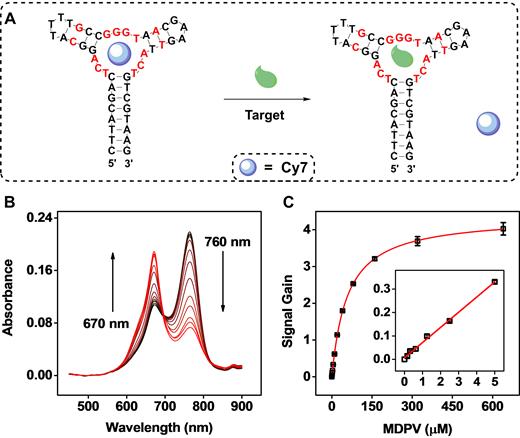
We then employed MA in a label-free colorimetric assay for the detection of MDPV, where target-induced displacement of Cy7 from the aptamer TWJ binding domain produces a change in dye absorbance (Figure 5A). Specifically, we added different concentrations of MDPV (0.1–640 μM) to a solution containing Cy7–MA complexes, and then immediately began recording the absorption spectra. Within seconds, we observed a progressive reduction of absorbance at 760 nm and enhancement of absorbance at 670 nm (Figure 5B). Using the absorbance ratio at 670/760 nm as an indicator, we obtained a linear range from 0 to 40 μM, with a low detection limit of 300 nM (Figure 5C).
Detection of MDPV with a label-free Cy7-displacement colorimetric assay. (A) Cy7 binds within the TWJ domain of MA (left). MDPV displaces Cy7 from the binding domain (right) to generate a change in the absorbance spectra of Cy7. (B) UV–vis spectra of 3 μM Cy7 premixed with 7 μM MA in the presence of varying concentrations of MDPV (0, 0.16, 0.31, 0.63, 1.25, 2.5, 5, 10, 20, 40, 80, 160, 320, 640 μM). The increasing concentrations (shown as black→red curves) resulted in decreased absorbance at 760 nm and increased absorbance at 670 nm. (C) A calibration curve based on the absorbance ratio at 670/760 nm. Inset shows assay performance at a low concentration range. Error bars show standard deviation from three measurements at each concentration.
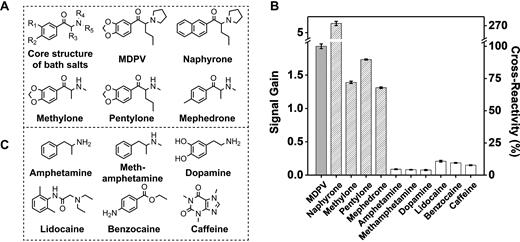
Notably, MA was highly cross-reactive to other synthetic cathinones (methylone, pentylone, naphyrone and mephedrone) that share the same beta-keto phenethylamine core structure as MDPV but with different side-chains (Figure 6A). We tested MA cross-reactivity to these four synthetic cathinones at concentrations of 50 and 250 μM in the Cy7-displacement assay. We observed cross-reactivity >65% and 80% for all four molecules at both concentrations (Figure 6B and Supplementary Figure S5). This suggests that MA specifically recognizes the core structure of synthetic cathinones rather than their side chain substituents. Interestingly, naphyrone showed exceptionally high cross-reactivity compared to other synthetic cathinones, possibly due to the hydrophobicity of its naphthalene moiety. The high cross-reactivity of MA is desirable for onsite detection of the synthetic cathinone family, a class of designer drugs for which new derivatives are continually being developed. In contrast, MA was not responsive to non-synthetic cathinone interfering agents. No specific signal was observed from any of our three counter-SELEX targets (Figure 6C) in our Cy7-displacement assay at concentrations of 50 μM (Figure 6B) or 250 μM (Supplementary Figure S5), despite their structural similarity to MDPV. We further tested the specificity of the assay against lidocaine, benzocaine, and caffeine, which are cutting agents commonly found in seized substances (Figure 6C), and likewise observed little cross-reactivity at either concentration (Figure 6B and Supplementary Figure S5). It should be noted that the counter-SELEX targets and cutting agents either do not contain all the moieties present in MDPV (a phenyl ring, a ketone and an amine group), or have all of these moieties but arranged in different positions. These results indicate that MA recognizes targets based on multiple specific interactions in a certain arrangement. We found that MA was somewhat cross-reactive to the structurally dissimilar compounds cocaine, DIS, and DOG at a concentration of 50 μM, with cross-reactivities of 50%, 18% and 40% respectively relative to MDPV (Supplementary Figure S6) in the Cy7-displacement assay. We believe this cross-reactivity occurs because counter-SELEX was not performed against these particular compounds, but it should be feasible to eliminate this unintended cross-reactivity with well-designed counter-SELEX procedures against additional structurally dissimilar compounds.
High cross-reactivity of MA to synthetic cathinone drugs and high specificity against other structurally similar or dissimilar interfering agents. (A) Chemical core structure and structures of individual synthetic cathinones. (B) Signal gain and cross-reactivity measurements from the Cy7-displacement assay with 50 μM MDPV, other synthetic cathinones, or interfering agents (structures shown in panel C). Error bars show standard deviations from three measurements of each compound.
Comparison of Cy7-displacement and strand-displacement assays
Finally, we compared the sensitivity of our Cy7-displacement colorimetric assay with a strand-displacement fluorescence assay (Supplementary Figure S7) based on the same MDPV-binding aptamer. We generated a fluorophore-modified version of MA (Supplementary Table S1, MA-F) and a quencher-modified complementary strand (Supplementary Table S1, cDNA-Q). In the absence of MDPV, MA-F forms a 15-bp duplex with cDNA-Q that situates the fluorophore in close proximity to the quencher (Supplementary Figure S7A). The fluorescence of 50 nM MA-F was almost completely quenched in the presence of 50 nM cDNA-Q (Supplementary Figure S8A). We subsequently titrated different concentrations of cDNA-Q into 50 nM MA-F and obtained a KD of 0.5 ± 0.2 nM (Supplementary Figure S8A, inset). In the presence of MDPV, the target induced a conformational change in MA-F, forming a TWJ structure that resulted in dissociation of cDNA-Q, generating a measurable fluorescence signal (Supplementary Figure S7B). We observed less than 50% fluorescence recovery upon adding various concentrations of MDPV up to 640 μM (Supplementary Figure S8B). We obtained a linear range of 0–20 μM, with a detection limit of 2.5 μM (Supplementary Figure S8C), eight-fold poorer than the Cy7-displacement assay. This is primarily because cDNA-Q has a much higher affinity for the aptamer than Cy7, which makes target-induced displacement of the complementary strand less energetically favorable. To further illustrate this point, we synthesized unmodified versions of MA-F and cDNA-Q (Supplementary Table S1, MA-L and cDNA) and performed ITC experiments in which we titrated 1 mM MDPV into 20 μM of MA-L either with or without 20 μM cDNA. We found that MA-L has an affinity for MDPV almost identical to that of MA (Supplementary Figure S9A, KD = 6.2 μM) in the absence of cDNA. However, in the presence of 20 μM cDNA, no MDPV binding was observed (Supplementary Figure S9B), confirming that MDPV could not efficiently displace the cDNA from the aptamer. It should be noted that strand-displacement efficiency was lower in the ITC experiment due to the higher concentration of aptamer-cDNA complex employed (20 μM), which greatly reduces the level of dissociation of this complex. As a control, we titrated 1 mM MDPV into 20 μM Cy7-MA-L (1:1), and found that the presence of Cy7 does not attenuate MDPV binding by MA-L (Supplementary Figure S9C, KD = 7.0 μM). These results clearly demonstrate that the Cy7-displacement assay is more sensitive than an equivalent strand-displacement assay for the detection of small-molecule targets.
DISCUSSION
In this report, we describe a general approach for the isolation of small-molecule-binding aptamers with intrinsic dye-displacement functionality that can be directly implemented in a label-free colorimetric assay. We first determined that Cy7 can bind to a randomized TWJ-structured DNA library pool in a sequence-independent manner. We then used a TWJ-structured library to isolate a DNA aptamer against MDPV, an emerging designer drug, and directly employed the isolated aptamer in a Cy7-displacement colorimetric assay without any further engineering. The assay showed high target sensitivity, detecting MDPV at concentrations as low as 300 nM, 20-fold lower than the KD of the aptamer, within seconds at room temperature. Our aptamer showed high specificity in terms of discriminating against structurally-similar non-synthetic cathinone compounds such as amphetamine, methamphetamine, and dopamine as well as common cutting agents such as lidocaine, benzocaine, and caffeine. At the same time, the isolated aptamer was highly cross-reactive to other synthetic cathinones such as naphyrone, methylone, pentylone and mephedrone, indicating that it could offer a broadly applicable yet specific assay for this particular class of designer drugs, based on recognition of shared core structural features.
Our determination that Cy7 can generally bind our DNA TWJs in a sequence-independent manner means that such dye-displacement assays can be generalized for the detection of other small-molecule targets using existing or future isolated TWJ-structured aptamers. Since we have previously demonstrated that the fluorescent dye ATMND can bind to the TWJ domain of a cocaine-binding aptamer (13), we anticipate that other hydrophobic small-molecule dyes may also serve as signal reporters in such dye-displacement assays. Our assay is simple, rapid, sensitive and cost efficient, as it can be performed by simply mixing the sample, Cy7 and unmodified aptamer. The Cy7 absorbance read-out can be quantified in seconds by a microplate-reader or portable photometer, allowing for high-throughput or on-site detection, respectively. Importantly, the absorbance of Cy7 at 670 nm and 760 nm exhibits minimal overlap with other small molecules (23) or sample matrices (24), potentially allowing for interference-free detection in complex specimens. Finally, our assay is eight-fold more sensitive than an equivalent strand-displacement fluorescence assay, because Cy7 is more efficiently displaced by small-molecule targets than a hybridized cDNA strand. Therefore, we believe that our SELEX strategy using our TWJ-structured library could offer a broad foundation for the development of sensitive and simple Cy7-displacement colorimetric assays for the detection of a wide variety of small-molecule targets.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We are grateful for Dr. Milan N. Stojanovic's guidance on aptamer isolation.
FUNDING
National Institutes of Health—National Institute on Drug Abuse (NIDA) [R15DA036821]; National Institute of Justice, Office of Justice Programs, U.S. Department of Justice Award [2015-R2-CX-0034]. Funding for open access charge: NIDA.
Conflict of interest statement. None declared.




Comments