-
PDF
- Split View
-
Views
-
Cite
Cite
Dominique Bréchemier-Baey, Lenin Domínguez-Ramírez, Jacques Oberto, Jacqueline Plumbridge, Operator recognition by the ROK transcription factor family members, NagC and Mlc, Nucleic Acids Research, Volume 43, Issue 1, 9 January 2015, Pages 361–372, https://doi.org/10.1093/nar/gku1265
Close - Share Icon Share
Abstract
NagC and Mlc, paralogous members of the ROK family of proteins with almost identical helix-turn-helix DNA binding motifs, specifically regulate genes for transport and utilization of N-acetylglucosamine and glucose. We previously showed that two amino acids in a linker region outside the canonical helix-turn-helix motif are responsible for Mlc site specificity. In this work we identify four amino acids in the linker, which are required for recognition of NagC targets. These amino acids allow Mlc and NagC to distinguish between a C/G and an A/T bp at positions ±11 of the operators. One linker position, glycine in NagC and arginine in Mlc, corresponds to the major specificity determinant for the two proteins. In certain contexts it is possible to switch repression from Mlc-style to NagC-style, by interchanging this glycine and arginine. Secondary determinants are supplied by other linker positions or the helix-turn-helix motif. A wide genomic survey of unique ROK proteins shows that glycine- and arginine-rich sequences are present in the linkers of nearly all ROK family repressors. Conserved short sequence motifs, within the branches of the ROK evolutionary tree, suggest that these sequences could also be involved in operator recognition in other ROK family members.
INTRODUCTION
A variety of DNA binding motifs have been described in prokaryotes, but the most ubiquitous in prokaryotic transcription factors is the helix-turn-helix (HTH) motif (1,2). The basic HTH motif consists of three alpha helices forming a bundle, of which the second and third helices are separated by the ‘turn’ while the third helix is in contact with the DNA and is usually referred to as the ‘recognition’ helix. The HTH DNA binding motifs are often found in separate domains at either the N-terminal (e.g. LacI family (3,4)) or C-terminal (e.g. CRP family (5)) of the protein. Some small monomeric DNA binding proteins consist of essentially only one domain carrying two DNA binding HTH motifs (6). However for the majority of HTH family members there is a second domain involved in oligomerization and/or effector binding and the true DNA binding motif is made up of the two HTH motifs of a dimer (1).
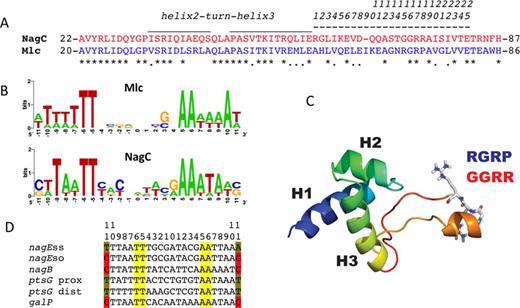
NagC and Mlc are paralogous members of the ROK (Repressors, Open reading frames (ORFs) and kinases) family of proteins (7), and are responsible for controlling use of amino sugars and uptake of glucose, respectively, in Escherichia coli (8). Despite very similar DNA operator sites (Figure 1B) and also very similar amino acid sequences of the recognition helix of the HTH motif (Figure 1A), there is no cross regulation between the NagC- and Mlc-controlled regulons in vivo, at least not with physiological levels of the proteins (9,10). Overexpressing the proteins from a plasmid does allow heterologous repression, confirming that the two proteins and their targets have a common origin but also demonstrating that the affinity of one protein for the other's target is lower (8). The Mlc and NagC binding sites are similar quasi-palindromes and their sequence logos are essentially indistinguishable with only four totally conserved positions TT/AA, at positions ±5,6 on either side of the center of symmetry (Figure 1B). However, as we noted before, high affinity NagC sites are characterized by C or G at positions ±11, whereas Mlc sites all have A or T at this position. The nucleotides at position ±11 are not necessarily palindromic (9–11).
(A) Comparison of the HTH and linker sequences of NagC and Mlc. Sequences corresponding to NagC are shown in red and to Mlc in blue. Identical amino acids are indicated by asterisks. The locations of the HTH motif and linker are shown by over-lining. The extended linker sequence corresponds to amino acids 59–82 in NagC and 57–81 in Mlc of E. coli. For convenience (and to avoid confusion since the aligned amino acids do not occupy the same numbered positions in Mlc and NagC proteins) these 25 amino acids are numbered linker positions 1–25 as shown. Note that this sequence is only 24 amino acids in NagC, one amino acid in NagC is missing in the alignment (at linker position 9, ΔI65). Here and elsewhere, NagC sequences are shown in red and Mlc sequences in blue. (B) Sequence logos derived from the known native Mlc (six sites) and high-affinity NagC targets (10 sites) (see (12) for list). Note that the TT and AA at positions −5,6 +5,6 around the center of symmetry (position 0) are the only completely conserved positions. Positions at −11, +11 are A or T in Mlc sites and mostly G or C in the high-affinity NagC sites, but they are not necessarily palindromic. The logo for all NagC sites is shown in Supplementary Figure S1B. (C) Model of the NagC DNA binding domain and linker (12). Helices 1, 2 and 3 of the DNA binding domain are indicated. The linker is predicted to form a finger like projection with a short alpha helix at the apex. The amino acids GGRR (red) in the NagC linker are shown in stick form. Replacing the GGRR motif of NagC with the RGRP motif (blue) of Mlc allows NagC to repress Mlc targets (12). (D) Sequences of relevant NagC and Mlc operators. The conserved TT/AA motif at positions −5,6 and +5,6 from the center of symmetry are highlighted in yellow. C or G at positions –11 and +11 of NagC sites are highlighted in red.
Previously, using in vivo repression assays of lacZ fusions, we showed that exchanging the 25 amino acids encompassing helices 2 and 3 of the HTH motif between Mlc and NagC (Figure 1A) did not change the specificity of repression by either Mlc or NagC, and had only a small effect on the level of repression (12). This strongly suggested that, in the case of both Mlc and NagC, the HTH motif is not important for discriminating between their binding sites. On the other hand replacing 16 amino acids of NagC protein from the C-terminal side of the HTH, with the corresponding amino acids of Mlc (linker amino acids numbered 10–25 in Figure 1A) to give a NagC-Mlc sandwich protein, NM66–81N, (previously called NMLM (12)), did allow NagC to repress ptsG, an Mlc target, ∼10-fold. NM66–81N still repressed nagEso, a specific NagC target but less efficiently than wild-type NagC (12). This replacement mutation thus exhibited both a change in specificity and a loss in specificity, since it now recognized both NagC and Mlc targets with comparable affinity. Dissecting the region replaced in NagC showed that of the 16 amino acids from Mlc, only two changes were necessary and sufficient for the phenotypic change to Mlc-like DNA binding specificity. These two amino acid replacements gave NagC(G72R,R75P) (subsequently called Nag102), which repressed nagEso and ptsG almost identically to NM66–81N (12)(these data are summarized in Supplementary Figure S1). The two linker amino acids changed correspond to linker positions 15 and 18 (Figure 1A). Moreover, we showed that these two amino acids of the linker were recognizing the A/T base pair (bp) at positions ±11 of the Mlc operator.
However, we were less successful in converting Mlc to a protein, which could repress NagC targets. Replacing the same 16 amino acids of Mlc with the equivalent amino acids of NagC (to give MN67–82M, previously called MNLM) did allow ∼3-fold repression of the NagC target, nagEso, but the replacement of just the two amino acids, Mlc(R71G,P74R) (subsequently called Mlc32), was completely inactive for repression of nagEso (Supplementary Figure S1) (12). In the present work, by mutagenesis of Mlc and in vivo repression assays, we have investigated which amino acids are required for recognition of a NagC target.
The 16 amino acid region we replaced previously had been described as an unstructured linker in Mlc (13), since no electron density corresponding to 14 of these amino acids was visible in the crystal structure of Mlc (pdb 1Z6R). However the crystal structure of the Mlc homolog from Vibrio cholerae (VC2007, pdb 1Z05), where these amino acids were visible, implied that the linker was longer and comprised of ∼25 amino acids. All the amino acids between the third, ‘recognition’ helix of the HTH motif and the alpha-helix at the beginning of the C-terminal oligomerization/effector binding domain (amino acids numbered 1–25 in Figure 1A) were present in an extended, basically unstructured, finger-like projection but with a short alpha helix near the apex. The modeled structures of the linker regions of NagC and Mlc based on that of VC2007 were compatible with this interpretation (12). The two amino acids replaced in NagC, which allow repression of Mlc targets, are near the apex of the projection, and change the sequence GGRR in NagC to RGRP as found in Mlc and VC2007 (Figure 1C).
While ROK protein family members are widely distributed among bacteria, they are notably absent from the Archaeal domain (14). Bacterial genomes are often equipped with several ROK paralogs reflecting the diversity and evolution of metabolic pathways for carbohydrate import and utilization (15). A wide survey of all prokaryotic genomes for ROK family repressor proteins is presented here in parallel to the mutational analysis of the E. coli NagC and Mlc transcription regulators and demonstrates that conserved glycine- and arginine-rich linker sequences are present in all ROK repressors. In particular an almost invariant GR pair is found at the equivalent of linker positions 16 and 17 in all ROK proteins (Figure 1A). The linker sequence could thus be an inherent part of the DNA binding motif in other members of this family and represent an extension to the classical model for specific operator recognition.
MATERIALS AND METHODS
Bacteriological methods
The strains used were described previously (12) and are given in Table 1. JM-G359 carries a ptsG-lacZ fusion, an Mlc target. As a target for NagC regulation we have used the nagEso promoter (Figure 1D) present in JM-Eso4 (8,12). The wild-type nagE operator is unusual in the sense that it has -11A,+11T unlike most NagC sites, which have C or G at positions ±11 (9). The wild-type nagE operator appears to have characteristics of both Mlc and NagC sites and relies on NagC binding co-operatively to the strong nagB operator via DNA loop formation, for its repression (9,16). Changing the operator from -11A,+11T to -11C,+11G produced nagEso (for super-operator) (Figure 1D), which is a ‘pure’ NagC target and allows strong repression by NagC even with just a single operator. JM-Ess4 carries the same nagE-lacZ fusion but with the single wild-type nagE NagC operator, it is not regulated by NagC (9,11). The JM-Gal32 strain carries a galP-lacZ fusion with a single NagC site, which is regulated ∼5-fold by NagC (17). All fusion-carrying strains are deleted for nagC and mlc.
Bacterial strains used
| . | Genotype . | Reference . |
|---|---|---|
| JM101 | F′(traD36 lacIq ΔlacZM15 proA+B+) supE thiD (lac-proAB) | Lab stock |
| JM-G359 | JM101 λ RS415/ptsG-lacZ ΔnagC::tc Δmlc::cat | (18) |
| JM-Ess4 | JM101 λ RS415/nagEss-lacZ nagC::cat mlc::tc | (9) |
| JM-Eso4 | JM101 λ RS415/nagEso-lacZ nagC::cat mlc::tc | (9) |
| JM-GalP32 | JM101 λ RS415/galP-lacZ ΔnagC::tc Δmlc::cat | (17) |
| . | Genotype . | Reference . |
|---|---|---|
| JM101 | F′(traD36 lacIq ΔlacZM15 proA+B+) supE thiD (lac-proAB) | Lab stock |
| JM-G359 | JM101 λ RS415/ptsG-lacZ ΔnagC::tc Δmlc::cat | (18) |
| JM-Ess4 | JM101 λ RS415/nagEss-lacZ nagC::cat mlc::tc | (9) |
| JM-Eso4 | JM101 λ RS415/nagEso-lacZ nagC::cat mlc::tc | (9) |
| JM-GalP32 | JM101 λ RS415/galP-lacZ ΔnagC::tc Δmlc::cat | (17) |
| . | Genotype . | Reference . |
|---|---|---|
| JM101 | F′(traD36 lacIq ΔlacZM15 proA+B+) supE thiD (lac-proAB) | Lab stock |
| JM-G359 | JM101 λ RS415/ptsG-lacZ ΔnagC::tc Δmlc::cat | (18) |
| JM-Ess4 | JM101 λ RS415/nagEss-lacZ nagC::cat mlc::tc | (9) |
| JM-Eso4 | JM101 λ RS415/nagEso-lacZ nagC::cat mlc::tc | (9) |
| JM-GalP32 | JM101 λ RS415/galP-lacZ ΔnagC::tc Δmlc::cat | (17) |
| . | Genotype . | Reference . |
|---|---|---|
| JM101 | F′(traD36 lacIq ΔlacZM15 proA+B+) supE thiD (lac-proAB) | Lab stock |
| JM-G359 | JM101 λ RS415/ptsG-lacZ ΔnagC::tc Δmlc::cat | (18) |
| JM-Ess4 | JM101 λ RS415/nagEss-lacZ nagC::cat mlc::tc | (9) |
| JM-Eso4 | JM101 λ RS415/nagEso-lacZ nagC::cat mlc::tc | (9) |
| JM-GalP32 | JM101 λ RS415/galP-lacZ ΔnagC::tc Δmlc::cat | (17) |
The single copy plasmids (R1 replicon) carrying the nagC and mlc genes, pXE/NagC and pXE/Mlc, have been described (18). NagC or Mlc are expressed from a weak constitutive promoter on the plasmid. Mutations in the nagC and mlc genes on the pXE plasmids were made by the ‘two-rounds-of-PCR’ method as described previously (12,18), using the oligonucleotides and templates listed in Supplementary Tables S1 and S2.
Bacteria were cultured in the synthetic Morpholinopropane sulfonate (MOPS) medium described by Neidhardt (19) supplemented with 0.4% glycerol, 0.5% cas amino acids and 50-μg/ml ampicillin, and β-galactosidase assays were carried out as described previously (18). β-galactosidase activities (Miller units (20)) were measured throughout exponential growth and values are reported for cultures with optical densities (A650) between 0.5 and 0.8. Values are the mean of at least two and generally more independent cultures.
N.B. The magnitude of repression by Mlc and NagC derivatives cannot be compared directly since the levels of expression of Mlc and NagC from the pXE1 single copy plasmids are not necessarily identical and NagC levels seem to be lower (18). However relative levels of repression from Mlc (or NagC) derived proteins in different strains are valid comparisons.
Bioinformatics techniques
Selection of ROK proteins for genome screening
In addition to Mlc and NagC, E. coli carries a third uncharacterized ROK repressor, YphH. The XylR repressor in Bacilli is the only other well-characterized ROK repressor (21–25). Kazanov et al. (15) noted that there were multiple ROK family paralogs in the Thermotogae. By bioinformatics and subsequent experimental validation, they could identify seven repressors for specific operator sequences associated with sugar utilization genes for xylose, chitobiose, inositol, β-glucosidase, mannose, trehalose and glucose (15). Other known ROK proteins are found in Streptomyces species (26,27) and by screening with NagC we found there were 14 ROK family repressors in Streptomyces coelicolor. In addition there are multiple ROK repressors in Bifidobacterium longum (six ROK repressors) (28,29) and Mycobacterium smegmatis (four ROK repressors) (30). Together these 35 proteins constituted a ‘seed’ set of ROK repressors from diverse phyla (see Supplementary Table S3 for a complete list).
Genomes selection
To avoid redundancy, single species were selected alphabetically for each prokaryotic genus present in the NCBI complete genome repository (∼3000 genomes) (ftp://ftp.ncbi.nih.gov/genomes/Bacteria/), with the exception of the genera used for the selection of the ‘seed’ proteins, where the seed protein species was used. The genome selection and processing was carried out on our existing local DNA genome database described previously (31) and updated daily from the NCBI repository. This resulted in a list of 662 genomes. The presence of ROK proteins was investigated by submitting the selected genomes to our ROKnRoll in-house program as follows. Each genome was submitted to TBLASTN analysis with 35 known ROK proteins (Supplementary Table S3). The Basic Local Alignment Search Tool (BLAST) bit score of all resulting hits was normalized using the seed protein as described (32). The absolute nucleotide position of each hit was then converted into the GenInfo identifier (GI) of the corresponding protein using GenBank genomic annotations. A normalized BLAST score was assigned to each protein-GI. GIs hit multiple times were rotated to retain only the highest score. The resulting list of unique GIs was then ranked by score and the proteins sequences with a normalized score ≥20% were extracted from the NCBI database in FASTA format using the following NCBI E-Utilities command: efetch.fcgi?db = protein&id = [GI number]&rettype = fasta&retmode = text.
Phylogeny
The 414 resulting protein sequences were aligned with the Clustal Omega online resource (http://www.ebi.ac.uk/Tools/msa/clustalo/). A phylogenetic tree was then generated with the neighbor-joining algorithm using the ClustalW2 Phylogeny resource (http://www.ebi.ac.uk/Tools/phylogeny/clustalw2_phylogeny/) and exported in Newick format. The tree was then visualized using the iTOL Web tool (http://itol.embl.de/) and exported in postscript format for further graphical processing.
Sequence logos
Aligned protein (or nucleotide) sequences were uploaded to the WebLogo online resource (http://weblogo.berkeley.edu/) and the resulting generated sequence logos were exported as portable network graphics (PNG) images for further graphical processing.
RESULTS
Linker amino acids required for recognition of nagEso NagC target
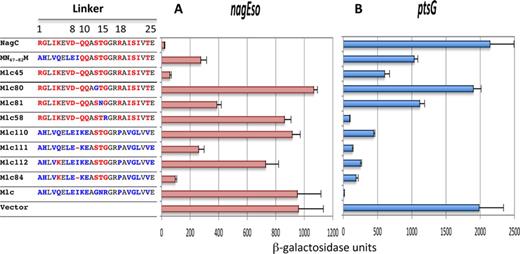
To investigate if the amino acids immediately adjacent to the C-terminal of the HTH motif, positions 1–9 of the extended linker in the VC2007 structure, had a role in NagC recognition of its targets, we replaced the whole linker sequence, positions 1–25 of Mlc, with those of NagC (Figure 1A) to give a hybrid protein Mlc45 (MN59–82M). Effectively Mlc45, with this larger replacement, repressed nagEso 16-fold compared to ∼3-fold for the previously studied MN67–82M (Figure 2A). Thus, NagC operator recognition implicates a longer linker sequence than that required for recognition of Mlc targets.
Amino acid replacements required to convert Mlc into a NagC-type repressor. The sequences of the linker in NagC, Mlc and the Mlc-derived proteins are shown. Linker positions are numbered as in Figure 1A. Amino acids specific to NagC are shown in red, those specific to Mlc in blue and those identical in the two linkers in black. All proteins are expressed from the single copy pXE plasmid. The ability of these Mlc derivatives to repress nagEso (A) and ptsG-lacZ (B) fusions is shown compared to the control vector without insert. Activities are the mean of at least two (and generally more) independent cultures with standard deviation. The complete set of Mlc derivatives tested is shown in Supplementary Figure S2.
To identify the amino acids important for NagC operator recognition, we made a series of replacements of one or two amino acids in the linker of the Mlc45 (MN59–82M) construct back to the amino acids found in Mlc to determine which positions were important for repression of nagEso. Amino acids near the center of the linker appeared to be essential for repression. Replacing one or more of amino acids in positions 13, 14, 15 (STG) with the GNR of Mlc produced complete or almost complete derepression of the nagEso fusion (constructs Mlc80, 81, 58; Figure 2A). Replacements of the amino acids in positions 1–12 or positions 16–25 of the linker had relatively small effects on nagEso repression (Supplementary Figure S2) even though the amino acids in the left hand half of the linker (positions 1–9 of NagC) are required for good repression (Mlc45) (Figure 2A).
We subsequently tried to find what was the minimum set of amino acid changes within the Mlc linker, which would allowed Mlc to repress nagEso. The presence of STG at positions 13–15 (Mlc110) was not sufficient for Mlc to repress nagEso but combined with the ΔI9 mutation (Mlc111) produced 4-fold repression (Figure 2A). Although the Q5K change with the STG replacement did not produce significant repression (Mlc112), when combined with ΔI9 and STG, it produced ∼10-fold repression (Mlc84). Including other exchanges (V4I and/or L22I) did not improve repression (Supplementary Figure S2A). The Mlc84 construct is only slightly less proficient at repressing nagEso than Mlc45, with the complete 24 amino acid NagC linker. Thus, we conclude that these five changes (Q5K, Δ9, G13S, N14T and R15G) in the Mlc linker are the minimum replacements necessary for Mlc to recognize the nagEso NagC target.
To verify that the amino acid changes identified here, which allow Mlc to repress nagEso, also allow repression of another NagC target, we tested the galP promoter (12,17). Mlc with the whole NagC linker (Mlc45) as well as constructs with just five amino acid replacements (Mlc84) repressed galP similarly to NagC (Supplementary Figure S3A and B).
The minimal Mlc derivatives, which repress nagEso, are less discriminatory
We tested the series of Mlc derivatives for their ability to repress ptsG (Figure 2B). In general the Mlc derivatives carrying parts or all of the NagC linker, with or without changes back to the equivalent amino acids from Mlc, were less capable of repressing ptsG than wild-type Mlc (Figure 2B and Supplementary Figure S2B). Certain plasmids were completely inactive, repressing neither nagEso nor ptsG (e.g. Mlc 80). A few however behaved similarly to wild-type Mlc and repressed ptsG strongly but not at all nagEso (e.g. Mlc58). However, most of the Mlc derivatives, which repressed nagEso well, were not as discriminatory as wild-type NagC. In fact several, including Mlc84 (with the minimal changes for NagC-type repression) and related constructs (Mlc85, 92, 93; Supplementary Figure S2), repressed both nagEso and ptsG rather well (∼10-fold). Thus the amino acid changes, which have allowed Mlc to repress nagEso, have resulted in a broadening of the target recognition and a loss of specificity for the original target (and presumably some affinity since the magnitude of the repression is lower). This is, in fact, comparable to the case for the G15,R18 change in NagC to give NagC(G72R,R75P) (= Nag102), which allowed NagC to repress ptsG but which continued to repress nagE (12) (Supplementary Figure S1).
The Mlc derivatives, which repress nagEso, recognize the C/G bp at ±11 of the operator
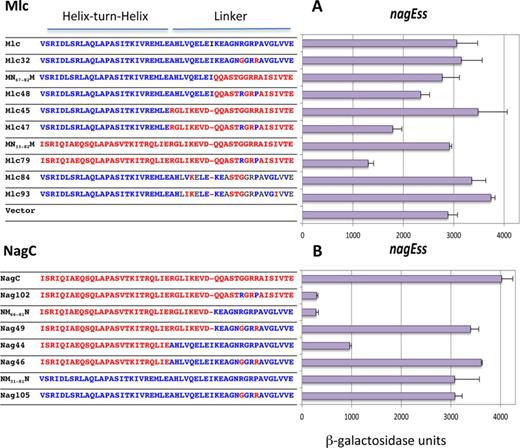
We also tested repression of the wild-type nagE promoter with -11A,+11T in its operator (called nagEss; Figure 1D). This promoter is not normally repressed by NagC in the absence of the nagB operator (9). However it is repressed by the NagC(G72R,R75P)(Nag102) mutant (Figure 3), demonstrating that these two amino acid replacements were allowing NagC to recognize the A/T base pairs at positions ±11 of the operator (12). Applying a similar logic to the Mlc derived constructs, since none of the Mlc constructs that repress nagEso were capable of repressing nagEss (e.g. Mlc45, Mlc84; Figure 3) and since the only difference between nagEso and nagEss is the nature of the base pair at position ±11 of the nagE operator, this confirms that the amino acid replacements in Mlc84 have allowed it to recognize the C/G base pairs at the extreme positions of the NagC operator.
Effect of mutations in the linker sequences of Mlc and NagC on nagEss-lacZ expression. Sequences of the HTH and linker regions of the proteins are shown. Blocks of amino acids derived from NagC are shown in red while those from Mlc are shown in blue. The body of the protein is derived from Mlc (A) or from NagC (B). The ability of Mlc and its derivatives and NagC and its derivatives to repress nagEss-lacZ is shown. Activities are the mean of at least two (and generally more) independent cultures with standard deviation.
Relative contributions of amino acids at linker positions 15 and 18 to specificity and repression
We noted that of the amino acid determinants required for NagC-type or Mlc-type operator recognition, only linker position 15 (G in NagC and R in Mlc) is crucial to the two proteins. However, the quantitative effect of changes in this position on repression depended upon the rest of the protein.
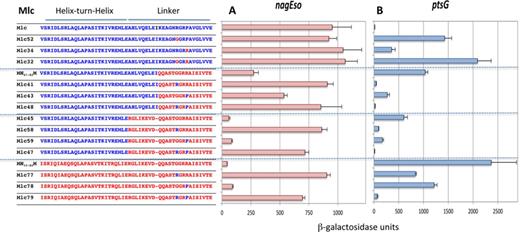
To investigate the context effects further we made a systematic analysis of the effects of different combinations of R or G at position 15 and of P or R at position 18 of the linkers of NagC and Mlc, within different contexts of the rest of the protein, i.e. origin of the HTH, linker and the body of the protein. We use the term ‘body’ to refer to Mlc- or NagC-derived proteins where all the rest of the protein, outside the HTH and linker sequences are supplied by Mlc (Figure 4A and B) or NagC (Supplementary Figure S4A and B). In many cases, exchanging just the amino acid at position 15 can have severe effects on the ability of the protein to repress.
Effect of exchanging G and R at position 15 and R and P at position 18 of the linkers in Mlc-derived constructs on nagEso-lacZ and ptsG-lacZ expression. Sequences of the HTH and linker regions are shown as described in Figure 3. The ability of Mlc and its derivatives to repress nagEso-lacZ (A) and ptsG-lacZ (B) are shown. Activities are the mean of at least two (and generally more) independent cultures with standard deviation. Note that Mlc32 and Mlc52 carry the sequence G13,T14,G15. This sequence seems to preclude repression of any target (see Supplementary Figure S4).
Context effects of linker mutations in Mlc on repression
In wild-type Mlc, mutations at either linker positions, R15G and/or P18R, reduced repression at ptsG, with loss of R15 having the greater effect (constructs Mlc52, 34, 32; Figure 4B). Likewise, in the Mlc derivatives with all or part of the NagC linker (MN67–82M, Mlc45) mutating either position 15 or 18 back to the Mlc-specific amino acids (G15R and/or R18P) improved repression of ptsG with R15 (constructs Mlc41,58) being more effective than P18 (constructs Mlc43,59). Perhaps surprisingly, the sequence RGRR within the NagC linker in Mlc (Mlc41 and Mlc58) produced better repression of ptsG than when the P18R mutation was introduced into wild-type Mlc (Mlc34). Similarly, GGRP in the NagC linker allowed better repression of ptsG than GGRP in wild-type Mlc (compare constructs Mlc43 and 59 with Mlc 52; Figure 4B), showing that depending on the context, R15 or P18 can be sufficient for ptsG repression. However when both the NagC HTH and linker are present in the Mlc body (MN33–82M), both G15R and R18P were necessary for strong repression of ptsG (Mlc79) (Figure 4B). This implies that the Mlc HTH does contribute to ptsG operator binding specificity in the constructs with just R15 within the NagC linker (Mlc41, Mlc58) and the HTH constitutes an alternative secondary specificity determinant. Two specificity determinants are thus required for ptsG repression: either R15 and P18 or the Mlc HTH and R15 (or P18).
The same series of Mlc-derived constructs was tested for repression of nagEso (Figure 4A). In constructs that carry the full-length NagC linker (and hence the secondary specificity determinants K5,Δ9,S13,T14, required for NagC-type repression), only the presence of G15 was required for repression (Mlc59,78). The presence or absence of R18 had little or no effect. Even the identity of the HTH sequence had no effect (compare Mlc45 and 59 with MN31–82M and Mlc78; Figure 4A). The pattern of repression at the galP promoter (another NagC target) was similar (Supplementary Figure S3). Thus G15 associated with K5,Δ9,S13,T14 in Mlc-derived proteins are necessary and sufficient conditions for repression of NagC targets, nagEso and galP.
The importance of the G or R at position 15 as the primary specificity determinant is emphasized when one compares certain pairs of proteins. Exchanging the G for an R at position 15 can switch a protein from NagC-type, which represses nagEso well and ptsG only weakly, to Mlc-type, repressing ptsG well and nagEso weakly (compare Mlc45 and Mlc58 or Mlc78 and Mlc79) (Figure 4A and B). Note that the secondary determinants for both NagC (K5,Δ9,S13,T14) and Mlc (P18 or the Mlc HTH) are present in all these constructs.
Context effects of linker mutations in NagC on repression
The pattern of ptsG repression by NagC-derived proteins reinforced the view that the contribution of the secondary specificity determinants is context dependent. In all NagC-derived proteins both R15 and P18 are necessary for good (∼10-fold) repression of ptsG. Loss of either R15 or P18 in any context (even with the full Mlc HTH and linker in NagC) resulted in complete loss of repression (Nag103,104,105; Supplementary Figure S4B). In this case, the NagC body, independent of the origin of the HTH, must also be playing a role and dictating that R15 is insufficient and both R15 and P18 are required for recognition of the Mlc operator of ptsG. The NagC body and/or HTH also play a role in repression of nagEso. Replacing G15 or G15,R18 in wild-type NagC does not result in complete loss of nagEso repression, 2- to 4-fold repression remains (Supplementary Figure S4A). This implies that the secondary determinants, K5,Δ9,S13,T14 together with the NagC HTH and the NagC body can partially compensate for the loss of G15.
Context effects of linker mutations on repression of nagEss
Analysis of the pattern of repression of the nagEss fusion (with ±11 AT) showed that the context effect of the linker is predominant. Only two constructs repressed nagEss strongly (∼10-fold), NM66–81N and Nag102 ( = NagC(R72,G75)) (Figure 3B) which we had analyzed previously (12). These two proteins have the NagC HTH and R15,P18, either within wild-type NagC (Nag102) or as part of the short Mlc linker in NagC, so that both proteins have linker positions 1–9 from NagC. Two other constructs with the NagC HTH and R15,P18 (Mlc79 and Nag44) also repressed nagEss, but less well, while Mlc47 and Mlc48, with the Mlc HTH and R15P18 within the NagC linker, produced less than 2-fold repression (Figure 3). These last four constructs however all allowed strong repression of ptsG (also with ±11A/T) (Figure 4 and Supplementary Figure S4).
The nagEss operator appears to function as a hybrid Mlc/NagC site in the sense that it has A/T at positions ±11 characteristic of Mlc operators, but at least in the presence of the nagB operator, it is preferentially regulated by NagC (9). This implies that other parts of the nagE operator sequence, outside the positions ±11, favor NagC binding. Since the best repression of nagEss was given by constructs that have the NagC HTH and NagC sequence in positions 1–9 of the linker, this could suggest that the secondary NagC determinants, present in positions 1–9 of the linker (K5 ΔI9), might be recognizing some other characteristic of NagC sites other than the C/G at positions ±11.
DISCUSSION
The Mlc and NagC linker is part of a winged HTH motif
The specificity determinants, which permit NagC and Mlc, paralogous members of the ROK family in E. coli, to recognize their binding sites on DNA, reside not in the canonical HTH DNA binding motif but in the adjacent amino acid sequences to the C-terminal side of the HTH. These ∼25 amino acids form an extended structure in the one crystal structure (1Z05) of an ROK family repressor where they were all visible. The crystallographic structures of three proteins of the repressor class of ROK proteins are available in the PDB: in addition to VC2007 from Vibrio cholera (1Z05), which is a homolog of Mlc, the structure of Mlc itself is known, both free (1Z6R) (13) and in complex with EIIBGlc (3BP8) (33), and also that of TM1224 from Thermotoga maritima (2HOE). In Mlc and TM1224 several amino acids are missing in the electron density map corresponding to the linker region. However, all three proteins are classified as winged HTH (wHTH) proteins in the SCOP database (http://scop.mrc-lmb.cam.ac.uk/scop/data/scop.b.b.j.e.gc.html) (34,35). Interestingly VC2007 (1Z05) and the modeled structures of Mlc and NagC linkers, based on VC2007, are predicted to have two short beta sheets at the bases of the linker extension, which is consistent with the extended wing concept (Supplementary Figure S5). In addition, a small alpha helix is observed between linker positions 9–15 of VC2007 and short alpha helices were predicted in similar positions in the modeled wild-type NagC protein and Nag102 (12), and also in the Mlc derivatives described here (Mlc84 and Mlc45) (Supplementary Figure S5). The amino acids that we have identified as required for specific Mlc-type repression (R15,P18) and NagC-type (K5,Δ9,S13,T14,G15) are all part of the predicted wing structure. The Mlc specificity determinants (R15,P18) are located at the apex of the finger-like projection in the modeled structures, while the NagC specificity determinants (S13,T14,G15) are situated at the C-terminal end of the short alpha helix (Supplementary Figure S5). The Δ9 requirement for NagC-type repression will mean that contacts made by the amino acids at positions 15–18 will be shifted compared to Mlc.
Operator site recognition
We previously postulated that the extended finger motif was stretching along the DNA helix and that the RGRP sequence, characteristic of Mlc-type repressors, was making a contact with the DNA near position +11 and −11 of the Mlc operator (12). This contact was expected to occur in the narrow, minor groove afforded by A-T base pairs. Minor groove recognition was predicted, since we had previously shown that the effect of a C-G or G-C base pair at position ±11 of the nagE operator was equivalent and different from A-T or T-A at the same two positions (11). The ability of arginines to discriminate between the electrostatic potential of minor grooves formed by C-G and A-T bp, as seen originally in certain homeodomain proteins, has been documented (36–40). The glycine to arginine switch at linker position 15, important for changing from NagC recognition to Mlc-type recognition, will crucially change the ability of the protein to read the electrostatic potential of the minor groove.
We used the DNAshape prediction program (41) to look for any inherent differences in the structures of Mlc and NagC operators (Supplementary Figure S6). For both Mlc and NagC sites the minor groove width is at its lowest around positions ±5–7 corresponding to the absolutely conserved TT-AA bp (Figure 1B). No significant differences between Mlc and NagC sites were predicted at positions ±11. The presence of the C-G base pairs at positions ±11, characteristic of NagC operators, especially those with higher affinity (9,11,42,43), must be altering some other property of the DNA, such as the flexibility (indicated by differences in propeller twist and roll (Supplementary Figure S6)) so that they are preferentially bound by NagC. Other examples exist in eukaryotic transcription factors of sequences flanking the operator consensus affecting DNA binding site specificity e.g. to orientate a heterodimeric repressor (44) or to distinguish a subset of closely related sites (45).
Determinants of operator specificity and affinity
Structures of several wHTH proteins in complex with DNA are known. The amino acids of the wings can be in direct contact with the DNA and have been shown by mutagenesis to contribute to DNA binding affinity (e.g. OhrR-ohrA (46)). However they have not been postulated to be involved in operator selection. The HTH motif is usually considered the primary specificity determinant. In the case of NagC and Mlc, the wings are determining operator site specificity and primarily by the presence of a glycine or an arginine at position 15 near the apex of the extended wing.
Nevertheless, the HTH is important for operator binding per se. Mutations in the absolutely conserved TT–AA at positions ±5,6 around the center of symmetry of the operator (shown in yellow in Figure 1B and D) lead to complete loss of repression by Mlc and NagC in vivo (10,11). Contacts between these positions and the HTH are likely to be identical or very similar in Mlc and NagC, due to the quasi-identity of the amino acids in their recognition helices (Figure 1A) and are presumed to occur in the major groove as observed in other HTH protein–DNA structures (e.g. (5,47)). This idea is supported by several observations; not least that exchanging the HTH motif (helices 2 and 3) of NagC and Mlc had no effect on operator recognition and only a small effect on affinity (12) (Supplementary Figure S1A). In the present study several constructs repress nagEso or ptsG similarly, irrespective of whether the HTH is derived from Mlc or NagC (e.g. Mlc45 and MN33–82M repressing nagEso (Figure 4A)) or NM66–81N and NM31–81N repressing ptsG (Supplementary Figure S4B). The HTH appears to be more important for contributing affinity rather than specificity. Although, as described above, in certain contexts the HTH can act as a secondary specificity determinant, for example when there is only R15 to define Mlc specificity (compare Mlc41 and Mlc58 to Mlc77 (Figure 4B)).
The analysis of the linker mutations within different contexts of the rest of the protein (Figure 4) has led us to identify at least three regions which can contribute to the final level of repression of the different promoters: the linker, the HTH and body of the protein. We propose that the specificity is primarily determined by the linker contacts and, in particular, by the amino acid at position 15, glycine or arginine, which constitutes the primary specificity determinant. Specific binding then requires other secondary contacts, e.g. the K5,Δ9,S13,T14 in NagC or P18 in Mlc from the linker. Depending on the context, the HTH or body can help to compensate for the absence of these secondary or even primary determinants.
Evolution of ROK family operator recognition
Mlc and NagC are essentially only found in a subset of gamma-proteobacteria, mostly Enterobacteriales and Vibrionales. Alignment of several NagC and Mlc homologs (selected by BLAST) from independent genera showed that the RGRP of Mlc and K5-Δ9-STGGRR of NagC linkers were almost totally conserved and were as well or even better conserved than the HTH motif.
ROK proteins are well distributed in numerous other bacteria but very few have been characterized. One class, which has been investigated, is the xylose repressors present in many Bacilli (21–25). Some XylR operators had been identified experimentally and a bioinformatics analysis identified others and allowed generation of a binding-site logo (25,48). The XylR operator sites resemble those of NagC and Mlc with the TT and AA at positions ±5,6 absolutely conserved. Moreover, as in the case of NagC, the majority of XylR operator sites have a C or a G at positions ±11 (48). Significantly, inspection of the aligned sequences of the XylR proteins from the corresponding Bacilli showed that their linker sequences resembled that of NagC (data not shown but see below).
This apparent conservation of a similar linker motif in the XylR group prompted us to look at other ROK proteins. We devised a method to retrieve ROK proteins from all prokaryotic genomes. This reiterated Blast procedure (ROKnRoll) used a broad set of 35 known ROK proteins (Supplementary Table S3) with which to individually screen a representative example of each genus in GenBank. This procedure gave us a set of 414 unique ROK proteins, which were used to construct a phylogenetic tree as described in the Materials and Methods section.
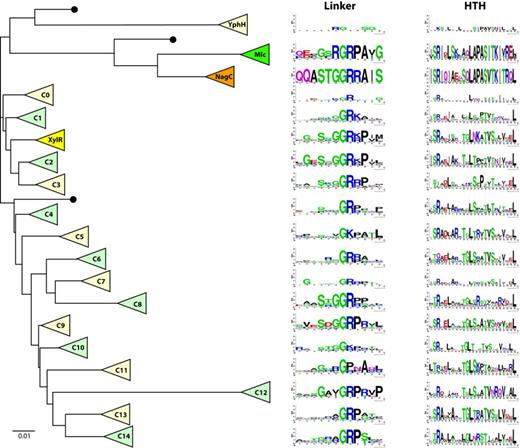
Reassuringly the Mlc, NagC and XylR proteins formed three distinct clusters within the ROK phylogenetic tree. Other clusters (C0-C14) of deeply rooted proteins were clearly visible. A simplified tree is shown in Figure 5. The complete tree is in Supplementary Figure S7. Proteins within each cluster were aligned and logos of the linker sequences created for each cluster. The linker sequences for NagC and Mlc were well conserved, especially the amino acids we had identified as necessary for specific Mlc and NagC binding to their operators. The XylR linker was also relatively well conserved and the sequence resembled that of the NagC linker with linker positions 13–17 as SsGGRr/k (compared to STGGRR of NagC) (Figure 5).
Evolution of ROK family linker sequences. In this schematic phylogenetic tree, each triangle represents a cluster of homologous ROK proteins, either previously identified (Mlc, NagC or XylR) or less characterized (YphH and C0 to C14). Adjacent to each cluster, 12aa and 24aa sequence logos represent the corresponding linker motif and helix-turn-helix motif, respectively. Black dots indicate clusters comprising single proteins or clusters with too few members to generate meaningful sequence logos. The scale bar corresponds to 0.01 amino acid substitutions per site. The YphH branch has been shortened for clarity. This simplified tree was expanded from a complete phylogenetic tree in order to display the lineage of each cluster. The detailed phylogenetic tree was obtained as indicated in the Materials and Methods section and is presented in Supplementary Figure S7. Possibly other ROK families exist, which have not been extracted with one of the 35 seeding proteins.
Encouraged by this we continued the analysis of the linker sequences of other branches of the tree. Quite remarkably short consensus sequences rich in glycine, arginine and often proline were found in the linker regions of all the clusters and a conserved GR (corresponding to positions 16,17 of the Mlc and NagC linkers) was identifiable in every cluster except three, C10, C11 and the small YphH group (Figure 5). In these clusters the central motifs are GK, GP and RG. GGR (as in NagC) and GRP (as in Mlc) were the most frequently found linker motif sequences. Sequences outside the central part of the logos were less well conserved, presumably reflecting the fact that these clusters contain uncharacterized proteins and could include repressors with different functions. For example the C11 cluster contains four of the Streptomyces proteins used as the seeding set and six of the seven Thermotoga proteins are part of cluster C1, consistent with the multiple duplication and functional diversification of ROK proteins described by Kazanov et al. (15). The other seeding proteins are more evenly distributed with two or three in most clusters. The NagC, Mlc, XylR and YphH clusters are defined by their nominal seeding protein and two clusters (C2 and C3) contain none.
To gain further insight into the significance of the conserved GR containing linker sequences, we examined another defined segment of the protein, and also aligned the HTH motifs (taking 24 amino acids based on clustal alignment with the NagC HTH motif) and created the equivalent logos (Figure 5 and Supplementary Figure S7). As expected most of the sequence of the HTH from the NagC and Mlc clusters is well conserved. The XylR HTH logo shows clearly less conservation. For all the other clusters a few positions are well conserved but they mostly correspond to the hydrophobic and uncharged amino acids, which form the basis of all HTH motifs, as recognized by the early alignment programs for identifying HTH motifs and referred to as a hydrophobic brace (2,49–51). Relatively few of the clusters show any significant conservation of the amino acids at the beginning of the recognition helix, which are generally found making specific contacts with the DNA (equivalent of PAS-TK in Mlc and NagC; Figure 1A). Comparison of the logos derived from the HTH sequence and the amino acids of the adjacent linker (Figure 5) demonstrates that the linker sequence and especially the central GR is a more strongly conserved motif than the HTH within the ROK phylogenetic tree.
To assess the importance of the conserved central GR motif for regulation by Mlc and NagC, we mutated these two positions (linker position G16 to R and A and position R17 to G and A). All the mutated proteins were essentially inactive for repression of their targets ptsG or nagEso (Supplementary Figure S8), demonstrating that these two positions are not just part of a region discriminating between the closely related Mlc and NagC binding sites but are essential for DNA binding per se.
CONCLUSION
The inevitable conclusion to draw is that the sequence adjacent to the HTH motif (the so-called linker motif) of these proteins represents a highly conserved sequence centered around glycine and arginine residues. Moreover, it implies that the contact between linker and minor groove of the operators that we have demonstrated in the case of Mlc and NagC is a fundamental property of the ROK family of transcription factors. For the paralogs, NagC and Mlc, these contacts are both essential for DNA binding and the major sites discriminating Mlc and NagC targets. It remains to be examined whether they have the same function in operator specificity and/or affinity in other ROK proteins.
We thank Annie Kolb and Miklos de Zamaroczy for their critical reading of the manuscript. L.D.-R. gratefully acknowledges the support of NVIDIA Corporation with the donation of the Tesla K40 GPU used for this research.
FUNDING
Centre National de Recherche Scientifique (CNRS) [to FRE3630]; Agence National de Recherche [ANR-09-Blanc 0399 (GRONAG)]; French “Initiative d’ Excellence” Program [ANR-11-LBX-0011-01 (DYNAMO)]. Funding for Open Access charge: CNRS.
Conflict of interest statement. None declared.
Present addresses:
Lenin Domínguez-Ramírez, Department of Chemical and Biological Sciences, School of Sciences, Universidad de las Américas Puebla, Santa Catarina Mártir Cholula, 72820 Puebla, Mexico.
Jacques Oberto, Institute for Integrative Biology of the Cell (I2BC), Université Paris-Saclay, CEA, CNRS, Université Paris-Sud, 91405 Orsay, France.




Comments