-
PDF
- Split View
-
Views
-
Cite
Cite
Ganesha P. Pitchai, Manuel Kaulich, Anna H. Bizard, Pablo Mesa, Qi Yao, Kata Sarlos, Werner W. Streicher, Erich A. Nigg, Guillermo Montoya, Ian D. Hickson, A novel TPR–BEN domain interaction mediates PICH–BEND3 association, Nucleic Acids Research, Volume 45, Issue 19, 2 November 2017, Pages 11413–11424, https://doi.org/10.1093/nar/gkx792
Close - Share Icon Share
Abstract
PICH is a DNA translocase required for the maintenance of chromosome stability in human cells. Recent data indicate that PICH co-operates with topoisomerase IIα to suppress pathological chromosome missegregation through promoting the resolution of ultra-fine anaphase bridges (UFBs). Here, we identify the BEN domain-containing protein 3 (BEND3) as an interaction partner of PICH in human cells in mitosis. We have purified full length PICH and BEND3 and shown that they exhibit a functional biochemical interaction in vitro. We demonstrate that the PICH–BEND3 interaction occurs via a novel interface between a TPR domain in PICH and a BEN domain in BEND3, and have determined the crystal structure of this TPR–BEN complex at 2.2 Å resolution. Based on the structure, we identified amino acids important for the TPR–BEN domain interaction, and for the functional interaction of the full-length proteins. Our data reveal a proposed new function for BEND3 in association with PICH, and the first example of a specific protein–protein interaction mediated by a BEN domain.
INTRODUCTION
Cell proliferation requires the genome to be replicated accurately and completely, and the resulting sister chromatids to be segregated evenly to the daughter cells in mitosis (1). Progression through mitosis presents numerous challenges to genome integrity. In particular, successful sister chromatid disjunction in anaphase requires not only the dissolution of sister chromatid cohesion (2–4), but also the removal of any DNA intertwinings (catenanes) that persist from interphase (5–7). Precocious or delayed sister chromatid disjunction can cause chromosomal instability, or even mitotic catastrophe, if not rectified prior to telophase. Defective chromosome segregation is conventionally analyzed by observing aberrant mitotic structures, such as DNA bridges, lagging chromatin, or micronuclei, using the DNA dye, DAPI (8–10). However, perhaps the most prevalent marker of sister chromatid non-disjunction in anaphase, the ultra-fine anaphase bridge (UFB), cannot be detected using DAPI (11,12). UFBs are present in most anaphase cells, and comprise histone-free threads of DNA (11–13). Currently, the only method to reveal UFBs is via detection of specific proteins that bind to them, including PICH, BLM, topoisomerases IIα and IIIα, and RIF1 (11,12,14–16). UFBs arise primarily from specific genomic loci, including centromeres, telomeres, rDNA loci and common fragile sites (11,12,17–19). These loci are characterized by their atypical patterns of nucleotide sequence, DNA replication and/or chromatin structure.
PICH is a multi-domain protein (Figure 1A) that is conserved in all metazoans, but is apparently absent from yeast (11). The most recognizable characteristic of PICH is an SNF2-type, adenine nucleotide-binding domain that mediates the hydrolysis of ATP required for dsDNA translocation (20). PICH also contains two tetratricopeptide repeat (TPR) motifs, and a so-called PICH family domain (PFD) of unknown function that is a defining feature of PICH homologs across species (11). TPR motifs are generally required to mediate interactions with other proteins (21,22); however, the specific TPR interaction partners for PICH have yet to be defined. Depletion of PICH from human cells causes a loss of association of BLM, topoisomerase IIIα and RIF1 from UFBs, suggesting that PICH regulates the localization of these proteins to UFBs either by direct recruitment or via structural changes to UFBs (14,16,18). PICH also possesses the unusual property of acting as a form of DNA ‘tension sensor’ by binding more stably to DNA that is stretched, such as would be expected for a UFB exposed to the pulling forces imposed by the mitotic spindle (20). It is important to note that the association of PICH with DNA can occur only following nuclear envelope breakdown in prometaphase, because PICH is excluded from the nucleus prior to this stage (11,23). Recent data indicate that one key role for PICH is to co-operate with topoisomerase IIα to promote decatenation of UFBs and hence facilitate faithful sister chromatid disjunction. A similar role has been proposed for its interaction with BLM and topoisomerase IIIα (7,12,16,24).
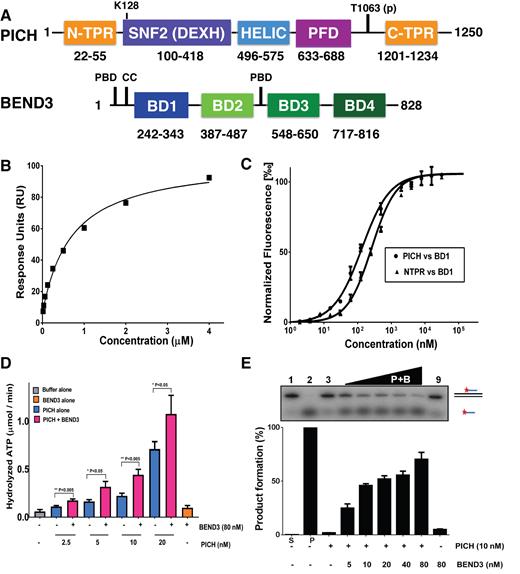
Binding of the N-TPR and BD1 domains and stimulation of the PICH activity by BEND3. (A) Schematic representation of the domain structure of PICH (upper) and BEND3 (lower). The numbers below each domain represent amino acid positions. The ATP (K128) and PLK1 (T1063) binding sites in PICH are indicated. The PBD and CC motifs marked in BEND3 denote putative Polo-box binding domains and a coiled-coil region, respectively. (B) Steady state binding curve for the PICH–BEND3 interaction using Biacore analysis. Responses at steady state were fitted with a simple binding model to obtain a Kd value of 0.7 ± 0.2 μM. (C) MST analysis of the interaction of the BD1 domain of BEND3 with either full length PICH or the isolated N-TPR domain, as indicated. The Kd values for the interaction with full length PICH and N-TPR were calculated to be 0.2 ± 0.25 μM and 1.2 ± 0.25 μM, respectively. Data points represent the mean of at least three independent experiments. Error bars denote SD. (D) dsDNA-dependent ATPase activity of different concentrations of PICH in the absence (blue bars) or presence (pink bars) of 80 nM BEND3 (monomer concentration; equivalent to 10 nM BEND3 octamers). The grey and orange bars denote control reactions with buffer or BEND3 alone, respectively. Statistical significance of pairwise interactions is indicated. (E) dsDNA translocase activity of PICH. Upper panel: representative polyacrylamide gel of the triplex DNA substrate and the ssDNA reaction product, as indicated diagrammatically on the right. The red asterisk denotes the radiolabeled end. Lower panel: quantification of the data from panel a, with protein monomer concentrations shown below the bars. Lanes marked S and P represent reactions using substrate alone and a heat-denatured DNA sample to define the position of the ssDNA product, respectively.
In this study, we examined the PICH ‘interactome’ in mitosis. This analysis identified a BEN domain-containing protein (BEND3) as a novel interaction partner of PICH. We have characterized the PICH–BEND3 interaction functionally, and have solved the structure of two of the interacting domains at high resolution. The crystal structure was used to identify key residues in PICH and BEND3 required for this functional interaction. Our data reveal an unexpected role of the evolutionarily conserved BEN domain in mediating a protein:protein interaction.
MATERIALS AND METHODS
Cell culture
HeLa cells were grown under standard conditions in Dulbecco's modified of Eagle's medium supplemented with 10% fetal bovine serum and antibiotics. Cells were cultured at 37°C in a humidified atmosphere containing 5% CO2.
Immunoprecipitation and mass spectrometry
Preparation of cell lysates was performed essentially as described previously (25) using Lysis Buffer (50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 1% IGEPAL CA-630 containing 20mM NaF, 20mM ß-glycerophosphate, 0.3 mM sodium vanadate, 20 μg/ml RNase A and phosphatase inhibitor cocktail; Sigma). Immunoprecipitations were carried out using 0.5 μg of the appropriate antibody together with 20 μl affiprep protein A agarose beads (Bio-Rad) for 2 h at 4°C, followed by extensive washing with Lysis Buffer and then phosphate buffered saline. For analysis of the PICH interactome, the immunoprecipitated PICH from HeLa cells arrested in prometaphase with nocodazole for 16 h was subjected to digestion with trypsin, and the resulting peptides were analyzed by mass spectrometry using an Orbitrap-Velos (ThermoFisher).
Purification of full length PICH
The full length PICH was expressed in insect cells and purified as described previously (20).
Purification of full length BEND3
The full length BEND3 was expressed in both HEK293 cells and Escherichia coli. For HEK cells, the BEND3 cDNA was cloned into a pCPR0053 (in-house) LIC vector containing 6XHis and strep-I tags. For E. coli, the cDNA was cloned into a pNIC28-Bsa4 LIC vector containing sequences encoding 6xHis and Strep-II tags. Following recombinant protein expression, the cells were resuspended in lysis buffer containing 100 mM Tris–HCl, pH 8.0, 150 mM NaCl, 1 tablet/50 ml Complete Inhibitor cocktail EDTA Free (Roche), 1 mM PMSF, 50 U/ml Benzonase, 10% glycerol and 0.5 mM TCEP. The cleared cell lysate was passed through a 5ml Strep-Tactin® column and the bound protein eluted with buffer containing 2.5 mM desthiobiotin (according to the IBA technologies manual). The eluted fractions were further purified using Heparin sepharose (GE Healthcare) and Superose 6 size exclusion columns (GE Healthcare) with the final buffer containing 25 mM Hepes–KOH, pH 7.5, 150 mM NaCl, 0.5 mM TCEP and 10% glycerol.
Purification of domains of PICH and BEND3
Fragments of the PICH and BEND3 cDNAs corresponding to different domains were cloned into pNIC28-Bsa4 LIC vector containing sequences encoding 6xHis and Strep-II tags at the N-terminus. The encoded domains were then expressed in E. coli Rosetta cells. The cells were grown to an OD 0.8 of 37°C, and expression was induced by 0.5 mM IPTG at 18°C for 26 h. Cells were then harvested and resuspended in lysis buffer (50 mM Na-phosphate, pH 7.5, 300 mM NaCl, 1 tablet/50 ml Complete Inhibitor cocktail EDTA-Free (Roche), 1× BugBuster (Novagen), 50 U/ml Benzonase, 10 mM Imidazole, 10% glycerol, 0.5 mM TCEP). The cell suspension was lysed using a French press, and the lysate was clarified by centrifugation. The cleared lysate was then purified using the combination of Ni-column chromatography, Strep tag and IMAC and further purified using heparin, anion and cation exchange chromatography. In each case, the final purification step was via size exclusion chromatography (Superdex 75 or Superdex 200; GE Healthcare). In each case, the column was equilibrated in 25 mM Hepes–KOH, pH 7.5, 150 mM NaCl, 0.5 mM TCEP and 10% glycerol. All purified proteins were stored in aliquots at –80°C, and were analyzed and validated using both SDS-PAGE and mass spectrometry. The correct folding of each of the isolated domains was also verified using circular dichroism.
Native gel electrophoresis
The oligomeric state of the purified BEND3 protein was analyzed using Blue Native PAGE. The full length BEND3 was purified from HEK293 cells. The purified protein sample (15 μl) was mixed with Native PAGE 4× sample buffer (5 μl). The sample was loaded onto a 4–16% Bis–Tris gel (Invitrogen). The gel was run at 4°C at a constant voltage of 150 V for 2 h. The gel was then fixed with 10% acetic acid for 30 min and stained using Coomassie blue.
Surface plasmon resonance (SPR)
Surface Plasmon resonance was performed using the BIAcore T100 (GE Healthcare) instrument at 25°C. Three different sets of experiments were performed using purified PICH and BEND3 protein at different time intervals. PICH (30–50 μg/ml) was immobilized to a level of 1500 and 3000 response units using amine-coupling chemistry onto a CM3 chip. The BEND3 analyte was then titrated against the immobilized PICH. To address mass-transfer effects, two different flow rates (30 and 60 μl/min) were used to study the protein:protein interaction, and HBS buffer (10 mM Hepes, pH 7.4, 150 mM NaCl, 0.5mM TCEP, 0.005% (vol/vol) Tween-20) was used for dissociation. The sensograms were globally analyzed with the analysis software BIA-evaluation 4.0.1 (Biacore AB). The equilibrium dissociation constant (Kd) was determined from the steady state binding curve. The experiment was performed on three different batches of purified proteins to validate the reproducibility.
Micro-Scale thermophoresis (MST)
The Protein sample was labeled using the RED-NHS Labeling kit (NanoTemper Technologies). The labeling reaction was performed according to the manufacturer's instructions in the supplied labeling buffer applying a concentration of 20 μM protein (molar dye:protein ratio ≈ 2:1) at room temperature for 30 min. Unreacted dye was removed using a P-10 desalting column equilibrated with MST buffer (25 mM Hepes pH 7.5, 150 mM NaCl, 0.5 mM TCEP and 0.05% Tween 20). The label:protein ratio was determined using photometry at 650 and 280 nm, and typically found to be 0.8. The labeled protein was adjusted to 20 nM with MST buffer supplemented with 0.05% Tween-20 (Sigma). The ligand was dissolved in MST buffer supplemented with 0.05% Tween-20 and a series of 16 capillaries with 1:1 dilutions was prepared using the identical buffer. For thermophoresis, each ligand dilution was mixed with one volume of labeled protein. After 10 min incubation, ∼5 μl of each solution was added to Monolith NT Standard Treated/Hydrophilic Capillaries (NanoTemper Technologies GmbH). Thermophoresis was measured using a Monolith NT.115 instrument (NanoTemper Technologies GmbH) at an ambient temperature of 25°C with 5 s/30 s/5 s laser off/on/off times, respectively. Instrument parameters were adjusted according to the signal from 20% to 50% LED power and 20–40% MST power. Data from three independently pipetted measurements were analyzed (NT.Analysis software version 1.5.41, Nano Temper Technologies) using the signal from Thermophoresis.
ATPase assays
The dsDNA-dependent ATPase activity of PICH was quantified using a commercial kit (Innova Biosciences). 500 ng dsDNA was added to all reactions. The samples were incubated in 25 mM Hepes–KOH, pH 7.5, 15 mM NaCl (final concentration), 1 mM ATP and 5 mM MgCl2 for 15 min at 37°C. The reaction was stopped by addition of 25 μl of ‘gold mix’ per reaction. After 2 min incubation, 10 μl of stabilizer was added. The reaction mixtures were transferred into 96-well plates and incubated for 15–30 min at room temperature before measuring absorbance at a wavelength of 590 nm.
DNA translocase assays
Two oligonucleotides (5′-CGCAAGAAAAGAAAGAAGAAAGAAACCGAGCT-3′ and 5′-CGGTTTCTTTCTTCTTTCTTTTCTTGCGGTAC-3′) were annealed and the resulting duplex with KpnI/SacI sites at the termini was cloned into the pBluescript KS+ vector. A 400 bp fragment containing the triplex-forming fragment obtained by restriction digestion of this construct with PvuII was then gel-purified. Following this, 10 pmol of the triplex forming oligonucleotide (5′-TTCTTTTCTTTCTTCTTT CTTT-3′) was 5′ end-labeled using γ32P-ATP and polynucleotide kinase, and was then annealed to an equimolar amount of the 400 bp fragment in buffer containing 10 mM MgCl2, 50 mM NaCl and 40 mM MES, pH 5.5. The reaction was incubated at 57°C for 15 min, and then allowed to cool down to room temperature overnight. The substrate was stored in aliquots at –20°C.
1 fmol of triplex substrate was used per translocase reaction, which was performed in buffer containing 20 mM Tris–HCl, pH 7.0, 100 mM NaCl, 2 mM MgCl2, 1 mM DTT, 2 mM ATP and 0.1 mg/ml BSA for 15 min at 37°C. Reactions were terminated by addition of a stop buffer to give a final concentration of components as follows: 3% SDS, 0.04 mg/ml proteinase K, 15% sucrose and 250 mM MES, pH 5.5. After incubation for an additional 15 min at 37°C, the reactions were loaded onto a 3–8% polyacrylamide gradient gel and run at 80V for 45 min in pre-cooled 2xTAM buffer (40 mM Tris/HAc, pH 5.5; 1 mM MgCl2). The gel was fixed in a solution containing 20% isopropanol and 10% acetic acid for 20 min and then washed two times with MilliQ water for 10 min. The gel was then dried and signals quantified using a PhosphorImager.
Selenomethionine-derived BD1 purification
A selenium-labeled BD1 derivative was produced as described previously (26).
NTPR–BD1 crystallization
Crystallization was performed as described previously (26).
Protein structure determination, model building and refinement
Crystals were mounted on CryoLoops (Hampton Research) and flash-cooled in liquid nitrogen. For data collection under cryogenic conditions, crystals were briefly soaked in a universal cryosolution, consisting of mother liquor supplemented with 20% (w/v) glycerol. The structure of the BD1–NTPR1 complex was determined by the multiple wavelength anomalous diffraction (MAD) technique from a selenium derivative. Three data sets, peak (PK, 0.9785 Å), inflection point (IP, 0.9787 Å) and remote (RM, 0.9709 Å), were collected at the Se K edge from the same SeMet-containing crystal (see Table 1 for data-collection details and statistics). The MAD data were collected from a single frozen crystal at 100 K using a PILATUS detector at the PXI-XS06 beamline (SLS Villigen, Switzerland). Data processing and scaling were accomplished with XDS (27). All methionines were substituted by Seleno-methionine and the two possible Se sites were identified using SHELX package (28). Initial phases were calculated at 2.5 Å resolution using the Autosolve program included in PHENIX (29). These initial phases were extended to 2.2Å resolution using a new data set with the PHENIX Autobuild routine. Data collection, phasing and refinement statistics are summarized in Table 1. The Ramachandran plot showed 98% and 2% of the residues in the favoured/allowed and disallowed, regions, respectively. The identification and analysis of the hydrogen bonds and van der Waals contacts was performed with the Protein Interfaces, Surfaces and Assemblies service (PISA) and LIGPLOT at the European Bioinformatics Institute (http://www.ebi.ac.uk/msdsrv/ prot_int/pistart.html). Figures were generated using PyMOL.
X-Ray data. Data collection, MAD phasing and refinement statistics for the BD1-N-TPR structure. PDB code 5JNO
| . | . | Crystal phasing . | . | Crystal refinement . |
|---|---|---|---|---|
| Data collection | ||||
| Space group | P6122 | P6122 | ||
| Number of crystals | 1 | 1 | ||
| Beamline | X06SA SLS | XS06SA SLS | ||
| Cell dimensions | ||||
| a, b, c (Å) | 47.67, 47.67, 430.70 | 47.28, 47.28, 431.58 | ||
| α, β, γ (°) | 90, 90, 120 | 90, 90, 120 | ||
| Peak | Inflection | Remote | High Resolution | |
| Wavelength | 0.9785 | 0.9787 | 0.9709 | 1.000 |
| Temperature (K) | 100 | 100 | 100 | 100 |
| Resolution (Å) | 2.5–50 | 2.5–50 | 2.5–50 | 2.2–50 |
| Rsym | 6.1 (22.6) | 5.9 (26.4) | 6.4 (37.6) | 11.2 (68.1) |
| I/σI | 8.3 (2.2) | 8.7 (2.1) | 8.3 (1.7) | 3.7 (1.1) |
| Completeness (%) | 99.8 (99.8) | 99.9 (99.3) | 99.8 (98.7) | 99.1 (94.3) |
| Redundancy | 16.6 (17.5) | 16.5 (17) | 16.1 (15.3) | 30 (16.3) |
| Refinement | ||||
| Resolution (Å) | 2.2–40 | |||
| No. of reflections | 14907 | |||
| Rwork/Rfree | 22.1/23.7 | |||
| No. of atoms | ||||
| Protein | 1259 | |||
| Ligand/ion | 6 | |||
| Water | 18 | |||
| B-factors | ||||
| Protein BD1/NTPR | 30.0/40.2 | |||
| Ligand/ion | 65.9 | |||
| Water | 51.8 | |||
| R.m.s. deviations | ||||
| Bond lengths (Å) | 0.013 | |||
| Bond angles (°) | 1.58 | |||
| Ramachandran plot | ||||
| Residues in preferred and allowed regions | 145 (98%) | |||
| Outliers | 3 (2%) | |||
| . | . | Crystal phasing . | . | Crystal refinement . |
|---|---|---|---|---|
| Data collection | ||||
| Space group | P6122 | P6122 | ||
| Number of crystals | 1 | 1 | ||
| Beamline | X06SA SLS | XS06SA SLS | ||
| Cell dimensions | ||||
| a, b, c (Å) | 47.67, 47.67, 430.70 | 47.28, 47.28, 431.58 | ||
| α, β, γ (°) | 90, 90, 120 | 90, 90, 120 | ||
| Peak | Inflection | Remote | High Resolution | |
| Wavelength | 0.9785 | 0.9787 | 0.9709 | 1.000 |
| Temperature (K) | 100 | 100 | 100 | 100 |
| Resolution (Å) | 2.5–50 | 2.5–50 | 2.5–50 | 2.2–50 |
| Rsym | 6.1 (22.6) | 5.9 (26.4) | 6.4 (37.6) | 11.2 (68.1) |
| I/σI | 8.3 (2.2) | 8.7 (2.1) | 8.3 (1.7) | 3.7 (1.1) |
| Completeness (%) | 99.8 (99.8) | 99.9 (99.3) | 99.8 (98.7) | 99.1 (94.3) |
| Redundancy | 16.6 (17.5) | 16.5 (17) | 16.1 (15.3) | 30 (16.3) |
| Refinement | ||||
| Resolution (Å) | 2.2–40 | |||
| No. of reflections | 14907 | |||
| Rwork/Rfree | 22.1/23.7 | |||
| No. of atoms | ||||
| Protein | 1259 | |||
| Ligand/ion | 6 | |||
| Water | 18 | |||
| B-factors | ||||
| Protein BD1/NTPR | 30.0/40.2 | |||
| Ligand/ion | 65.9 | |||
| Water | 51.8 | |||
| R.m.s. deviations | ||||
| Bond lengths (Å) | 0.013 | |||
| Bond angles (°) | 1.58 | |||
| Ramachandran plot | ||||
| Residues in preferred and allowed regions | 145 (98%) | |||
| Outliers | 3 (2%) | |||
Highest resolution shell is shown in parenthesis.
| . | . | Crystal phasing . | . | Crystal refinement . |
|---|---|---|---|---|
| Data collection | ||||
| Space group | P6122 | P6122 | ||
| Number of crystals | 1 | 1 | ||
| Beamline | X06SA SLS | XS06SA SLS | ||
| Cell dimensions | ||||
| a, b, c (Å) | 47.67, 47.67, 430.70 | 47.28, 47.28, 431.58 | ||
| α, β, γ (°) | 90, 90, 120 | 90, 90, 120 | ||
| Peak | Inflection | Remote | High Resolution | |
| Wavelength | 0.9785 | 0.9787 | 0.9709 | 1.000 |
| Temperature (K) | 100 | 100 | 100 | 100 |
| Resolution (Å) | 2.5–50 | 2.5–50 | 2.5–50 | 2.2–50 |
| Rsym | 6.1 (22.6) | 5.9 (26.4) | 6.4 (37.6) | 11.2 (68.1) |
| I/σI | 8.3 (2.2) | 8.7 (2.1) | 8.3 (1.7) | 3.7 (1.1) |
| Completeness (%) | 99.8 (99.8) | 99.9 (99.3) | 99.8 (98.7) | 99.1 (94.3) |
| Redundancy | 16.6 (17.5) | 16.5 (17) | 16.1 (15.3) | 30 (16.3) |
| Refinement | ||||
| Resolution (Å) | 2.2–40 | |||
| No. of reflections | 14907 | |||
| Rwork/Rfree | 22.1/23.7 | |||
| No. of atoms | ||||
| Protein | 1259 | |||
| Ligand/ion | 6 | |||
| Water | 18 | |||
| B-factors | ||||
| Protein BD1/NTPR | 30.0/40.2 | |||
| Ligand/ion | 65.9 | |||
| Water | 51.8 | |||
| R.m.s. deviations | ||||
| Bond lengths (Å) | 0.013 | |||
| Bond angles (°) | 1.58 | |||
| Ramachandran plot | ||||
| Residues in preferred and allowed regions | 145 (98%) | |||
| Outliers | 3 (2%) | |||
| . | . | Crystal phasing . | . | Crystal refinement . |
|---|---|---|---|---|
| Data collection | ||||
| Space group | P6122 | P6122 | ||
| Number of crystals | 1 | 1 | ||
| Beamline | X06SA SLS | XS06SA SLS | ||
| Cell dimensions | ||||
| a, b, c (Å) | 47.67, 47.67, 430.70 | 47.28, 47.28, 431.58 | ||
| α, β, γ (°) | 90, 90, 120 | 90, 90, 120 | ||
| Peak | Inflection | Remote | High Resolution | |
| Wavelength | 0.9785 | 0.9787 | 0.9709 | 1.000 |
| Temperature (K) | 100 | 100 | 100 | 100 |
| Resolution (Å) | 2.5–50 | 2.5–50 | 2.5–50 | 2.2–50 |
| Rsym | 6.1 (22.6) | 5.9 (26.4) | 6.4 (37.6) | 11.2 (68.1) |
| I/σI | 8.3 (2.2) | 8.7 (2.1) | 8.3 (1.7) | 3.7 (1.1) |
| Completeness (%) | 99.8 (99.8) | 99.9 (99.3) | 99.8 (98.7) | 99.1 (94.3) |
| Redundancy | 16.6 (17.5) | 16.5 (17) | 16.1 (15.3) | 30 (16.3) |
| Refinement | ||||
| Resolution (Å) | 2.2–40 | |||
| No. of reflections | 14907 | |||
| Rwork/Rfree | 22.1/23.7 | |||
| No. of atoms | ||||
| Protein | 1259 | |||
| Ligand/ion | 6 | |||
| Water | 18 | |||
| B-factors | ||||
| Protein BD1/NTPR | 30.0/40.2 | |||
| Ligand/ion | 65.9 | |||
| Water | 51.8 | |||
| R.m.s. deviations | ||||
| Bond lengths (Å) | 0.013 | |||
| Bond angles (°) | 1.58 | |||
| Ramachandran plot | ||||
| Residues in preferred and allowed regions | 145 (98%) | |||
| Outliers | 3 (2%) | |||
Highest resolution shell is shown in parenthesis.
QuikChange mutagenesis
Site-directed mutagenesis of the N-TPR domain was performed using the QuikChange II site-directed mutagenesis kit. The list of primers used were as follows:
ERCC6L T73g_a74c (FW1) 5′ gtccggaacaggcagcacatgctctgcgttatgttaaaga 3′
ERCC6L T73g_a74c (RV1) 5′ tctttaacataacgcagagcatgtgctgcctgttccggac 3′
ERCC6L a44c_c49g_t50c (FW2) 5′ gttttccggaagcagcagcagcgagtccggaacaggc 3′
ERCC6L a44c_c49g_t50c (RV2) 5′ gcctgttccggactcgctgctgctgcttccggaaaac 3′
The mutants were expressed and purified as described above for the wild type N-TPR.
RESULTS
Determining the PICH interactome in mitosis
To identify binding partners of PICH in mitotic human cells, we precipitated PICH from HeLa cells arrested in prometaphase (Supplementary Figure S1). We then used mass spectrometry to identify co-precipitating proteins (a full list of proteins identified is shown in Supplementary Table S1). This analysis identified several known PICH-interacting factors, such as PLK1, as well as many of the proteins shown previously to bind to UFBs in a PICH-dependent manner (RlF1, BLM and topoisomerase IIIα). Selected proteins identified and their functions are shown in Supplementary Table S2. A prominent hit from the screen was BEND3, a BEN domain-containing protein identified previously in screens for factors associated with mitotic chromatin (30–32). BEND3 contains four BEN domains (Figure 1A), which generally mediate interactions with DNA (33,34). Aside from these BEN domains, there are no obvious functional domains in BEND3. Nevertheless, previous work showed that BEND3 localizes to heterochromatin and may mediate transcriptional repression (35).
PICH and BEND3 interact in vitro
We first addressed whether PICH and BEND3 are able to interact directly in the absence of DNA or any other proteins. For this, we purified recombinant PICH from insect cells, as described previously (20), and recombinant BEND3 from human HEK293 cells (Supplementary Figure S2A and B). Using two independent biophysical techniques, surface plasmon resonance (SPR) and microscale thermophoresis (MST), we showed that PICH and BEND3 can interact directly (Figure 1B–C; Supplementary Figure S3A and B). Using SPR, we determined the Kd of the PICH–BEND3 interaction to be 0.7 ± 0.3 μM. It should be noted that the interaction with PICH was also evident when using a bacterially expressed version of BEND3, making it unlikely that post-translational modifications on BEND3 are essential for the interaction (data not shown).
PICH and BEND3 interact via a TPR–BEN domain interface
To map the regions of interaction on PICH and BEND3, we purified several recombinant fragments of each protein (Supplementary Figure S2A and B). Using MST, we showed that full length PICH is able to bind to BEN domains 1 and 3 (BD1/BD3) of BEND3, but not to BEN domains 2 and 4 (BD2/BD4) (Figure 1C; Supplementary Figure S3C and D; data not shown). Similarly, we observed that the N-terminal TPR motif of PICH (N-TPR), but not the C-terminal TPR (C-TPR), could bind full length BEND3 (Figure 1C; Supplementary Figure S3C and D). Hence, we analyzed whether the N-TPR motif interacts directly with BD1 and/or BD3. We observed that the N-TPR and BD1 domains interact directly, with a Kd determined to be 1.2 ± 0.25 μM (Figure 1C; Supplementary Figure S3C). We also observed that BD3 interacted with the catalytic SNF2/HELIC region of PICH, with a Kd of 1.5 ± 0.3 μM. (Supplementary Figures S2A and B and S3C and D). In contrast, we did not detect any interaction between the other domains of PICH and BEND3 (Supplementary Figures S2A and B and S3C and D). Although it is possible that the interaction between BD3 of BEND3 and the SNF2/HELIC region of PICH is of functional significance, we focused in this article on the role of the BD1/N-TPR interaction due to the well-established role of TPR domains in mediating protein:protein interactions.
BEND3 enhances the ATPase and translocase activities of PICH in vitro
BEND3 has no obvious biochemical function that can be analyzed in vitro. However, PICH is a DNA-dependent ATPase, and an ATP-dependent translocase on dsDNA (20). To investigate whether BEND3 might influence these activities, we first determined the oligomeric state of BEND3 to better define the relative molar ratios of each protein to be used in subsequent biochemical assays. For this, we utilized gel filtration and native gel electrophoresis (Supplementary Figure S4A and B). Using either method, we observed that BEND3 exists in a homogeneously higher-order form that has a molecular mass of ∼800 kDa, suggestive of an octamer. The isolated N-terminal region of BEND3 (residues 1–534) was similarly found to be homogeneously oligomeric (Supplementary Figure S4B), indicating that an oligomerization domain of BEND3 is present in this region. It might be significant in this regard that the N-terminal domain contains a putative coiled-coil region, a feature utilized commonly for the oligomerization of proteins (36). Previous work has demonstrated that PICH is largely monomeric in solution (20), and hence we considered a PICH:BEND3 protein ratio of 1:8 to be equimolar. We observed that addition of an equimolar amount of BEND3 stimulates both the ATPase activity (Figure 1D) and the translocase activity of PICH (Figure 1E). Therefore, the interaction between PICH and BEND3 leads to enhanced PICH activity, suggesting that BEND3 might regulate PICH function in vivo. In parallel, we investigated whether the isolated BD1 domain might stimulate the ATPase activity of PICH, but the results were negative (data not shown).
The crystal structure of the TPR–BEN domain complex
Next, we examined in more detail the interaction of PICH with BEND3. Because one of the interaction regions was mapped to the well-defined BD1 and N-TPR domains, we crystallized and solved the structure of a complex of these two domains. The phase problem was solved using the multiple anomalous dispersion (MAD) method after substituting the methionine residues of BD1 with selenomethionine (26). Using the initial map, a model of the complex was built and through iterative model building and refinement cycles was refined to 2.2 Å with an Rwork/Rfree of 0.22/0.23 (Table 1).
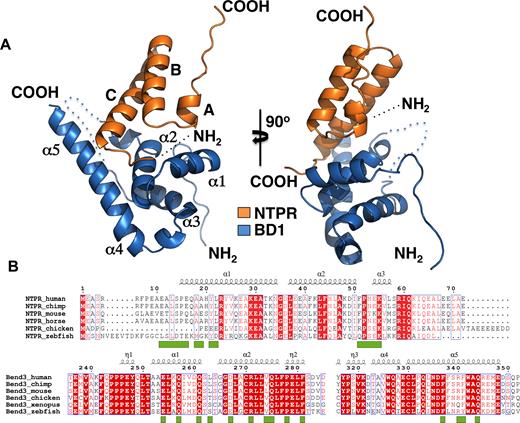
The crystal structure of the complex revealed that BD1 forms a heart-shaped domain. In contrast to previously described BEN domain structures (33,34), BD1 is exclusively composed of α-helices connected by loops and turns, with no β-strands (Figure 2A and B). The N-terminal region of BEND3 is connected via a long loop to the initial α1 of the BD1 core. A short loop then connects α1 to α2, which is joined to α3 via another loop. In the published insv-BEN and Bsg25A BEN domain structures (33,34), a much longer loop running between these helices is involved in DNA binding, but this arrangement is not conserved in either BD1 or in the other BEN domains of BEND3 (Figure 3A; Supplementary Figure S5). The α3 helix of BD1 is then connected to a long C-terminal helix (α4/5) by a turn. The α-helices 1–3 of BD1 are organized into a spiral structure that comprises the central core of the domain. The C-terminal α-helix then flanks the spiral core.
Structure of the N-TPR-BD1 complex. (A) Left: ribbon representation of the N-TPR domain (orange), which is composed largely of α-helices, and the BD1 domain (blue) which is composed of α1-α5 helices packed together to form a heart-shaped structure. A loop not visible in the structure is represented in dotted lines. Right; the N-TPR–BD1 complex has been tilted 90° to give an insight into the interaction interface. (B) Sequence alignment of N-TPR domains from different PICH proteins (upper) and BEN domain 1 from different BEND3 proteins (lower) from different species. The alignment was created using ESPRIPT online software. Identical residues are shown in white text in red boxes. Similar residues are in red text. The key residues involved in the interaction are represented by green squares below.
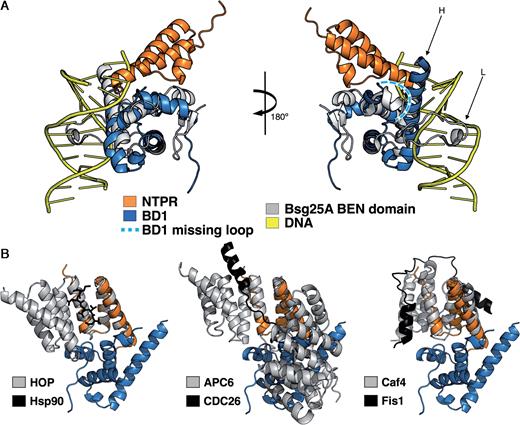
Superimposition of available PDBe structures with N-TPR and BD1 complex. (A) (Left) The Bsg25A BEN domain structure (grey) together with dsDNA (yellow) is superimposed with human BEND3 BD1 (blue). (Right) The superimposed structures have been tilted 180° to see the DNA binding region of the Bsg25A BEN domain. The missing loop from BD1 is highlighted in pale blue with white dots. (B) The N-TPR–BD1 complex structure (blue and orange, respectively) is superimposed with the Hop–Hsp90 structure (gray and black). Note that the helix A of N-TPR is distorted.
TPR domains are also generally composed of α-helical elements, and are involved in protein–protein interactions and polynucleotide recognition (37). The N-TPR domain of PICH consists of three α-helices (denoted A–C) that pack together to form a bundle; however, the N-terminal helix A is distorted in our structure, and only a turn can be discerned clearly (Figure 2). The residues responsible for the interaction with BD1 reside in a loop positioned between helices A and B, and also at the tip of helix C in the C-terminal loop, thus defining a new mode of interaction among TPR domains. Several other modes of protein interaction have been defined previously with TPR modules (38). A typical example is that of an extended short peptide binding conformation that can be observed in the Hop–Hsp90 interaction (39). There, the TPR2A domain of the Hop adaptor protein binds the five C-terminal amino acids of the Hsp90 chaperone in the concave side of the TPR module (Figure 3B). In other representative cases, either a long peptide adopts an extended helical conformation and the TPR domain forms a solenoid structure surrounding it (as in the complex of APC6 and CDC26) (40), or the peptide ligand forms a U-fold interacting with both the concave and convex sides of the TPR, as observed in the interaction between Fis1 and a 46 residue Caf4 peptide (41) (Figure 3B).
Although BD1 does not appear to dimerize in solution, it is interesting to note that the N-terminal 15 residues of the α3 helix of BD1 make a crystal contact with the corresponding region of an adjacent monomer in an interlocked hook configuration (Supplementary Figure S6). This interaction could potentially be involved in the oligomerization of BEND3 and its N-terminal domain (Supplementary Figure S4). If true, that would promote an antiparallel orientation of the BD1 domain pairs.
The interaction surface of N-TPR and BD1
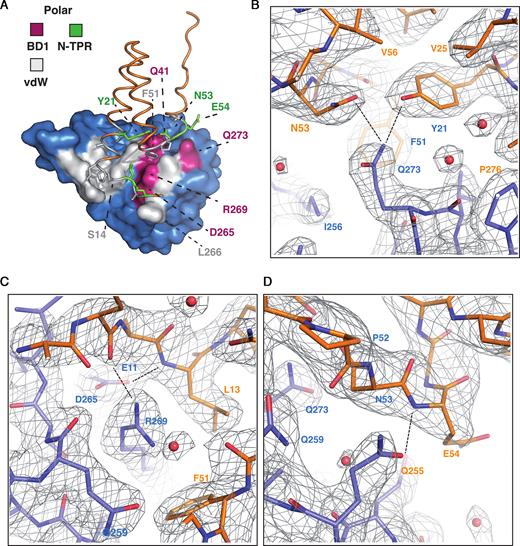
A concave surface on the upper part of the heart-shaped BD1 domain, which is moulded mainly from the α1 and α2 helices, is where the PICH N-TPR domain docks to form the binding interface (Figure 4, Supplementary Figure S7). Thus, the TPR interaction occurs in a region of BEND3 that is different from that used in BEN domains for mediating interactions with DNA (Figure 3A; Supplementary Figure S5). The buried surface between the TPR and BEN domains is 2868.7 Å2, which is in the typical range for many protein–protein interaction surfaces. The surface is characterized by a combination of polar and hydrophobic contacts, which seem to be organized in an alternating pattern on the docking surface of BD1 (Figure 4A; Supplementary Figure S7). Side chains of BD1 that make polar contacts with N-TPR do so via the main chain of the N-TPR domain (Figure 4B–D); the only exception being Gln273, whose side chain can form hydrogen bonds with either the hydroxyl group of Tyr21 or the carbonyl of Asn53 of N-TPR (Supplementary Figure S7). The remainder of the polar interactions engage the side chains of Gln255, Asn265 and Arg269 in BD1 with the main chain amides of Leu13, Glu54 and the carbonyl of Glu11, respectively, of N-TPR (Figure 4A–D; Supplementary Figure S7).
Residues in the N-TPR domain required for interaction with BD1. (A) A surface model of the N-TPR/BD1 interaction. The N-TPR domain (orange) is shown sitting on top of the heart-shaped BD1 domain (blue). The interaction interfaces are highlighted; green (for N-TPR) and magenta (for BD1) represent surfaces involved in polar interactions. Grey represents the van der Waals interaction interface in both domains. The N-TPR domain is in orange and the residues involved in the binding are represented in different colors. The side chains of interacting residues are shown as sticks in N-TPR. Green represents the amino acids involved in polar contacts, and grey represents hydrophobic interactions from N-TPR. (B–D) Detailed view of the key residues involved in the interaction of the N-TPR and BD1 complex shown as a 2Dfo-mfc electron density map contoured at 1σ. The N-TPR is shown in orange and BD1 in blue. The key residues are depicted with their single letter code and corresponding amino acid position. Dashed lines represent the hydrogen bonds between the amino acids (see Supplementary Figure S6 for a schematic representation).
The interaction surface of BD1 is well conserved in other vertebrate BEND3 homologs (Figure 2B; Supplementary Figure S8A). Moreover, there is a high degree of evolutionary conservation of amino acid positions in BD1 and other BEND3 homologs in the key positions involved in the polar contacts with N-TPR. In contrast, the more highly conserved residues in N-TPR are those responsible for the helix–loop–helix arrangement of the domain (Figure 2B; Supplementary Figure S8B), rather than for making polar interactions with BD1. A notable exception to this is Tyr21, which is very well conserved. The contacts on the N-TPR surface are mainly hydrophobic and the hydrogen bonds are made with the main chain of the polypeptide (Figure 4A–D, Supplementary Figure S7). Those residues whose side chains are associated with BD1 residues are well conserved or are substituted by amino acids that can engage in similar hydrogen bonding.
Mutations in key binding residues affect how BEND3 stimulates PICH
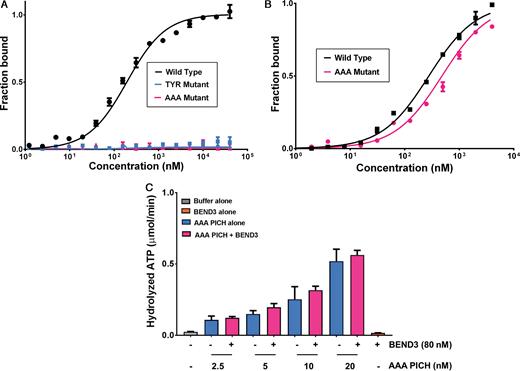
To confirm the role of key residues involved in the N-TPR and BD1 domain interactions, we generated two mutant derivatives of the N-TPR motif: one with Tyr-21 mutated to Alanine, and one with three Alanine substitutions at residues Glu11, Leu13 and Tyr21 (termed the ‘AAA mutant’). We first confirmed that both PICH mutant variants were folded correctly using circular dichroism (data not shown). We then tested for any alterations in their ability to bind to BD1. MST analysis indicated that both mutant forms failed to bind BD1, confirming the importance of these residues for mediating interactions with BD1 (Figure 5A; Supplementary Figure S7). Consistent with the finding that PICH also binds to BD3 of BEND3 (Supplementary Figure S3C and D), we observed that full length BEND3 interacts with full length PICH containing the AAA substitutions in the N-TPR domain (Figure 5B). Nevertheless, the AAA substitutions reduced the Kd of the interaction by 1.5-fold (which was a statistically significant decrease; P < 0.05), as determined using MST analysis. Despite the N-TPR substitutions having only a limited effect on the ability of PICH to associate with BEND3, the AAA-PICH mutant protein was refractory to stimulation of ATPase activity by BEND3 (Figure 5C; for comparison with Figure 1D).
MST binding and activity assays of the mutant versions of N-TPR (A) A comparison of the interaction of either wild type or mutant versions of N-TPR domain with BD1 using MST analysis. Normalized thermophoresis is plotted against the concentration of the unlabeled ligand. (B) As panel (A), except using full length BEND3 together with either full length PICH or the PICH-AAA mutant, as indicated. Data points represent the mean of at least three independent experiments. Error bars denote SD. (C) dsDNA-dependent ATPase activity of different concentrations of mutant AAA-PICH in the absence (blue bars) or presence (pink bars) of 80 nM BEND3 (monomer concentration; equivalent to 10 nM BEND3 octamers). The grey and orange bars denote control reactions with buffer or BEND3 alone, respectively. The ATPase activity of the PICH-AAA protein was not stimulated by BEND3 at any protein concentration tested (P > 0.05). Data points represent the mean of at least three independent experiments. Error bars denote SD.
DISCUSSION
We have identified BEND3 as a new interaction partner for PICH in mitosis, and have defined the residues within a novel TPR–BEN domain interface that mediate this interaction. Although TPR motifs are well established as mediators of protein–protein interactions, the proposal that BEN domains might mediate protein–protein interactions in addition to protein-nucleic acid interactions is derived largely from computational predictions. Our study provides the first structural view of a protein–protein interaction mediated by a BEN domain. Moreover, to our knowledge, this is the first characterization of a specific protein–protein interaction being mediated through a TPR–BEN interface. In many cases, TPR motifs occur tandemly in proteins in order to mediate self-association (22,42,43). Because PICH can exist in monomeric or dimeric forms (20), it is possible that the two TPR motifs promote PICH dimerization. If true, it will be interesting to determine how the dynamic association (and potential competition) of N-TPR with either the C-TPR of PICH or BD1 of BEND3 is regulated.
BEN domains are proposed to function as adaptors to promote the assembly of higher-order chromatin, or to regulate chromatin remodeling during transcription (44–47). Indeed, the Drosophila melanogaster Elba2 protein has been shown to functionally substitute for histone H1 if the natural linker histone is depleted from cells (48). The structures of the insv-BEN and the Bsg25A BEN domains from Drosophila (33,34) have revealed that the association of BEN domains with DNA is generally accomplished through the insertion of both the loop located between the α2 and α3 helices and the long C-terminal helix into two consecutive major grooves of the nucleic acid molecule (Figure 5A; Supplementary Figure S6A and B). Based on our observations, we propose that BEN domains offer the potential for two distinct association surfaces: one for the recognition of nucleic acids, and the other for protein–protein interactions. Indeed, sequence and structural comparisons of BD1 with the BEN domains of Bsg25A and insv bound to DNA, suggest that it should be possible to predict those BEN domains that are likely to be involved primarily in binding either nucleic acids or proteins. We propose that the insertion in the loop connecting α2 and α 3 defines those BEN domains involved in DNA binding. Although we have no evidence for DNA binding in the case of the isolated BD1 domain, it remains conceivable that a single BEN domain could associate alternately with nucleic acid and protein.
It is likely that the interaction of these proteins is more complex than our current study has revealed, because BEND3 also contacts the SNF2/HELIC region of PICH via BD3, thus explaining why the disruption of the BD1-N-TPR interaction does not affect binding of the full length PICH and BEND3 proteins more severely. Further studies will be required to evaluate the possible functional significance of the interaction of the SNF2/HELIC region of PICH with BEND3. Even if this interaction were to be of significance in some contexts, we showed that the stimulation of PICH catalytic activity was abolished by substitution of key residues in the N-TPR motif that mediate binding to BD1 of BEND3, indicating that the 1.5-fold decrease in the Kd causes a greater impact on PICH activity than it does on complex formation per se. Nevertheless, it should be noted that the roles of BEND3 are very unlikely to be restricted to the modulation of PICH function. This is because PICH apparently plays a role exclusively in mitosis due to its nuclear exclusion prior to mitosis (11,24), whereas BEND3 is known to influence the transcriptional activity of heterochromatic loci during interphase (35).
UFBs arise from specific loci, even in an unperturbed mitosis, and it is clear that cells frequently enter anaphase with persistent catenation of centromeric and ribosomal DNA (11,16,19). Moreover, UFB formation at these loci is enhanced strikingly following inhibition of Topoisomerase II with ICRF-193 (12,16,19). In the context of the previously proposed role of BEND3 in modulating locus-specific transcription, the possibility that PICH might influence, for example, the synthesis of rRNA or centromeric non-coding RNAs should be considered. Transcription is known still to be occurring at these loci in early mitosis (49,50) and this has been proposed to influence the timing of DNA condensation. Further work will be required to address the validity of this speculation.
PICH was initially identified as a PLK1 interacting factor (11). It will be interesting, therefore, to address whether PLK1 regulates the functional association of PICH and BEND3. In this regard, it might be significant that BD1 of BEND3 contains a strong PLK1 phosphorylation site consensus sequence (Supplementary Figure S9A–C). This region is located at the interface of N-TPR with BD1, and therefore its phosphorylation would be predicted to influence this interaction. Interestingly, the PICH interaction region of the BD1 domain of BEND3 also has a putative connection to human disease biology because mutations affecting the Asp265 residue have been detected in gastric cancers (www.cBioportal.org). Our structural analysis revealed that Asp265 is one of the key residues in BD1 for mediating interactions with the TPR domain PICH, and therefore this mutation could interfere with PICH function and hence chromosome stability. Future studies should be aimed at investigating this possibility.
DATA AVAILABILITY
The structure has been deposited in the Protein Data Bank (http://www.rcsb.org/pdb/) under accession number 5JNO.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank members of the Hickson, Montoya and Nigg groups for helpful discussions, Dr H.W. Mankouri for helpful comments on the manuscript, the Proteomics Core facility at the Basel Biozentrum for help with the mass spectrometry, and the beamline staff of X06SA at the Swiss Light Source (Paul Scherrer Institut, Villigen, Switzerland) for support during data collection.
FUNDING
Danish National Research Foundation [DNRF115]; European Research Council, Nordea Foundation, and the Danish Medical Research Council (to K.S. and I.D.H.); NovoNordisk Foundation [NNF14CC0001]; Danish Cancer Society (to G.M.); Swiss National Science Foundation [310030B 149641 to E.A.N.]; European Community FP7 [283570] (BioStruct-X, providing access to the Swiss Light Source and the MAX-Lab). Funding for open access charge: Danmarks Grundforskningsfond.
Conflict of interest statement. None declared.
Present addresses:
Ganesha P. Pitchai, Department of Biochemistry, University of Oxford, UK.
Manuel Kaulich, Institute of Biochemistry II, Goethe University, Frankfurt, Germany.
Werner W. Streicher, Novozymes A/S, Bagsvaerd, Denmark.
REFERENCES




Comments