-
PDF
- Split View
-
Views
-
Cite
Cite
Akihiro Ueda, Weiming Shi, Kazutsuka Sanmiya, Mariko Shono, Tetsuko Takabe, Functional Analysis of Salt-Inducible Proline Transporter of Barley Roots, Plant and Cell Physiology, Volume 42, Issue 11, 15 November 2001, Pages 1282–1289, https://doi.org/10.1093/pcp/pce166
Close - Share Icon Share
Abstract
We cloned a cDNA encoding Hordeum vulgare Proline Transporter (HvProT) from salt-stressed barley roots by differential display. HvProT was 2,161 bp long and had an open reading frame encoding 450 amino acids. The deduced amino acid sequence of HvProT was similar to those of proline transporter proteins of rice (65.7%), Arabidopsis (57.7%) and tomato (42.0%). Northern blot analysis showed that the transcript level of HvProT was induced in roots at 30 min after 200 mM NaCl treatment and its peak was observed at 3 h. However, the transcript level was very low in leaves and did not increase by salt stress. The expression level of Δ1-pyrroline-5-carboxylate synthetase (P5CS), encoding a key enzyme of proline synthesis, was induced later than HvProT by salt stress. A transport assay using a yeast with mutation in proline uptake revealed that HvProT was a transporter with high affinity for l-proline (Km = 25 µM). HvProT was found to be a unique transporter with high affinity for l-proline. Since its transport activity was dependent on the pH gradient, HvProT was suggested to be a H+/amino acid symporter. In situ hybridization analysis showed that the HvProT mRNA was strongly expressed in root cap cells under salt stress. HvProT might play an important role in the transport of proline to root tip region urgently upon salt stress.
(Received June 27, 2001; Accepted September 10, 2001).
Introduction
High salinity lowers the productivity of crops by disturbing ion-homeostasis and inhibiting water uptake. Overaccumulation of sodium ion in cells results in inhibition of various essential activities for growth (Cachorro et al. 1993, Guerrier 1996, Tattini et al. 1997). Moreover osmotic stress is also another aspect of salt stress. Generally, under abiotic stresses, osmoprotectants such as proline, glycinebetaine and sugar polyols are synthesized in plants (Hanson and Hintz 1982, Ishitani et al. 1993). In particular, the synthesis and degradation of proline have been well studied (Delauney and Verma 1993, Yoshiba et al. 1995, Kiyosue et al. 1996). Transgenic tobacco overaccumulating proline acquired enhanced tolerance to salt stress (Kavi Kishor et al. 1995). Besides osmoregulation, proline protects the membrane and proteins (Schobert and Tschesche 1978), scavenges radicals (Smirnoff and Cumbes 1989) and decreases the Tm of DNA (Rajendrakumar et al. 1997) under salt stress. On the other hand, excess proline has been reported to suppress ribulose 1,5-bisphosphate carboxylase/oxygenase activity (Sivakumar et al. 1998).
Little is known about the transport of proline in higher plants. In microorganisms such as Salmonella typhimurium and Escherichia coli, the proline transport system has been well characterized (Stalmach et al. 1983, Cairney et al. 1985a, Cairney et al. 1985b). In S. typhimurium, three kinds of proline transporters are known. Namely putP is specific for proline, whereas proP and proU transport both proline and glycinebetaine (Cairney et al. 1984, Cairney et al. 1985a, Cairney et al. 1985b). cDNAs encoding proline transporter (ProT) have been cloned from Arabidopsis, tomato and rice (Rentsch et al. 1996, Schwacke et al. 1999, Igarashi et al. 2000). Although both Arabidopsis and tomato are non-accumulators of glycinebetaine, both Arabidopsis ProT2 (AtProT2) and tomato ProT1 (LeProT1) use choline, γ-amino butyric acid (GABA) and glycinebetaine as efficient substrates in addition to l-proline (Breitkreuz et al. 1999, Schwacke et al. 1999). AtProT and LeProT have a broad substrate specificity, but the possible function of ProT to tolerate abiotic stress and the relationship between ProT and glycinebetaine remain unknown.
Previously, we cloned many salt-inducible genes in barley roots by differential display (Ueda et al. submitted). Among the many candidate genes related to environmental stresses, we cloned a ProT homologue. In barley, both glycinebetaine and proline are accumulated in leaves in response to salt stress. In the present study, we characterized HvProT and the localization of its mRNA to clarify the function of ProT in salt stress tolerance and elucidate the mechanism of proline transport in a glycinebetaine-accumulator, barley.
Results
Cloning and sequence analysis of HvProT
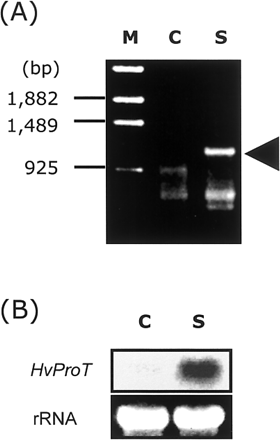
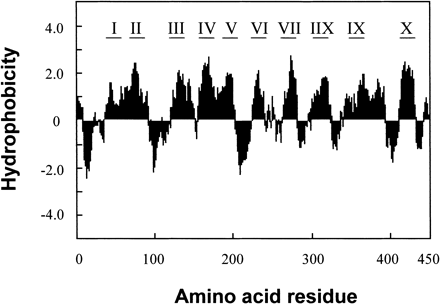
We obtained a highly salt inducible 1.2 kbp PCR product by differential display RT-PCR using the D39 primer (Fig. 1A). DNA sequencing and searching on the database of its fragment revealed that its amino acid sequence showed 61% homology to proline transporter 2 protein in Arabidopsis (AtProT2). After screening a cDNA library of 1×105 plaques, we isolated four positive clones. The largest cDNA clone was sequenced and it was 2,161 bp in length and contained a single putative open reading frame (Fig. 2). We performed 5′-rapid amplification of cDNA ends and obtained a sequence identical to the 5′-region. Since the deduced amino acid sequence of full-length cDNA had high homology to ProT in Arabidopsis, tomato and rice (Table 1), it was considered to encode ProT in barley. By hydropathy plotting, HvProT appeared to have ten transmembrane regions as in Arabidopsis and tomato (Fig. 3) (Rentsch et al. 1996, Schwacke et al. 1999).
Expression pattern of HvProT under salt stress
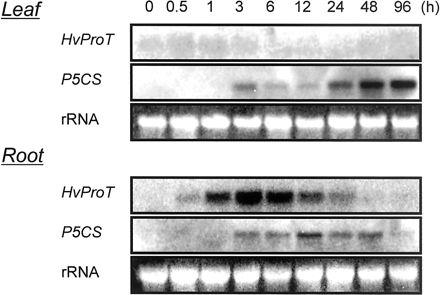
Under 200 mM NaCl stress, the expression of HvProT was quickly enhanced within 30 min in barley roots (Fig. 4). The level of HvProT mRNA was the highest in the roots at 3 h after stress treatment and dropped to the control level after 48 h. This result indicated that the expression patterns were different although the expression of HvProT was induced by both mild salt stress of a long term (as detected by differential display, Fig. 1B) and severe salt stress of a short term (as detected by Northern blot analysis). In barley leaves, in contrast with roots, HvProT was not induced at all. Although the Δ1-pyrroline-5-carboxylate synthetase (P5CS) gene, which encodes the key enzyme of proline synthesis, was induced in both leaves and roots, interestingly its expression occurred later than that of HvProT.
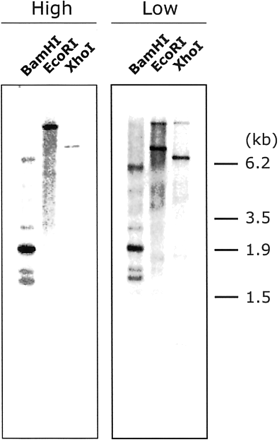
By Southern blot analysis, many bands were observed under a low stringency condition using a full length cDNA as a probe (Fig. 5). Under high stringency conditions using a 0.5 kb fragment of the 3′ untranslate region as a probe, one band was detected in each lane (data not shown). These results showed that HvProT has a family in barley.
Substrate specificity of HvProT by yeast complementation
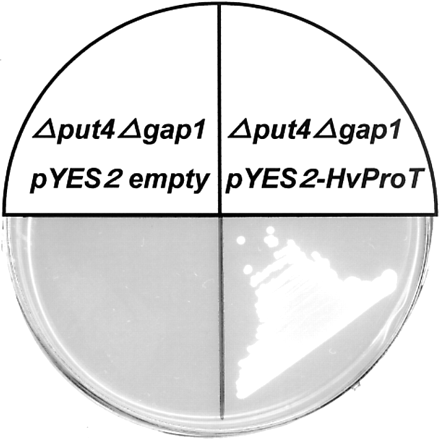
A yeast strain with a mutation of ProT was provided from ATCC. Moreover general amino acid permease (GAP1), which also transports proline, was disrupted (refer to Materials and Methods). Thus, a double mutant (Δput4Δgap1) was constructed and used for the experiments. The mutant did not grow on low proline medium as a sole nitrogen source (Fig. 6). So its phenotype was similar to that reported previously (Lasko and Brandriss 1981). In order to perform the functional analysis of HvProT, the pYES2-HvProT plasmid was introduced into a yeast double mutant. Only transformed yeast grew on the low proline medium (Fig. 6). To determine the substrate specificity of HvProT, a yeast double mutant complemented by pYES2-HvProT was used for the [14C]l-proline uptake experiment. We found that its Km value was very low (Km = 25.1 µM), strongly suggesting a specific proline transporter, and the [14C]l-proline uptake was dependent on pH gradient (Table 2). Moreover, treatment with carbonyl cyanide m-chlorophenyl-hydrazone (CCCP), a reagent to collapse the pH gradient, decreased significantly the [14C]l-proline uptake rate. Generally, the amino acid transporter in plants is coupled with H+ transport (Delrot et al. 2000). These results indicated that HvProT belongs to the H+/amino acid transporter family. It appeared to have low or no affinity to amino acid species other than proline in a competition test (Fig. 7). l-proline analogues, d-proline and d/l-3,4-dehydroproline inhibited to some extent the uptake of [14C]l-proline. GABA and quaternary ammonium compounds (choline, betaine aldehyde and glycinebetaine) are not efficient substrates for HvProT expressed in yeast. In the experiment using [14C]GABA as a substrate, the net uptake of GABA was not observed in HvProT transformed yeast (data not shown). So far the substrate specificity has also been examined for AtProT1, 2 and LeProT1. However, the uptake of [14C]l-proline by the yeast mutant expressing those ProTs was reported to be severely inhibited by quaternary ammonium compounds and GABA (Breitkreuz et al. 1999, Schwacke et al. 1999). However, we did not observe such inhibition by either quaternary ammonium compounds or GABA in our experiments with HvProT. Thus it was suggested that the substrate specificity of HvProT is quite different from those of AtProT and LeProT so far examined.
Localization of HvProT mRNA in roots
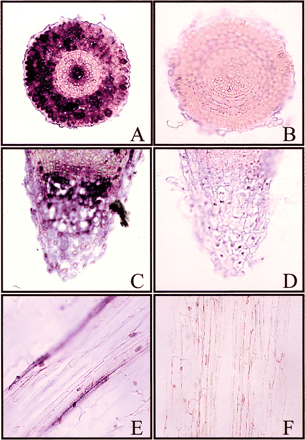
Since HvProT mRNA was highly induced in only roots under salt stress (Fig. 4), it was interesting to examine its localization in root tissues treated with 200 mM NaCl for 3 h by in situ hybridization. When a sense probe was used, no signals were detected (Fig. 8B, D, F). Using an antisense probe, we found HvProT mRNA to be abundant in root cap columella cells (Fig. 8A). Moreover the signals were observed in the cortex layer, stele and phloem (Fig. 8C, E). On the other hand, no signals were observed in root apical meristem or epidermis cells.
Discussion
The general idea is that plants sense the signals firstly in roots under salt and drought stresses. Quick sensing and response in roots is essential for survival under stress conditions. Synthesis and transport of proline would be a useful mechanism for salt tolerance. We found that HvProT is a high affinity transporter for proline (Km = 25.1 µM) but not for quaternary ammonium compounds or GABA. In Arabidopsis and tomato, glycinebetaine-non accumulators, their ProTs (AtProT1, 2 and LeProT1) are transporters for GABA, choline and glycinebetaine in yeast (Breitkreuz et al. 1999, Schwacke et al. 1999). Since barley has a gene family of ProT (Fig. 5), other ProTs might transport quaternary ammonium compounds or GABA. Secondly, we found that the expression of HvProT was induced much more rapidly than that of P5CS under salt stress (Fig. 4). This observation was in contrast to the case of Arabidopsis (Rentsch et al. 1996). These results suggest a unique characteristic of HvProT compared to ProTs previously reported. Proline, is constitutively and ubiquitously present in plant cells, to be quickly transported to specific cells or tissues during synthesis of other osmoprotectants in vivo under stress conditions. Thirdly, in situ hybridization analysis revealed that HvProT mRNA is mainly localized in the root tip region (Fig. 8). In the plant root, cells in the root tip region, especially apical meristem cells are most active. However, no signal was observed in meristem cells. Alternatively, HvProT mRNA is abundant in the root cap and cortex cells. Generally, the major role of proline accumulation is considered to be osmoregulation under stress conditions. It can be suspected that proline accumulation occurs quickly to adjust the osmotic pressure within the root cap and cortex cells. Among the plants that accumulate proline under stress conditions, barley is a low-proline accumulator (Delauney and Verma 1993). In barley, the proline transport system in the root through HvProT may contribute to not only osmoregulation but also other functions. One possible function could be as a component of stress-related protein. Transgenic Arabidopsis transformed with antisense P5CS to decrease proline synthesis showed changes in morphogenesis of leaves in parallel with reduction of proline and hydroxyproline contents in cell wall fraction (Nanjo et al. 1999). Moreover we isolated the cDNA coding two proline rich proteins by differential display under salt stress (Ueda et al. submitted). Proline-rich protein increases in cell walls under salt stress, but its function is still unknown (Deutch and Winicov 1995). Now we are studying the possible contribution of ProT to the cell structure.
Materials and Methods
Plant material and stress treatment
Seeds of barley (Hordeum vulgare L. cv Haruna-nijyo) were germinated at 25°C in dark conditions. Seedlings were hydroponically grown with Hoagland nutrient solution (Hoagland and Arnon 1950) in a growth chamber under 13 h light period (100 µmol m–2 s–1 25°C humidity 70%)/11 h dark period (22°C humidity 75%) conditions. For differential display, 2-week-old plants were treated with Hoagland solution containing 100 mM and 200 mM NaCl for 3 d each. Then, roots were harvested and stored at –80°C.
Differential display
Total RNA was extracted by the SDS-phenol method and mRNA was purified using Oligotex-dT30 Super (Takara). RT-PCR and differential display were performed as previously described (Muramoto et al. 1999). To detect HvProT from barley roots by differential display RT-PCR, we used the D39 primer (Bex; 5′-GTCGACGATGCT-3′). The salt-inducible PCR product was cloned into TOPO TA-cloning vector (Invitrogen).
Screening from cDNA library and DNA sequencing
A cDNA library of salt-stressed barley roots was constructed in λZAPII (Stratagene) (as described in Plant material and stress treatment). The library was screened from 1×105 plaques using the PCR fragment detected by differential display as a probe. The probe was prepared using Megaprime DNA labeling systems (Amersham Pharmacia Biotech). Lambda ZAPII clones were excised with pBluescript SK (–) and sequenced using a Big Dye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems) and a DNA Sequencer (ABI PRISM 373A).
Genomic Southern blot analysis
Genomic DNA was incubated with the restriction endonucleases, BamHI, EcoRI and XhoI. Digested 15 µg DNA fragments were separated on 1% agarose gel and blotted onto a nylon membrane (Hybond-N+; Amersham Pharmacia Biotech). Hybridization was performed in a solution containing 6× SSC, 5× Denhardt’s solution, 0.1% SDS and 0.1 mg ml–1 denatured herring sperm DNA at 65°C. After exposure overnight, the signals were analyzed using a BAS2000 Bioimage Analyzer (Fuji).
Northern blot analysis
For northern blot analysis, total RNA was extracted from the leaves and roots by the ATA-method (Nagy et al. 1988). Fifteen µg total RNA was separated on 1.2% agarose gel containing 0.66 M formaldehyde and blotted onto a nylon membrane (Hybond-N; Amersham Pharmacia Biotech). The membrane was washed in 6× SSC twice for 30 min. All procedures were performed at 65°C.
Yeast strain and transport assay
Saccharomyces cerevisiae (strain BY4741; MATa, his3D1, leu2D0, met15D0, ura3D0, Δput4) was provided from American Type Culture Collection (ATCC). To disrupt general amino acid permease gene (gap1) also, we designed two primers, gap1HISF (5′-CCAAGGACAGCAACATTTATAAGAAACAAAAAAAAGAAATAAAAAATGACAGAGCAGAAA-3′) and gap1HISR (5′-TGATTATCTAAAAAATAAAGTCTTTTTTTGTCGTTGTTCGATTCACTACATAAGAACACC-3′). For disruption, the DNA fragment was prepared by PCR using the pRS413 vector (Stratagene). Transformation was carried out by the lithium acetate method. After confirmation of the mutation by Southern blot analysis, the yeast double mutant (Δput4Δgap1) was used for the transport assay in this study.
In order to complement proline transport in the yeast double mutant, we constructed the pYES2-HvProT. The pYES2 vector (Invitrogen) and pBluescript-HvProT were digested with EcoRI and XhoI and they were ligated. The yeast mutant was complemented with pYES2-empty and pYES2-HvProT. For transport assay, yeast cells were grown in synthetic dextrose medium containing 2% galactose and appropriate amino acids at 30°C and harvested at OD600 = 0.5–0.8. After washing twice in water, the cells were resuspended in assay buffer (50 mM potassium phosphate, 0.6 M sorbitol, pH 4.5) to final OD600 = 3. Following addition of 50 mM galactose and incubation at 30°C for 10 min, to start the reaction, the cell suspension was added to the same volume of the transport assay buffer containing 10 µM [14C]l-proline (0.5 µCi; Amersham Pharmacia Biotech). The transport assay was stopped at 30, 60 120 and 240 s by transferring to ice-cold assay buffer. Cells were filtrated on filter (GF/C glass fiber filter; Whatman), washed with 10 ml of ice-cold assay buffer once and additional 5 ml of the same buffer. A competition test was performed by adding a 10-fold excess of each competitor (final 50 µM). [14C]l-proline uptake rate was measured by liquid scintillation counter. The transport activity was linear until 240 s.
In situ hybridization
For in situ hybridization, plants were treated with 200 mM NaCl for 3 h. The root sample was fixed in 4% paraformaldehyde and 0.25% glutalaldehyde solution overnight at 4°C. After dehydration in ethanol/buthanol series and embedding into Paraplus (New England Co.), the sample was sliced 10 µm in thickness. Sense and antisense probes were prepared using a digoxygenin (DIG) RNA labeling system (Boehringer). Probes were degraded to a mean length of 150 bp by incubation in alkali at 60°C. The sample was hybridized in a solution (50% deionized formamide, 0.3 M NaCl 1× Denhardt’s solution 10% dextran sulfate, 30 mM DTT 10 mM TE) at 50°C overnight. After it was washed and treated with anti-DIG-alkaline phosphatase treatment, the sample was stained at room temperature overnight.
Acknowledgements
This research was supported by a grant from a JSPS project “Research for Future”.
Corresponding author: E-mail, h44854a@nucc.cc.nagoya-u.ac.jp; Fax, +81-52-789-5209.
Fig. 1 (A) Comparison of gene expression between control and salt-stressed barley roots detected by DD RT-PCR using the D39 primer. Salt stress treatment was carried out as described in Materials and Methods. Arrowhead shows salt-inducible DNA fragment corresponding to HvProT. (B) Northern blot analysis of HvProT in salt-stressed roots. Roots were treated with 100 mM and 200 mM NaCl for 3 d each. M, markers; C, control; S, salt stress.
Fig. 2 Nucleotide sequence and deduced amino acid sequence of HvProT. Sequences indicated by black box show annealing region by the D39 primer. Asterisk shows stop codon. Single underline indicates specific probe for Northern analysis and in situ hybridization. Bold underlines show putative transmembrane regions.
Fig. 3 Hydropathy plot of HvProT protein according to the method of Kyte and Doolittle (1982). Window size is 10. I-X show putative transmembrane regions.
Fig. 4 Northern blot analysis of HvProT and HvP5CS in leaves and roots under 200 mM NaCl stress. The 3′-untranslated region as described in Fig. 2 was used as a specific probe for HvProT.
Fig. 5 Southern blot analysis of HvProT. Fifteen µg of DNA was digested by BamHI, EcoRI or XhoI. A full-length cDNA was used as a probe. Washing was performed in 0.5× SSC and 0.5% SDS (high stringency) and in 5× SSC and 1% SDS (low stringency) at 65°C.
Fig. 6 Complementation of the proline uptake in a yeast double mutant (Δput4Δgap1) transformed with the HvProT. Yeast transformed with pYES2-HvProT grew on medium containing 2% galactose and 0.001% proline as a sole nitrogen source, whereas yeast transformed with pYES2-empty did not grow at all.
Fig. 7 Substrate specificity of HvProT using a yeast double mutant transformed with the HvProT. [14C]l-proline (5 µM) was used for the experiments. The uptake rate was determined in the presence of various competitors. Control (black bar), proline analogues (open bars), other amino acid species (grey bars), GABA and quaternary ammonium compounds (cross-hatched bars). Data shows the average of four replications ±SE. 100% proline uptake rate was 24.0±0.8 pmol min–1 (mg FW)–1.
Fig. 8 Localization of HvProT mRNAs in salt-stressed barley roots by in situ hybridization. Experiments were carried out as described in Materials and Methods. Antisense (A, C, E) and sense (B, D, F) probes are labeled using DIG. Transverse sections are shown in A, B, E and F and longitudinal sections are shown in C and D. Signals are detected in root cap columella cells (A), cortex layer and stele (C) and phloem (E).
Comparison of amino acid sequence of HvProT with those of other plants
| Accession No. | Identity (%) | |
| OsProT | T50690 | 65.7 |
| AtProT1 | T50692 | 48.1 |
| AtProT2 | T47713 | 57.7 |
| LeProT1 | T50689 | 37.1 |
| LeProT2 | T50688 | 40.5 |
| LeProT3 | T50687 | 42.0 |
| Accession No. | Identity (%) | |
| OsProT | T50690 | 65.7 |
| AtProT1 | T50692 | 48.1 |
| AtProT2 | T47713 | 57.7 |
| LeProT1 | T50689 | 37.1 |
| LeProT2 | T50688 | 40.5 |
| LeProT3 | T50687 | 42.0 |
Os, Oryza sativa; At, Arabidopsis thaliana; Le, Lycoperison esculentum.
Comparison of amino acid sequence of HvProT with those of other plants
| Accession No. | Identity (%) | |
| OsProT | T50690 | 65.7 |
| AtProT1 | T50692 | 48.1 |
| AtProT2 | T47713 | 57.7 |
| LeProT1 | T50689 | 37.1 |
| LeProT2 | T50688 | 40.5 |
| LeProT3 | T50687 | 42.0 |
| Accession No. | Identity (%) | |
| OsProT | T50690 | 65.7 |
| AtProT1 | T50692 | 48.1 |
| AtProT2 | T47713 | 57.7 |
| LeProT1 | T50689 | 37.1 |
| LeProT2 | T50688 | 40.5 |
| LeProT3 | T50687 | 42.0 |
Os, Oryza sativa; At, Arabidopsis thaliana; Le, Lycoperison esculentum.
pH dependence of[14C]l-proline uptake rate in yeast mutant complemented with HvProT
| pH | [14C]l-proline uptake rate (pmol min–1 (mg FW)–1) | Percentage of pH 4.5 (%) |
| 4.5 | 20.4±0.8 | 100 |
| 5.5 | 4.2±1.1 | 21 |
| 6.5 | 1.6±0.1 | 8 |
| 4.5 + CCCP a | 0.3±0.2 | 1 |
| pH | [14C]l-proline uptake rate (pmol min–1 (mg FW)–1) | Percentage of pH 4.5 (%) |
| 4.5 | 20.4±0.8 | 100 |
| 5.5 | 4.2±1.1 | 21 |
| 6.5 | 1.6±0.1 | 8 |
| 4.5 + CCCP a | 0.3±0.2 | 1 |
Yeast was harvested in log phase (OD600 = 0.5–0.8) and resuspended in assay buffer to final OD600 = 3 (50 mM potassium phosphate, 0.6 M sorbitol, appropriate pH). Yeast was incubated at 30°C for 10 min with 50 mM galactose and uptake was started by adding same volume of [14C]l-proline (final concentration was 5 µM). Uptake was stopped at 30, 60 120 and 240 s. Data show the average of four replications ± SE.
a Final concentration of CCCP was 1 mM.
pH dependence of[14C]l-proline uptake rate in yeast mutant complemented with HvProT
| pH | [14C]l-proline uptake rate (pmol min–1 (mg FW)–1) | Percentage of pH 4.5 (%) |
| 4.5 | 20.4±0.8 | 100 |
| 5.5 | 4.2±1.1 | 21 |
| 6.5 | 1.6±0.1 | 8 |
| 4.5 + CCCP a | 0.3±0.2 | 1 |
| pH | [14C]l-proline uptake rate (pmol min–1 (mg FW)–1) | Percentage of pH 4.5 (%) |
| 4.5 | 20.4±0.8 | 100 |
| 5.5 | 4.2±1.1 | 21 |
| 6.5 | 1.6±0.1 | 8 |
| 4.5 + CCCP a | 0.3±0.2 | 1 |
Yeast was harvested in log phase (OD600 = 0.5–0.8) and resuspended in assay buffer to final OD600 = 3 (50 mM potassium phosphate, 0.6 M sorbitol, appropriate pH). Yeast was incubated at 30°C for 10 min with 50 mM galactose and uptake was started by adding same volume of [14C]l-proline (final concentration was 5 µM). Uptake was stopped at 30, 60 120 and 240 s. Data show the average of four replications ± SE.
a Final concentration of CCCP was 1 mM.
Abbreviations
- CCCP
carbonyl cyanide m-chlorophenyl-hydrazone
- DIG
digoxygenin
- GABA
γ-amino butyric acid
- P5CS
Δ1-pyrroline-5-carboxylate synthetase
- ProT
proline transporter.
The nucleotide sequence of HvProT has been submitted to DDBJ, EMBL and GenBank under accession number AB073084.
References
Breitkreuz, K.E., Shelp, B.J., Fischer, W.N., Schwacke, R. and Rentsch, D. (
Cachorro, P., Ortiz, A. and Cerda, A. (
Cairney, J., Booth, I.R. and Higgins, C.F. (
Cairney, J., Booth, I.R. and Higgins, C.F. (
Cairney, J., Higgins, C.F. and Booth, I.R. (
Delauney, A.J. and Verma, D.P. (
Delrot, S., Atanassova, R. and Maurousset, L. (
Deutch, C.E. and Winicov, I. (
Guerrier, G. (
Hanson, A.D. and Hintz, W.D. (
Hoagland, D.R. and Arnon, D.I. (
Igarashi, Y., Yoshiba, Y., Takeshita, T., Nomura, S., Otomo, J., Yamaguchi-Shinozaki, K. and Shinozaki, K. (
Ishitani, M., Arakawa, K., Mizuno, K., Kishitani, S. and Takabe, T. (
Kavi Kishor, P.B., Hong, Z., Miao, G.H., Hu, C.A.A. and Verma, D.P. (
Kiyosue, T., Yoshiba, Y., Yamaguchi-Shinozaki, K. and Shinozaki, K. (
Kyte, J. and Doolittle, R.F. (
Lasko, P.F. and Brandriss, M.C. (
Muramoto, Y., Watanabe, A., Nakamura, T. and Takabe, T. (
Nagy, F., Kay, S.A. and Chua, N.H. (
Nanjo, T., Kobayashi, M., Yoshiba, Y., Sanada, Y., Wada, K., Tsukaya, H., Kakubari, Y., Yamaguchi-Shinozaki, K. and Shinozaki, K. (
Rajendrakumar, C.S., Suryanarayana, T. and Reddy, A.R. (
Rentsch, D., Hirner, B., Schmelzer, E. and Frommer, W.B. (
Schobert, B. and Tschesche, H. (
Schwacke, R., Grallath, S., Breitkreuz, K.E., Stransky, E., Stransky, H., Frommer, W.B. and Rentsch, D. (
Sivakumar, P., Sharmila, P. and Saradhi, P.P. (
Smirnoff, N. and Cumbes, Q.J. (
Stalmach, M.E., Grothe, S. and Wood, J.M. (
Tattini, M., Lombardini, L. and Gucci, R. (
Ueda, A., Shi, W., Nakamura, T. and Takabe, T. (
Yoshiba, Y., Kiyosue, T., Katagiri, T., Ueda, H., Mizoguchi, T., Yamaguchi-Shinozaki, K., Wada, K., Harada, Y. and Shinozaki, K. (




![Fig. 7 Substrate specificity of HvProT using a yeast double mutant transformed with the HvProT. [14C]l-proline (5 µM) was used for the experiments. The uptake rate was determined in the presence of various competitors. Control (black bar), proline analogues (open bars), other amino acid species (grey bars), GABA and quaternary ammonium compounds (cross-hatched bars). Data shows the average of four replications ±SE. 100% proline uptake rate was 24.0±0.8 pmol min–1 (mg FW)–1.](https://oup.silverchair-cdn.com/oup/backfile/Content_public/Journal/pcp/42/11/10.1093_pcp_pce166/2/m_pce16607.gif?Expires=1716509665&Signature=kY~sQbcC8ojsl3c6X8tqK1s1f17yG2rgdlboAmd9Dpyyf5ETIBhePxbA7owk~75bvBjggpHd28-GJhDRFh50hFVH45EiMLHgn8Gz8VIwz9pGheauZpv6Gc55MrqDqHzt79~0FeIYaoPqPAAFI9EwsXnS6cDPqQLmZvoLRo530JY1~~a8QsQMvlLNFfcb-WgB3AHW9JAIOWim1alQYM48nosVk7KgXy07Muxp0dnXx1mS4aJJEOuKuJtgCjBUZTflX0UpiHvYHlSVMZzJTsK256bEu2dlkdaXjXHybL5zkOU25FhK0NTsg3H8eHzZqWKkRbuMOK2pnkaan4rSVU8lGA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)