-
PDF
- Split View
-
Views
-
Cite
Cite
Kenichi Tsuda, Toshihiro Tsuji, Susumu Hirose, Ken-ichi Yamazaki, Three Arabidopsis MBF1 Homologs with Distinct Expression Profiles Play Roles as Transcriptional Co-activators, Plant and Cell Physiology, Volume 45, Issue 2, 15 February 2004, Pages 225–231, https://doi.org/10.1093/pcp/pch017
Close - Share Icon Share
Abstract
Multiprotein bridging factor 1 (MBF1) is known to be a transcriptional co-activator that mediates transcriptional activation by bridging between an activator and a TATA-box binding protein (TBP). We demonstrated that expression of every three MBF1 from Arabidopsis partially rescues the yeast mbf1 mutant phenotype, indicating that all of them function as co-activators for GCN4-dependent transcriptional activation. We also report that each of their subtypes shows distinct tissue-specific expression patterns and responses to phytohormones. These observations suggest that even though they share a similar biochemical function, each MBF1 has distinct roles in various tissues and conditions.
(Received June 11, 2003; Accepted November 28, 2003)
Transcriptional regulation plays a major role in expression of the genomic information during complex biological processes. The effect of binding of transcription factors to cis-elements must be transmitted to RNA polymerase to ensure transcription initiation and maintain active transcription. In humans and yeast, it has been reported that transcriptional co-activators activate gene expression by connecting a transcription factor with components of the basal transcriptional machinery (Kwok et al. 1994, Ge and Roeder 1994, Knaus et al. 1996). However, the function of co-activators from plants has been poorly understood.
One co-activator, multiprotein bridging factor 1 (MBF1), was first purified from posterior silk gland extracts of the silkworm, Bombyx mori. MBF1s from insects stimulate transcription from the fushi tarazu promoter in vitro through its binding to the TATA-box binding protein (TBP) and a nuclear receptor FTZ-F1 (Li et al. 1994, Takemaru et al. 1997). MBF1 also mediates transcriptional activation by bridging between a basic region/leucine zipper (bZIP)-type transcriptional activator and TBP in yeast (Takemaru et al. 1998), human (Kabe et al. 1999) and Drosophila (Liu et al. 2003). In plants, it has been shown that ER24 gene, a tomato counterpart of MBF1 (LeMBF1), is immediately and transiently induced in ethylene-treated late immature fruit (Zegzouti et al. 1999). Transcription of potato MBF1 (StMBF1) is also up-regulated during fungal attack and upon wounding (Godoy et al. 2001). Furthermore, it has been reported that phosphorylation of StMBF1 is promoted by the treatment with hyphal cell wall components derived from Phytophthora infestans both in vitro and in vivo (Zanetti et al. 2003). Tobacco MBF1 (NtMBF1a) and two Arabidopsis MBF1s (AtMBF1a and AtMBF1b) have been shown to interact with the tomato mosaic virus movement protein (Matsushita et al. 2002). These observations suggest that, so far, all plant MBF1 homologs reported are involved in defense responses to pathogen.
In this study we tested whether MBF1s from Arabidopsis thaliana are functional for complementation of a yeast mutant lacking MBF1 function. We did the test because the function of plant MBF1 as a multiprotein bridging factor has never been demonstrated. We found three candidates of MBF1 homologs (AtMBF1a, AtMBF1b, and AtMBF1c) in a public database of the A. thaliana genome whose deduced amino acid sequences had considerable identities to those of MBF1s from other eukaryotes. So far existence of two subtypes of MBF1 has been reported only in humans. The two proteins share identical amino terminal regions (129 aa), but differ slightly in their carboxyl-terminal sequences (Kabe et al. 1999). Only one gene for MBF1 has been reported in all other eukaryotes. A. thaliana, which has three subtypes of MBF1 genes, is the first such example among all organisms. It is interesting whether each MBF1 gene has a distinct biological role. This study is also intended to reveal a tissue-specific expression pattern of each subtype of MBF1s in A. thaliana. We also tested effects of addition of phytohormones and precursor on expression of each gene in vivo.
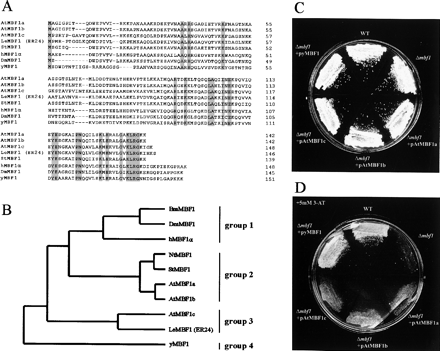
It has been shown that amino acid sequences of MBF1 proteins are widely conserved among eukaryotes (Takemaru et al. 1997). To characterize three subtypes of MBF1s from A. thaliana, the deduced amino acid sequences were compared with those of MBF1s from other eukaryotes (Fig. 1A) and a phylogenic tree was constructed with full-length amino acid sequences (Fig. 1B). All MBF1s are broadly classified into four groups (groups 1–4); plant MBF1s are classified into two groups (groups 2 and 3) among these four (Fig. 1B). The extent of amino acid sequence identity between AtMBF1a and AtMBF1b is 92%; these MBF1s belong to the same group (group 2) as StMBF1 reported by Godoy et al. (2001). Amino acid sequence identities of StMBF1 with AtMBF1a and AtMBF1b are 81% and 83%, respectively. AtMBF1c resides in the group 3 and shares 67% amino acid identities with LeMBF1 reported by Zegzouti et al. (1999), while it shows a lower degree of identity with that of AtMBF1a or AtMBF1b (50% or 53%, respectively).
It has been described that the yeast MBF1 (yMBF1) mediates the GCN4-dependent transcriptional activation of the HIS3 gene by bridging between GCN4 and TBP. Each yeast mutant lacking either MBF1 or GCN4 is reported to be viable, but sensitive to 3-aminotriazole (3-AT), an inhibitor of the HIS3 gene product (Hope and Struhl 1986, Takemaru et al. 1998). We tested the function of each Arabidopsis MBF1 as a co-activator by replacing yMBF1 with each subtype of AtMBF1 in the yeast mutant (Δmbf1 strain, KT131; Takemaru et al. 1998) in the presence of 3-AT in the histidine-free medium, which contains 1% galactose and 1% raffinose as carbon sources. While the Δmbf1 strain transformed with an empty vector, pYES2, (Invitrogen, Carlsbad, CA, U.S.A.) was sensitive to 3-AT, WT strain (KT130; Takemaru et al. 1998) and the Δmbf1 strains transformed with plasmid containing yeast MBF1 (pyMBF1) grew on a histidine-free medium in the presence of 3-AT (Fig. 1D). Expression of every AtMBF1 cDNA partially recovered the growth of the Δmbf1 strain upon histidine starvation (Fig. 1D) as in the case of Drosophila MBF1 (Liu et al. 2003), indicating that three AtMBF1s all play a role of co-activator for GCN4-dependent transcriptional activation in place of the yeast MBF1 in yeast cells. All yeast strains used in this experiment showed essentially the same normal growth rate on a histidine-free medium in the absence of 3-AT (Fig. 1C).
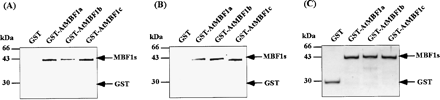
To demonstrate direct bindings of AtMBF1s to yeast GCN4 and yeast TBP (yTBP), we employed far-Western analysis. Fig. 2A shows direct bindings of all purified glutathione S-transferase (GST)-AtMBF1 fusion proteins to purified recombinant yTBP. Fig. 2B shows direct bindings of all purified GST-AtMBF1 fusion proteins to purified recombinant GCN4. GST polypeptide itself did not show any interaction to yTBP and GCN4 in both analyses as negative control.
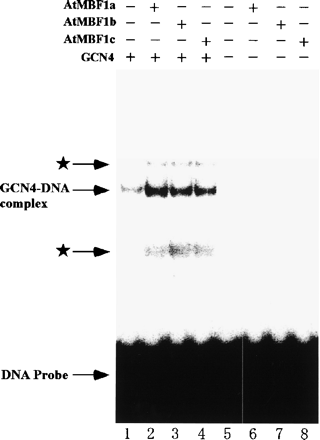
We also analyzed interaction between AtMBF1s and yeast GCN4 by electrophoresis mobility shift assay (EMSA). A significant elevation of DNA-binding activity of GCN4 to its target DNA sequence was observed when each purified recombinant AtMBF1 was added to the binding reaction mixtures (Fig. 3, lanes 1 and 2–4). But we have never detected any band corresponding to a ternary complex containing AtMBF1, GCN4, and its target DNA. Although we have detected two faint bands indicated by asterisks in Fig. 3, appearances of these bands were not dependent on AtMBF1 addition to the reaction mixture (data not shown). It has been reported that no interaction of GCN4 with yMBF1 occurred in the absence of Mg2+ and the complex that formed in the presence of Mg2+ dissociated immediately upon removal of Mg2+ (Takemaru et al. 1998). Therefore the ternary complex including AtMBF1 cannot be expected to be observed because of immediate dissociation of the GCN4-AtMBF1 complex in the electrophoresis buffer without Mg2+. We observed no significant difference in the effect of enhancement of DNA binding activity among three types of AtMBF1. Specificity of GCN4 binding to the labeled oligo DNA was confirmed by observing a reduction of GCN4-DNA complex formation by the addition of unlabeled oligo DNA as a competitor (data not shown). Similar elevation of bZIP type transcription factor binding to DNA fragment has been reported when yMBF1 (Takemaru et al. 1998) or hMBF1 (Kabe et al. 1999) was added to the binding reaction mixtures of EMSA. These observations suggest that contacts of AtMBF1s with the DNA-binding domain of GCN4 induce a conformational change in GCN4, allowing increased binding to its target DNA sequence. A database search by Jakoby et al. (2002) has revealed 75 distinct members of the bZIP family in Arabidopsis genome. Positions of four arginine residues in the basic region of the yeast GCN4 that are considered to be necessary for its binding to yMBF1 (Takemaru et al. 1998), are all conserved in the corresponding regions of some of the bZIP proteins from A. thaliana, including OBF4/TGA4, an ocs element binding protein (Buttner and Singh 1997, Zhang et al. 1993, Zhang et al. 1995) and ABI5, a factor involved in ABA signal transduction pathway (Carles et al. 2002, Lopez-Molina et al. 2001, Lopez-Molina et al. 2002, Finkelstein and Lynch 2000). These transcription factors will be possible candidates for partner of AtMBF1s in Arabidopsis.
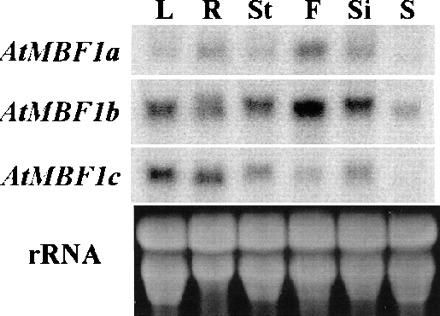
The mRNA expression pattern of each subtype of AtMBF1 was also investigated by Northern blot analysis using total RNA from different tissues of Arabidopsis plants (4–6 weeks old) or whole seedlings (8 d old). As shown in Fig. 4, we observed a similar expression pattern for AtMBF1a and AtMBF1b. While these two genes show relatively higher expression in flower, AtMBF1c shows lower expression in the same tissue. Expression of all AtMBF1 mRNAs was observed in almost all mature plant tissues. These observations suggest that similar promoters control expression of both AtMBF1a and AtMBF1b. However, the expression of AtMBF1c gene is controlled in a totally different manner from other two genes.
We tested whether mRNA levels of each AtMBF1 were affected by ethylene, salicylic acid (SA), or other phytohormones because it has been reported that the expression of MBF1 from tomato (Zegzouti et al. 1999) or potato (Godoy et al. 2001) are stimulated by ethylene or its precursor and SA, respectively. Ten-day-old seedlings were transferred into a liquid medium each containing phytohormones or precursor; then, mRNA accumulation of each AtMBF1 was determined by Northern blot analysis. As a control experiment, change in mRNA level of each gene after 24 h was investigated. mRNA level of AtMBF1c significantly increased in the presence of 0.1% ethanol (solvent for phytohormones) alone, while such prominent change was not observed in mRNA levels of AtMBF1a and AtMBF1b (Fig. 5, Control). This accumulation of AtMBF1c mRNA was also observed even in the absence of ethanol (data not shown). Since seedlings should be soaked into liquid medium during the phytohormone treatment, this increase is thought to be caused by physical stress or anaerobic stress. Several GT-motifs (AAACCA), which are known to be necessary for low oxygen induction of the Arabidopsis alcohol dehydrogenase gene (ADH1; Dolferus et al. 1994, Hoeren et al. 1998), were found in the promoter of AtMBF1c (positions –51 bp to –46 bp, –202 bp to –197 bp, –218 bp to –213 bp, and –662 bp to –657 bp relative to the transcription start site). No treatment tested in this study significantly changed accumulation of AtMBF1a and AtMBF1b mRNA (Fig. 5). In contrast, the AtMBF1c mRNA level significantly increased by treatment with ABA. Consistent with the observation, the AtMBF1c promoter contains a sequence similar to ABA-response element (ABRE) (Giraudat et al. 1994, Hobo et al. 1999, Shen and Ho 1995, Shen et al. 1996) at positions between –115 bp and –120 bp in the opposite orientation. However, up-regulation of AtMBF1c expression by ABA treatment may possibly be due to combinational effects of ABA, physical stress, and/or anaerobic stress. Further analyses to identify the effect of ABA are ongoing. Although the LeMBF1 gene has been reported to be induced after treatment with ethylene gas in fruits, roots and leaves of mature plants (Zegzouti et al. 1999) and StMBF1 is up-regulated following treatments with ethephon and SA in tubers (Godoy et al. 2001), mRNA levels of all AtMBF1s in 10-day-old seedlings were not significantly increased by these chemicals. In contradiction to previous observations, mRNA level of AtMBF1c was reduced by SA treatment (Fig. 5). However, we cannot exclude the possibility that response of promoters of each AtMBF1 to these chemicals may depend on type of tissue, growth stage, and experimental conditions.
In summary, the present study revealed that three MBF1s from Arabidopsis can mediate transcriptional activation by bridging between a bZIP-type transcriptional activator GCN4 and TBP in yeast. This result suggests that AtMBF1s may also function as a bridging factor between a bZIP-factor and TBP in A. thaliana. The distinct tissue-specific expression patterns and different responses of three AtMBF1s to phytohormones and stress suggest that each of them has a distinct biological role. Although no difference in the effect of GCN4 binding to DNA was observed, significant differences at the amino acid level between group 2 (containing AtMBF1a and AtMBF1b) and group 3 (containing AtMBF1c) imply that these AtMBF1s may have distinct interacting partners in A. thaliana. Studies on characterization of AtMBF1s binding to AtTBP and finding of a real partner of AtMBF1s in A. thaliana are in progress.
The Columbia ecotype of A. thaliana (L.) Heynh. was used for all experiments in this report. Seeds were sterilized with 1.5% (v/v) sodium hypochlorite and 0.02% Triton X-100 for 3 min with vigorous shaking, then washed five times with sterile water, and plated on half-strength MS medium (Murashige and Skoog 1962) solidified with 0.8% phytoagar (Wako Pure Chemical Inds., Osaka, Japan) and supplemented with half-strength Gamborg B5 vitamins (Gamborg et al. 1968) and 1% sucrose. After the plates were placed at 4°C for 3 d in the dark to break residual dormancy, plants were grown under continuous white light at 22°C. Seeds were sown on 1 : 1 (v/v) mixture of vermiculite and Metromix 250 (Hyponex, Marysville, OH, U.S.A.) irrigated with 0.1% Hyponex (Hyponex) in a pot and grown under continuous light at 22°C for analysis of tissue-specific expression patterns. Seedlings were grown on MS solid medium for 10 d for treatments with phytohormones and precursor.
Total RNA was isolated using TRIZOL® Reagent as described by the manufacturer (Invitrogen). For Northern blot analysis, total RNA was separated on a 1.2% agarose gel containing 2.2 M formaldehyde, then transferred to Hybond-N+ nylon membranes (Amersham Biosciences, Piscataway, NJ, U.S.A.). The RNA on membranes were hybridized with 32P-labeled DNA probes in 0.5 M Na2HPO4 (pH 7.2), 7% (w/v) SDS, and 1 mM EDTA at 60°C for 16 h. Each 32P-labeled gene specific DNA probe was produced from each corresponding PCR product using the Megaprime DNA Labelling System (Amersham Biosciences). Sequences of specific primers used for preparation of specific probes were: 5′-ACTGATGTAGCAAGTAACAAGAATC-3′ and 5′-CAACTATGTGATGAAAAGACCCAAG-3′ for AtMBF1a probe, 5′-AAGTGTAGAACAAAGCTCTTAAAGG-3′ and 5′-ATAATGACAAAAGGTTCCAAACAGC-3′ for AtMBF1b, 5′-TGTTCCTTTCTCTCAATTCATCGAC-3′ and 5′-CATTTATCAAACAAAACAACAAGAC-3′ for AtMBF1c. Membranes were washed once in 1× SSC containing 0.1% (w/v) SDS at room temperature for 10 min, then washed three times in 0.5× SSC containing 0.1% (w/v) SDS at 68°C for 10 min.
The three AtMBF1 and yMBF1 cDNAs were isolated from 4-week-old Arabidopsis plants and yeast WT strain KT130 by RT-PCR, then cDNA fragments were ligated into pGEM-T Vector (Promega, Madison, WI, U.S.A.), and their nucleotide sequences were analyzed. For preparing plasmid DNAs used in yeast transformation for yeast complementation test, the four DNA fragments corresponding to AtMBF1 and yMBF1 open reading frame (ORF) were amplified by PCR using three pairs of primers (for AtMBF1a, 5′-GGGAATTCAAGCTTATGGCCGGAATTGGAC-3′ and 5′-GGCTCGAGTCACTTCTTTCCACG-3′; AtMBF1b, 5′-GGGAATTCCATATGGCCGGAATTGGAC-3′ and 5′-GGCTCGAGCATATGCTACTTCTTTCCACG-3′; AtMBF1c, 5′-GGGAATTCCATATGCCGAGCAGATACC-3′ and 5′-GGCTCGAGCATATGTCATTTCCCAATTTT-3′; yMBF1, 5′-GGGAATTCCATATGTCTGACTGGGAT-3′ and 5′-GGCTCGAGCATATGTCATTTCTTCTTTGG-3′), then resultant DNA fragments were cloned into EcoRI and XhoI sites of pYES2 vector (Invitrogen). The plasmid DNAs containing AtMBF1 and yMBF1 ORFs were introduced into yeast Δmbf1 competent cells using a lithium acetate method (Ito et al. 1983).
For far-Western analysis we purified recombinant proteins. EcoRI–XhoI fragment (described above) containing each ORF of AtMBF1s were cloned into EcoRI and XhoI sites of pGEX-4T-1 vector (Amersham Biosciences). Proteins were overproduced in Escherichia coli strain BL21(DE3)pLysS as fusion proteins containing GST polypeptide in their amino terminus. GST and GST fusion proteins extracted from the cells were purified by Glutathione Sepharose® 4B (Amersham Biosciences) chromatography. ORF of GCN4 was cloned into NdeI site of 6His-pET15b (Novagen, Madison, WI, U.S.A.). This protein was overproduced in E. coli strain BL21(DE3)pLysS as fusion protein containing histidine tag in its amino terminus. Recombinant protein extracted from the cells was purified using Ni-affinity column (His-bind resin, Novagen). Histidine-tagged yTBP was produced as described previously (Takemaru et al. 1998).
For EMSA each ORF of GCN4 or AtMBF1s was cloned into NdeI site of 6His-pET15b. These proteins were overproduced in E. coli strain BL21(DE3)pLysS. Recombinant proteins extracted from the cells were purified using Ni-affinity column, and their N-terminal histidine tags were removed by digestion with thrombin (Novagen).
Acknowledgments
We thank Dr. Kenji Washio (Hokkaido University) and Dr. Kotaro T. Yamamoto (Hokkaido University) for their valuable discussions.
Corresponding author: E-mail, nahoken@ees.hokudai.ac.jp; Fax, +81-11-706-4522.
Fig. 1 Comparison of deduced amino acid sequences of three AtMBF1 subtypes with those of other MBF1s, and a functional complementation assay of AtMBF1s in yeast. (A) Deduced amino acid sequences of MBF1s were aligned using multiple alignment program of the Genetyx software (Genetyx, Tokyo, Japan). Identical amino acids among all MBF1s are shadowed. Dashes indicate gaps in the amino acid sequence introduced to optimize alignment. (B) Phylogenic tree of MBF1s. Deduced amino acid sequences of MBF1s were applied to create alignment with the multiple alignment program (Genetyx software); then, the dendrogram was created using clustering with the Unweighted Pair Group Method with Arithmetic Mean (UPGMA). Branch length indicates the relative evolutionary distances. The GenBank accession numbers of each nucleotide sequence of the MBF1 are AF370280 (AtMBF1a, A. thaliana), AF326909 (AtMBF1b, A. thaliana), AU235871 (AtMBF1c, A. thaliana), AF096246 (LeMBF1/ER24, L. esculentum), AB072698 (NtMBF1, N. tabacum), AF232062 (StMBF1, S. tuberosum), AB002282 (hMBF1α, H. sapiens), AB001078 (BmMBF1, B. mori), AB031273 (DmMBF1, D. melanogaster), and AB017593 (yMBF1, S. cerevisiae). (C and D) Rescue of the phenotype of yeast mutant lacking yMBF1 function by introduction of pAtMBF1a, pAtMBF1b, or pAtMBF1c. Cells of the yeast wild type (WT, KT130), the Δmbf1 strain (KT131) (Takemaru et al. 1998) transformed with the empty vector, and the Δmbf1 strain transformed with pAtMBF1a, pAtMBF1b, pAtMBF1c, or pyMBF1 were grown on a synthetic drop-out medium (SD-HIS-URA, 1% raffinose, 1% galactose, and 2% agar) in the absence (for 4 d) (C) or presence (for 6 d) (D) of 5 mM 3-AT at 30°C.
Fig. 2 Binding of GCN4 and yTBP to AtMBF1s. Binding of bacterially expressed and affinity-purified His-GCN4 and His-yTBP to GST-AtMBF1s were analyzed by far-Western analysis. (A and B) Equal amounts (2 µg) of affinity-purified GST-AtMBF1s and GST were separated on 12% polyacrylamide gel containing SDS and transferred onto a nitrocellulose membrane (Hybond-ECL; Amersham Biosciences) using a semi dry blotter (ATTO Bioscience, Tokyo, Japan). The blotted membrane (20 cm2) was incubated in TBST (20 mM Tris-HCl pH 7.5, 150 mM NaCl, and 0.1% Tween-20) containing 5% skim milk at room temperature for 1 h and then incubated in reaction mixture (3 ml) containing 20 mM HEPES-KOH (pH 8.0), 6 mM MgCl2, 60 mM KCl, 1 mM dithiothreitol, and 0.1% Nonidet P-40 in the presence of 30 µg of His-yTBP at 4°C for 8 h (A) or in the presence of 300 µg of His-GCN4 at 4°C for 16 h (B). The membrane was washed three times for 30 min in the same reaction mixture in the absence of protein at room temperature, and then His fusion proteins bound to AtMBF1 were fixed tightly for 25 min onto the membrane by capillary action driven by laminated paper towel fixed at the back side of the membrane in a blotting buffer containing 100 mM Tris-base, 192 mM glycine, and 20% methanol. The surface of the original membrane was protected by wrapping in one sheet of a new nitrocellulose membrane (Hybond-ECL) to prevent diffusion of interacting proteins from the original membrane. The original membrane was incubated in TBST containing 5% skim milk for 1 h at room temperature, and then His-yTBP and His-GCN4 bound to AtMBF1s were recognized by anti-His·Tag® Monoclonal antibody (Novagen). The membrane was incubated with substrate for alkaline phosphatase (ECF substrate; Amersham Biosciences) after further incubation with a second antibody, goat anti-mouse IgG alkaline phosphatase conjugate (Novagen), and His-yTBP and His-GCN4 were visualized by detecting chemiluminescence using fluorImager 595 (Molecular Dynamics, Sunnyvale, CA, U.S.A.). (C) Equal amount (2 µg) of affinity-purified GST-AtMBF1s and GST were separated on 12% polyacrylamide gel containing SDS and stained by Coomassie brilliant blue.
Fig. 3 Effects of addition of AtMBF1s on complex formation between GCN4 and its cis-element. Binding of bacterially expressed GCN4 to a 32P-labeled oligonucleotide carrying the GCN4 binding sequence of HIS3 gene promoter was analyzed by an EMSA. Twenty ng of purified recombinant GCN4 was incubated in reaction mixtures (10 µl) containing 12 mM HEPES-KOH (pH 8.0), 5 mM MgCl2, 60 mM KCl, 1 mM dithiothreitol, 10% glycerol, 250 ng of poly(dI-dC)/poly(dI-dC) (Amersham Biosciences), 2.5 µg of bovine serum albumin (BSA), and 1 ng of the 32P-labeled DNA probe in the presence (lanes 2–4 and 6–8) or absence (lanes 1 and 5) of 50 ng of purified recombinant AtMBF1s at 30°C for 30 min and separated at 100 V for 2.5 h on a 4% polyacrylamide gel in 0.5× Tris-borate-EDTA at room temperature. The DNA probe sequence was as described previously (Takemaru et al. 1998). The gel was exposed to a Fuji imaging plate (Fuji Photo Film, Tokyo, Japan) for 4 h and visualized using STORM 860 (Molecular Dynamics).
Fig. 4 Tissue-specific mRNA accumulation patterns of three AtMBF1s. Total RNA was isolated from different tissues of Arabidopsis wild-type plants grown on soil (L, R, St, F, and Si) and MS-agar medium (S). The accumulation of each AtMBF1 mRNA was determined by Northern blot analysis. Twenty-five µg of total RNA were used for each lane. Washed membranes were exposed to a Fuji imaging plate for 4 h and visualized using STORM 860. The ethidium bromide-stained rRNA of each sample is shown for loading control. L, leaves from 4-week-old plants; R, roots from 4-week-old plants; St, stems from 4-week-old plants; F, flowers from 4- to 6-week-old plants; Si, immature siliques from 4- to 6-week-old plants; S, 8-day-old seedlings grown on MS-agar medium. The patterns of mRNA accumulation are reproducible in three repeated experiments with different RNA preparations.
Fig. 5 Effects of addition of chemicals on mRNA accumulation of each AtMBF1. Ten-day-old Arabidopsis seedlings were placed in 50 mM sodium phosphate buffer (pH 7.0) that contained half-strength MS salt, half-strength Gamborg B5 vitamins, 1% sucrose, and a different phytohormone or precursor [100 µM 2,4-D (2,4-D), 100 µM 1-aminocyclopropane-1-carboxylic acid (ACC), 100 µM methyl jasmonate (MeJA), 2 mM salicylic acid (SA), or 100 µM ABA] for the time indicated above each lane. Ethanol 0.1% (v/v) was used as a solvent for the chemicals; it was also added to a control medium. Twenty µg of total RNA was used for each lane. Washed membranes were exposed to a Fuji imaging plate for 24 h and visualized using STORM 860. The ethidium bromide-stained rRNA is shown for loading control. The pattern of mRNA accumulation was reproducible in three experiments with different RNA preparations.
Abbreviations
- 3-AT
3-aminotriazole
- ABRE
ABA-response element
- bZIP
basic region/leucine zipper
- EMSA
electrophoresis mobility shift assay
- GST
glutathione S-transferase
- MBF1
multiprotein bridging factor 1
- ORF
open reading frame
- SA
salicylic acid
- TBP
TATA-box binding protein.
References
Buttner, M. and Singh, K.B. (
Carles, C., Bies-Etheve, N., Aspart, L., Leon-Kloosterziel, K.M., Koornneef, M., Echeverria, M. and Delseny, M. (
Dolferus, R., Jacobs, M., Peacock, W.J. and Dennis, E.S. (
Finkelstein, R.R. and Lynch, T.J. (
Gamborg, O.L., Miller, R.A. and Ojima, K. (
Ge, H. and Roeder, R.G. (
Giraudat, J., Parcy, F., Bertauche, N., Gosti, F., Leung, J., Morris, P.C., Bouvier-Durand, M. and Vartanian, N. (
Godoy, A.V., Zanetti, M.E., San Segundo, B. and Casalongue, C.A. (
Hobo, T., Asada, M., Kowyama, Y. and Hattori, T. (
Hoeren, F.U., Dolferus, R., Wu, Y., Peacock, W.J. and Dennis, E.S. (
Hope, I.A. and Struhl, K. (
Ito, H., Fukuda, Y., Murata, K. and Kimura, A. (
Jakoby, M., Weisshaar, B., Dröge-Laser, W., Vicente-Carbajosa, J., Tiedemann, J., Kroj, T. and Parcy, F. (
Kabe, Y., Goto, M., Shima, D., Imai, T., Wada, T., Morohashi, K., Shirakawa, M., Hirose, S. and Handa, H. (
Knaus, R., Pollock, R. and Guarente, L. (
Kwok, R.P., Lundblad, J.R., Chrivia, J.C., Richards, J.P., Bächinger, H.P., Brennan, R.G., Roberts, S.G., Green, M.R. and Goodman, R.H. (
Li, F.-Q., Ueda, H. and Hirose, S. (
Liu, Q.-X., Jindra, M., Ueda, H., Hiromi, Y. and Hirose, S. (
Lopez-Molina, L., Mongrand, S. and Chua, N.H. (
Lopez-Molina, L., Mongrand, S., McLachlin, D.T., Chait, B.T. and Chua, N.H. (
Matsushita, Y., Miyakawa, O., Deguchi, M., Nishiguchi, M. and Nyunoya, H. (
Murashige, T. and Skoog, F. (
Shen, Q. and Ho, T.H. (
Shen, Q., Zhang, P. and Ho, T.H. (
Takemaru, K., Harashima, S., Ueda, H. and Hirose, S. (
Takemaru, K., Li, F.-Q., Ueda H. and Hirose S. (
Zanetti, M.E., Blanco, F.A., Daleo, G.R. and Casalongue, C.A. (
Zegzouti, H., Jones, B., Frasse, P., Marty, C., Maitre, B., Latché, A., Pech, J.-C. and Bouzayen, M. (
Zhang, B., Chen, W., Foley, R.C., Buttner, M. and Singh, K.B. (



![Fig. 5 Effects of addition of chemicals on mRNA accumulation of each AtMBF1. Ten-day-old Arabidopsis seedlings were placed in 50 mM sodium phosphate buffer (pH 7.0) that contained half-strength MS salt, half-strength Gamborg B5 vitamins, 1% sucrose, and a different phytohormone or precursor [100 µM 2,4-D (2,4-D), 100 µM 1-aminocyclopropane-1-carboxylic acid (ACC), 100 µM methyl jasmonate (MeJA), 2 mM salicylic acid (SA), or 100 µM ABA] for the time indicated above each lane. Ethanol 0.1% (v/v) was used as a solvent for the chemicals; it was also added to a control medium. Twenty µg of total RNA was used for each lane. Washed membranes were exposed to a Fuji imaging plate for 24 h and visualized using STORM 860. The ethidium bromide-stained rRNA is shown for loading control. The pattern of mRNA accumulation was reproducible in three experiments with different RNA preparations.](https://oup.silverchair-cdn.com/oup/backfile/Content_public/Journal/pcp/45/2/10.1093_pcp_pch017/2/m_pch01705.gif?Expires=1716339229&Signature=uHvQ9vP41iOIpmDXjZadSSmR5XxPupfzPmkt75-GmCgtT3wn4GEMvqf2t2V9-KmH8zTHWG-FXhM5ZRpJ6aLMVvKXjUj0BBQvKK3AWwqR-i1eO0e0g3Qpe4GxQsfrG2TWV4HYALuY8NC-B4BfXvILMBRJMK0ov9bJ6op4kr4ocs5~cTgwnkBRNpSbX0eVn-Tef2Znz9eqN~5VrlwiDJmgnwwxKivYdCtQ08lACHToaa~ShZLZEHZyCcCg3QSBNCz9smY-LdOCiz3kD8rq-xQ4svRlF8QOrCuA-WyD793L8-XsKxage9ZLF3N03VTITuqJNkvmnYLBeWa7qAMciB8pdg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
