-
PDF
- Split View
-
Views
-
Cite
Cite
Min Xu, Ling Zhu, Huixia Shou, Ping Wu, A PIN1 Family Gene, OsPIN1, involved in Auxin-dependent Adventitious Root Emergence and Tillering in Rice, Plant and Cell Physiology, Volume 46, Issue 10, October 2005, Pages 1674–1681, https://doi.org/10.1093/pcp/pci183
Close - Share Icon Share
Abstract
Auxin transport affects a variety of important growth and developmental processes in plants, including the regulation of shoot and root branching. The asymmetrical localization of auxin influx and efflux carriers within the plasma membrane establishes the auxin gradient and facilitates its transport. REH1, a rice EIR1 (Arabidopsis ethylene insensitive root 1)-like gene, is a putative auxin efflux carrier. Phylogenetic analysis of 32 members of the PIN family, taken from across different species, showed that in terms of evolutionary relationship, OsPIN1 is closer to the PIN1 family than to the PIN2 family. It is, therefore, renamed as OsPIN1 in this study. OsPIN1 was expressed in the vascular tissues and root primordial in a manner similar to AtPIN1. Adventitious root emergence and development were significantly inhibited in the OsPIN1 RNA interference (RNAi) transgenic plants, which was similar to the phenotype of NPA (N-1-naphthylphalamic acid, an auxin-transport inhibitor)-treated wild-type plants. α-naphthylacetic acid (α-NAA) treatment was able to rescue the mutated phenotypes occurring in the RNAi plants. Overexpression or suppression of the OsPIN1 expression through a transgenic approach resulted in changes of tiller numbers and shoot/root ratio. Taken together, these data suggest that OsPIN1 plays an important role in auxin-dependent adventitious root emergence and tillering.
Introduction
The phytohormone auxin regulates a wide variety of developmental processes of plant growth. This includes changes in growth direction, in shoot and root branching, and in vascular differentiation (Laskowski et al. 1995, Leyser 2001, Paquette and Benfey 2001, Reinhardt et al. 2000). Auxin is thought to be synthesized in young apical tissues and then transported downward to the maturing stem and the roots by a polar transport system (Crozier et al. 2000). Auxin transportation accomplished by the auxin influx and efflux carrier proteins is strictly directional (Muday and DeLong 2001). It has been reported that the cell-to-cell polar auxin transport could be mediated via the asymmetric localization of auxin-efflux carriers (Friml and Palme 2002a). A number of auxin influx carriers, e.g. the AUX1 and LAX (AUX1-like) gene families, and efflux carriers, e.g the PIN-FORMED (PIN) family, have been isolated and characterized (Bennett et al. 1996, Friml et al. 2002b, Friml et al. 2002c, Gälweiler et al. 1998, Luschnig et al. 1998). The finding that influx and efflux carriers distribute within the plasma membrane asymmetrically provides direct evidence for the chemiosmotic model of auxin polar transport (i.e. auxin moves along concentration and electrochemical gradients; Gälweiler et al. 1998, Müller et al. 1998, Swarup et al. 2001).
An Arabidopsis mutant developed knitting needle-like apices when the gene encoding a putative auxin efflux carrier was interrupted. The mutant was therefore called pin-formed or simply pin1 (Okada et al. 1991). Several other PIN-FORMED genes have been recently identified in various plant species, including Arabidopsis, Brassica juncea, Medicago truncatula and the hybrid Populus (Ni et al. 2002, Schnabel and Frugoli 2004, Schrader et al. 2003). The Arabidopsispin mutants fail to establish endogenous auxin gradients and show developmental disorders in the shoot (Reinhardt et al. 2003), flower (Vernoux et al. 2000), root (Benková et al. 2003) and embryo (Weijers and Jürgens 2005). The pin1 mutation in Arabidopsis affects leaf formation and the development of the inflorescence. The triple pin mutant pin1 pin3 pin4 displayed defects in primary root development. The pin1 pin3 pin4 pin7 quadruple mutants showed strong defects in embryonic development (Weijers and Jürgens 2005).
In contrast with the primary root system in Arabidopsis, rice produces numerous adventitious roots that are essential for maintaining normal plant development. Application of the polar auxin transport inhibitor N-1-naphthylphalamic acid (NPA) in rice root collars (the junction between root and stem) blocked the initiation and growth of adventitious and lateral roots in rice. This suggested that polar auxin transport was critical for the initiation and elongation of adventitious roots (Zhou et al. 2003). Recently, an auxin responsive transcription factor, ARL1, the target of auxin response factor (ARF), was reported to regulate the initiation of adventitious root primordia (Inukai et al. 2005, Liu et al. 2005).
Apically derived auxin is known to inhibit shoot branching by inhibiting the activity of the axillary meristem in Arabidopsis (Chatfield et al. 2000). Recent studies indicate that the distribution of auxin can influence bud development in various stages, from having a relatively minor effect in bud initiation to a major but indirect effect on bud activity. At least two different secondary hormonal messengers have been identified in this auxin signalling pathway (Leyser 2003). Rice plants develop tillers from the axillary buds. The role of auxin in the development of axillary buds in Arabidopsis implies that auxin and its signalling may be the key regulator in the development of tillers.
Despite the connection between auxin signalling and the formation and development of adventitious roots and tillers in rice, the molecules involved in auxin polar transport have not yet been identified. EIR1/AGR1/AtPIN2, an Arabidopsis root-specific protein, has been shown to act as an efflux carrier (Chen et al. 1998, Luschnig et al. 1998, Müller et al. 1998, Utsuno et al. 1998). In this report, we explore the characterization and functional analysis of the rice homolog of Arabidopsis EIR1/AGR1/AtPIN2, OsPIN1 (previously named REH1, GenBank accession no. AF056027). The findings of the study will clarify the role of PIN dependent auxin polar transports in the initiation and development of rice adventitious roots and tillers.
Results
REH1/OsPIN1 encodes a PIN1 family protein
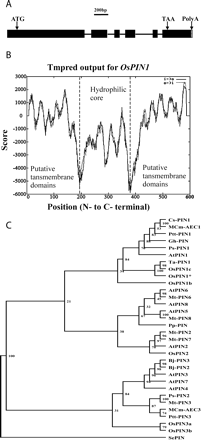
OsPIN1, previously named REH1, was proposed as an auxin transporter and a member of the PIN2 protein family (Luschnig et al. 1998). The full-length REH1 cDNA contains an open reading frame (ORF) of 1,785 nucleotides. The ORF encodes a protein of 595 amino acid residues with a calculated molecular mass of 65 kDa. Alignment between the cDNA and the genomic sequence of REH1 indicates that the REH1 gene has five introns (Fig. 1A). The deduced amino acid sequence of REH1 shows 69% sequence identity with Arabidopsis AtPIN1 (See supplemental data).
Hydropathy and transmembrane motif analyses of the deduced amino acid sequence revealed that the REH1 protein contains three characteristic regions, including a hydrophobic region with five transmembrane segments, a predominantly hydrophilic core, followed by another hydrophobic region with five transmembrane segments (Fig. 1B). This is consistent with the results of Luschnig et al. (1998). Alignment of members of the PIN1 family across different species revealed that the highly conserved portions of the family are uniformly located in the transmembrane domains (Fig. 1B). Phylogenetic analysis of 32 members of PIN family showed that REH1 was evolutionarily closer to the PIN1 family than to the PIN2 family (Fig. 1C). Based on the above result, we renamed REH1 as OsPIN1.
Expression pattern of OsPIN1
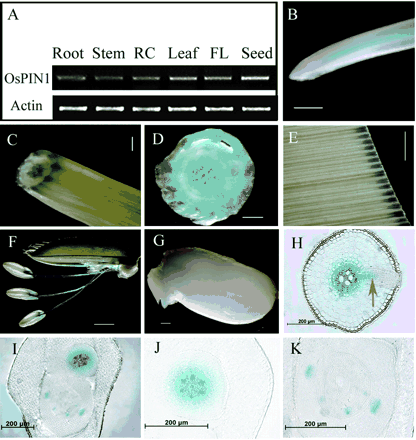
The expression of OsPIN1 in different tissues was analyzed by reverse transcriptase PCR (RT-PCR) and a β-glucuronidase (GUS) assay of transgenic plants expressing an OsPIN1 promoter/GUS fusion construct (Fig. 2). RT-PCR analysis showed that OsPIN1 mRNA was transcribed in all tissues tested. The expression of the gene was higher in leaf, flower and seed than in root, stem and root collar (Fig. 2A). To confirm the expression patterns, a binary vector containing the GUS gene driven by the OsPIN1 promoter was constructed and used for rice transformation. The transgenic plants showed GUS expression in the vascular tissues of root, stem, anther, leaf and embryo of the seed (Fig. 2B–G). Cross-sections showed that GUS is also expressed in the primordia of adventitious and lateral roots (Fig. 2D, H), suggesting that OsPIN1 may be involved in root development.
OsPIN1 is involved in polar auxin transport in rice
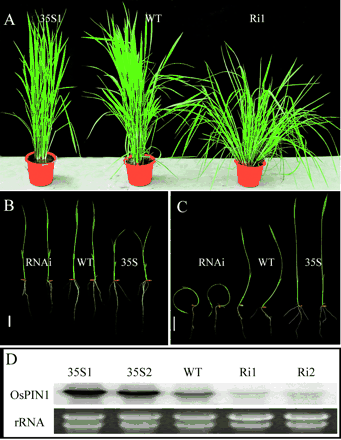
To investigate the function of OsPIN1 in rice, transgenic plants overexpressing or underexpressing OsPIN1 were produced using 35S-driven OsPIN1 and OsPIN1 RNA interfering (RNAi) constructs. Two transgenic lines were selected to represent the overexpressing or underexpressing groups. They were designated as 35S1 and 35S2, and RNAi1 and RNAi2, respectively (Fig. 3C). At the seedling stage, 14 days after germination, plant height, primary root length, adventitious root number and lateral root number on the primary root were investigated in wild-type and transgenic plants. The tiller number was counted at the maximum tillering stage (80 days after germination) under field conditions. The results showed that overexpression of the OsPIN1 gene significantly increased the primary root length and lateral root number, while underexpression of OsPIN1 resulted in a reduced number of adventitious roots and a significantly increased number of tillers (Table 1). Moreover, the tiller angles in the RNAi plants exceeded 30 degrees, which was much higher than the wild-type plants (Fig. 3A).
To verify whether OsPIN1 is involved in polar auxin transport, seedlings of the wild-type and transgenic plants were exposed to 0.1 µM α-naphthylacetic acid (α-NAA) and 0.5 µM NPA. The numbers of adventitious roots at the 7, 14 and 21-day-old seedling stages were recorded (Table 2). Results showed that the suppression of the OsPIN1 gene inhibited the formation of adventitious roots. Thus the number of adventitious roots in the OsPIN1 RNA interference (RNAi) transgenic plants was significantly less than that in wild-type plants (Table 2). NPA treatment had a similar effect on the wild-type plants (Table 2), suggesting that underexpression of the OsPIN1 gene may have affected the auxin transport process. Exogenously applied NAA on the RNAi transgenic plants partially complemented the inhibition effect of the underexpression of OsPIN1, confirming that OsPIN1 is involved in polar auxin transport.
Fig. 3B and C show that the expression level of OsPIN1 was inversely correlated with the sensitivity of the plants to the NPA treatment. In NPA treatment, the adventitious root numbers in transgenic plants overexpressing OsPIN1 (35S1and 35S2) were significantly higher than in wild-type seedlings (Table 2). This suggests that the higher level of OsPIN1 expression had partially overcome the inhibition effect of the NPA treatment on the formation of adventitious roots. On the other hand, the OsPIN1 RNAi plants were hypersensitive to the treatment of NPA. Application of NPA to the OsPIN1 RNAi transgenic seedlings resulted in these seedlings losing their apical dominance in shoot growth (Fig. 3B), and eventually dying after a 14-day treatment (Table 2). These results provided further evidence that OsPIN1 is involved in polar auxin transport.
OsPIN1 is involved in the emergence of adventitious root primordia
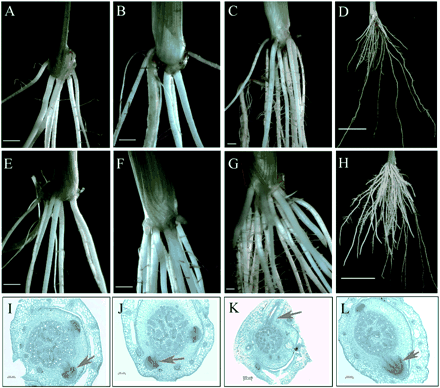
Five adventitious roots were formed at the coleoptile node of 7-day-old seedlings in both the RNAi transgenic and the wild-type plants (Fig. 4A, E). In the second week, numbers of adventitious roots developed from the first node of the wild-type plants, while no new adventitious roots were initiated until the third week in the RNAi transgenic plants (Fig. 4B, F). Moreover, the number of adventitious roots in the RNAi transgenic plants was significantly less than in the wild-type plants (Table 2, Fig. 4C, D, G, H).
To investigate whether the delayed development of the adventitious roots from the first node resulted from the arrest of primordia initiation or primordia emergence, transverse sections of the first node at different time points were examined. Results showed that adventitious root primordia were initiated in 7-day-old seedlings of both wild-type and OsPIN1 RNAi transgenic plants (Fig. 4 I, J), indicating that the formation of adventitious root primordia at the first node was not affected by the underexpression of OsPIN1. However, in the 14-day-old seedlings, new adventitious roots had developed in the wild-type plants (Fig. 4K), while they were not visible in the OsPIN1 RNAi transgenic plants. The adventitious root primordia in these RNAi plants stayed at the same size as those in the 7-day-old seedlings (Fig. 4L), indicating that the emergence of adventitious root primordia was arrested in these RNAi seedlings. Itoh et al. (2005) divided the process of adventitious root development into seven stages. Our data showed that the development of adventitious roots in RNAi seedlings ceased at the sixth stage.
Discussion
Phylogenetic analysis showed that the putative auxin efflux carrier, PIN1, is a highly conserved gene family, which may play a key role in polar auxin transport (Friml and Palme 2002a). The PIN1 family often contains a hydrophilic region interconnecting two blocks of transmembrane segments (Palme and Gälweiler 1999). The OsPIN1 protein contains all of these conserved transmembrane segments. It has been reported that the expression of AtPIN1 was localized at the basal side of auxin-transport-competent cells within root and shoot vascular tissues (Gälweiler et al. 1998) and narrowed down to provascular cells in germinated seeds (Palme and Gälweiler 1999). Our results showed an expression pattern in OsPIN1 similar to that in AtPIN1 (Fig. 2), which confirmed that OsPIN1 is evolutionarily closer to the PIN1 than the PIN2 family as previously reported (Luschnig et al. 1998).
That OsPIN1 acts as an auxin effux is confirmed by similar phenotypes in NPA-treated wild-type plants and OsPIN1 suppression transgenic plants. NPA is a well-characterized inhibitor of the polar auxin transport, which strongly inhibits auxin efflux (Luschnig 2001). NPA treatment of wild-type Arabidopsis plants phenocopied the mutant phenotype of pin1 in Arabidopsis (Vernoux et al. 2000). Application of an exogenous auxin to the naked inflorescence apex complemented the defects of flower development in NPA-treated plants or pin1 mutants (Reinhardt et al. 2000). In this study, the inhibition effect of OsPIN1 suppression on the development of adventitious root primordia was similar to that of NPA-treatment. Application of exogenous NAA rescued the defective phenotype in RNAi transgenic plants (Table 2). Our data supported that, acting like PIN1 in Arabidopsis, OsPIN1 is an auxin efflux carrier in rice.
Arabidopsis PIN1 efflux carriers were shown to express in a specific pattern in the shoot apical meristem. They are involved in the apical-basal axis formation of the embryo (Friml et al. 2003), development of lateral root primordia (Benková et al. 2003), and regulation of cell fate at the periphery of the shoot apical meristem (Vernoux et al. 2000). Adventitious roots and tillers are very important organs in rice. In this study, the transgenic plants that underexpress OsPIN1 display a significant decrease in adventitious root numbers. The decrease of adventitious root numbers in OsPIN1 RNAi plants was caused by the arrest of the primordia emergence. Our data support the hypothesis that auxin and its polar transport are critical for the development of adventitious roots (Zhou et al. 2003).
Unlike the ArabidopsisPIN1 family, there are three auxin efflux carriers in the rice PIN1 family (Fig. 1E), OsPIN1, OsPIN1b and OsPIN1c. Studies have shown that AtPIN1 is essential in the initiation and development of lateral root primordia in Arabidopsis (Benková et al. 2003). In the present study, suppression of OsPIN1 did not affect the development of lateral roots (Table 1). It is possible that the three OsPIN1 genes in rice are functionally complementary in the controlling of the development of later roots.
Studies in several species has shown that auxin does not act directly in the axillary bud, since apically applied IAA is not transported into inhibited buds (Hall and Hillman 1975, Morris 1977). It must, therefore, act elsewhere to control the synthesis, transport or metabolism of one or more of the secondary messengers, such as cytokinin and some yet unidentified substances (Ward and Leyser 2004). As a non-cell autonomous signal, auxin often interacts with other signalling pathways through its polar transport to regulate plant developmental processes. It has been reported that the endogenously applied auxin affects shoot branching (Rogg et al. 2001, Romano et al. 1991). Transgenic plants with a reduction of OsPIN1 gene expression display an increase in tiller number as well as till angle. The phenotype may result from the disturbance of auxin-mediated bud inhibition. Further research is needed to clarify the intrinsic reason. The difference in the regulation effect of OsPIN1 on the tiller promordia compared with the adventitious promordia may be caused by different effects of local auxin concentration in the different parts of the plants. More direct evidence, however, is needed to confirm this hypothesis.
Functional redundancy among PIN proteins has been found in Arabidopsis (Benková et al. 2003, Friml et al. 2003). For instance, the AtPIN1 protein localization was changed in Atpin3pin4pin7 triple mutants (Blilou et al. 2005). The highly conserved portions of PIN family proteins in the transmembrane domains, may be further evidence that they are, at least in part, functionally redundant. To understand the effect of OsPIN1 on the expression of other OsPIN genes, we analyzed the expressions of other putative auxin efflux carriers OsPIN1b, OsPIN1c, OsPIN2 and OsPIN3a in OsPIN1 RNAi transgenic plants. It was found that the expressions of these putative auxin efflux carrier genes were not altered by the underexpression of OsPIN1 (data not shown), indicating that these auxin efflux carriers may act independently.
In conclusion, our results proved that rice OsPIN1 acts as a polar auxin transport. The evidence for this conclusion came from the predicted OsPIN1 protein structure, the preferential expression of OsPIN1 in vascular tissues, and the alteration of the sensitivities to NAA and NPA in transgenic plants that over- or underexpressed the OsPIN1 gene. The finding that OsPIN1 is involved in auxin-dependent adventitious root emergence and tillering provides a new insight into the function of the PIN1 family in rice.
Materials and Methods
Structure and phylogenetic analysis
The hydropathy plot analysis was performed with Protscale (http://us.expasy.org/tools/protscale.html). The membrane-spanning regions and their orientation were predicted by the Tmpred method (http://www.ch.embnet.org/software/TMPRED_form.html). The phylogenetic tree was generated based on minimal evolution criterion using the neighbor-joining method with 100 bootstrap. SEPIN (gi:57636762) from Staphylococcus epidermidis was selected as the outgroup. Putative OsPIN1b (gi:32988390), OsPIN1c (gi:51535183), OsPIN2 (gi:52077371), OsPIN3a (gi:55297110) and OsPIN3b (gi:50932811) were identified through a TBLASTN search of Oryza sativa cDNA sequences in GenBank and named according to the phylogenetic analysis. AtPIN1 (gi:15219501), AtPIN2 (gi:42558886), AtPIN3 (gi:42558887), AtPIN4 (gi:42558871), AtPIN5 (gi:42558885), AtPIN6 (gi:42558888), AtPIN7 (gi:42558877) and AtPIN8 (gi:42558880) were from Arabidopsis thaliana (thale cress). Gh-PIN (gi:25140423), Ta-PIN1 (gi:42556526), Cs-PIN1 (gi:25956262), MCm-AEC1 (gi:33339148) and MCm-AEC3 (gi:33339152), Ptt-PIN1 (gi:10441744) and Ptt-PIN3 (gi:21435938), Ps-PIN1 (gi:28301753) and Ps-PIN2, Pp-PIN (gi:55859521), Bj-PIN2 (gi:15485153) and Bj-PIN3 (gi:15485155), Mt-PIN2 (gi:28204657), Mt-PIN3 (gi:25986775), Mt-PIN6 (gi:49035694), Mt-PIN7 (gi:49035696) and Mt-PIN8 (gi:58339253) were from Gossypium hirsutum (upland cotton), Triticum aestivum (bread wheat), Cucumis sativus (cucumber), Momordica charantia (balsam pear), Populus tremula x Populus tremuloides, (gi:39840930), Pisum sativum (pea), Physcomitrella patens, Brassica juncea and Medicago truncatula (barrel medic), respectively.
Construction of vectors and plant transformation
A 2.1 kb promoter region of the OsPIN1 gene was amplified by PCR using the upper primer 5′-TTTTAGGAGCACGCTTTAGCATTGTTCATCG-3′ and lower primer 5′-ATAGGTACCGGAGGAGGAGCATTTGATTGG-3′. The PCR product was then inserted upstream of the 5′ end of the GUS gene (gusA) in pCAMBIA1391Z to create a POsPIN1-GUS fusion construct vector. For the RNA interfering vector construct, a 219 bp cDNA fragment between positions 1136 and 1354 of the OsPIN1 cDNA, was amplified using the upper primer 5′-CGAAGGACAGGGAGGACTACGTGG-3′ and lower primer 5′-TGTTCGGGTTGCGGATGAG-3′. The second intron of NIR1 from maize was inserted between the sense and antisense cDNA fragments to form an intron-containing hairpin RNA. It was then inserted into pCAMBIA1301–35S. The resulting construct was named pOsPIN1-RNAi. The OsPIN1 cDNA was inserted into the binary plant vector pCAMBIA1301–35S. The resulting plasmid was named 35S::OsPIN1. The above constructs were transformed into a callus initiated from mature wild-type plant (Oryza sativa L ssp. Japonica, Nipponbare) seeds by Agrobacteriumtumefaciens (strain EHA105)-mediated transformation (Chen et al. 2003).
Histochemical analysis and GUS assay
Histochemical GUS analysis was performed as described (Jefferson et al. 1987). Transgenic plant samples were incubated with X-gluc buffer overnight at 37°C. The stained tissues were photographed using a LEICA MZ95 stereomicroscope with a color CCD camera (Leica Instrument, Nusslosh, Germany). After being stained, the tissues were rinsed and fixed in formalin acetic acid-alcohol (FAA) fixation solution at 4°C overnight, embedded in paraffin, and then sectioned. Sections of thickness, 8–10 µm were mounted on slides and photographed using a Zeiss Axiocam HRC digital color camera (Zeiss, Jena, Germany).
RT-PCR analysis
Total RNA from the roots, stems, root-collar, leaves, flowers, and immature seeds of wide-type plants were isolated using Trizol reagent (Invitrogen, Carlsbad, CA, USA) according the manufacturer’s instruction. The first-strand cDNA was synthesized by using Superscript II (Invitrogen, Carlsbad, CA, USA) and total RNA was used as the RT-PCR template. RT-PCR was performed using the gene-specific primers 5′-TCTCGCTCGGACATCTACTCCC-3′ and 5′-AGTCCTCCCTGTCCTTCGCTC-3′.
RNA gel blotting analysis
RNA gel blotting analysis was performed as described (Church and Gilbert 1984). Briefly, 20 µg of the total RNA isolated using Trizol was separated on 1% agarose gels containing 1× MOPS and 0.66 M formaldehyde, transferred onto a Hybond-N+ nylon membrane (Amersham Pharmacia, Piscataway, NJ, USA) and hybridized in a solution containing 0.5M sodium phosphate (PH 7.0), 1mM EDTA, 1% BSA, and 7% SDS for 20h at 65°C. The probe was prepared using a random priming method. After hybridization, the blot was washed twice with a solution containing 0.1× SSC and 0.1% SDS for 5 min at room temperature, followed by two washes of the same solution at 55°C for 20 min. Hybridization was performed with a [α-32P] dCTP-labeled probe by the random priming method (Sambrook et al. 1989). Hybridized membranes were scanned and analyzed with a Typhoon 1600 PhosphorImager scanner (Molecular Dynamics, Sunnyvale, CA USA).
Seedling growth conditions and hormone treatments
The germinated rice seeds were planted in a hydroponic culture containing a half-complete nutrient solution in the first week and a complete nutrient solution beyond that point. The hydroponic system was kept in the greenhouse with 12 h photoperiodic conditions and 28/24°C thermoperiods (12h/12h), and 80% humidity. The hydroponic solution contained 3.0 mM NH4NO3, 1.0 mM CaCl2, 0.32 mM NaH2PO4, 0.51 mM K2SO4, 1.65 mM MgSO4, 3.13 mM MnCl2, 1.52 mM (NH4)6Mo7O24, 1.5 mM H3BO3, 1.5 mM ZnSO4, 1.6 mM CuSO4, 35 mM FeCl3, and 70 mM C6H8O7. The pH of the solution was adjusted to 5.0. Wild-type and T2 transgenic plants were used for the analyses of the effects of exogenously applied α-NAA at 10–7 M and NPA at 5×10–7 M on adventitious root growth. The root collars of wild-type and OsPIN1 transgenic plants were photographed using a LEICA MZ95 digital camera. The lateral root number on the primary root was measured using used the scanner-based image-analysis system WinRhizo (Regent Instruments, Quebec, Canada).
Supplementary material
Supplementary material mentioned in the article is available to online subscribers at the journal website www.pcp.oupjournals.org.
Acknowledgements
The authors would like to thank Prof. Christopher Wood for critical reading of the manuscript. This study was supported by the National Basic Research Program (2005CB120900), Bureau of Science and Technology of Zhejiang Province in China, and the Natural Science Foundation of Zhejiang Province in China (M303053). The OsPIN1 cDNA was a kind gift from Dr. Christian Luschnig (Center of Applied Genetics, University of Agricultural Sciences Vienna, Austria).
Fig. 1 Gene structure and characterization of OsPIN1. (A) Gene structure of OsPIN1; bar = 200 bp. (B) The predicted membrane-spanning region and its orientation in the OsPIN1 protein using Tmpred. (C) Phylogenetic analysis of PIN families of Arabidopsis thaliana (At), Oryza sativa L. (Os), Gossypium hirsutum (Gh), Triticum aestivum (Ta), Cucumis sativus (Cs), Momordica charantia (MCm), Populus tremula x Populus tremuloides (Ptt), Pisum sativum (Ps), Physcomitrella patens (Pp), Brassica juncea (Bj) and Medicago truncatula (Mt). *, OsPIN1 gene studied. SEPIN (gi:57636762) from Staphylococcus epidermidis was selected as the outgroup. The phylogeny was constructed using the neighbor-joining method with 100 bootstrap replicates.
Fig. 2 OsPIN1 expression in different tissues. (A) Tissue-specific expression pattern revealed by RT-PCR. 28 and 25 running cycles for OsPIN1 and actin, respectively. RC, root collar, the junction between shoot and root. FL, flower. (B–K) GUS expression of different tissues in pOsPIN1-GUS transgenic plants. (B) Adventitious root. (C) Stem. (D) Hand cross-section of the root collar. The arrow points to a primordium of the adventitious root at the first node. (E) Leaf. (F) Flower. (G) Germinated seed. (H) Root cross-section of the segment arrow-pointed in Fig. 1B. Arrow here indicates GUS expression of young lateral root. (I) GUS staining of the germinating seeds. (J–K) Enlarged pictures of Fig. 1I. (B–G) Bar = 0.5 mm. (H–K) Bar = 200 µm.
Fig. 3 Phenotypes of OsPIN1 transgenic plants. (A) The 80-day-old (the maximum tillering stage) 35S-OsPIN1 (left), wild-type (WT) (middle) and RNAi (right) transgenic plants grown in the greenhouse. (B) and (C) WT, OsPIN1, RNAi event1 and event2, and 35S-OsPIN1 event 1 and event 2 were grown in rice nutrient solution (B) and the solution with 0.5 µM NPA (C) for 7 days. Bar = 2 cm. (C) Northern blot analysis for OsPIN1 in the WT, two transgenic lines overexpressing OsPIN1 (35S1 and 35S2) and two transgenic lines underexpressing OsPIN1 (Ri1 and Ri2).
Fig. 4 Adventitious roots in OsPIN1 transgenic plants. (A–D) Roots of the transgenic plants underexpressing OsPIN1 by RNA interference grown under solution culture for 7, 14, 21 and 50 days. (E–H) Roots of wild-type plants grown under solution culture for 7, 14, 21 and 50 days. (I–M) Transverse sections at the first node of the RNAi1 and wild-type (WT) plants sampled at 7 days old (WT, I; Ri, J) and 14 days old (WT, K; Ri, L). Red arrows point to the adventitious root primordial (I, J, L) or mature adventitious root (K).
Phenotypes of OsPIN1 transgenic and wild-type seedlings
| Genotypes | Shoot length (cm) | Primary root length (cm) | Shoot/root ratio | Number of adventitious roots | Number of lateral roots | Number of tillers |
| WT | 30.9 ± 1.5A | 10.2 ± 1.1C | 3.1 ± 0.3A | 13.6 ± 1.1A | 146.8 ± 9.8B | 24.0 ± 3.8C |
| RNAi-1 | 29.5 ± 1.1B | 10.1 ± 0.6C | 2.9 ± 0.2A | 5.7 ± 0.8B | 138.7 ± 10.4B | 46.2 ± 4.8A |
| RNAi-2 | 29.3 ± 1.0B | 9.9 ± 0.9C | 3.0 ± 0.3A | 5.9 ± 0.8B | 142.3 ± 9.6B | 40.9 ± 5.4B |
| 35S-1 | 21.7 ± 1.0C | 13.6 ± 1.1A | 1.6 ± 0.1B | 13.5 ± 1.5A | 163.0 ± 11.3A | 23.7 ± 4.2C |
| 35S-2 | 22.1 ± 1.2C | 12.9 ± 0.9B | 1.7 ± 0.1B | 13.5 ± 0.6A | 157.1 ± 10.1A | 23.9 ± 4.5C |
| Genotypes | Shoot length (cm) | Primary root length (cm) | Shoot/root ratio | Number of adventitious roots | Number of lateral roots | Number of tillers |
| WT | 30.9 ± 1.5A | 10.2 ± 1.1C | 3.1 ± 0.3A | 13.6 ± 1.1A | 146.8 ± 9.8B | 24.0 ± 3.8C |
| RNAi-1 | 29.5 ± 1.1B | 10.1 ± 0.6C | 2.9 ± 0.2A | 5.7 ± 0.8B | 138.7 ± 10.4B | 46.2 ± 4.8A |
| RNAi-2 | 29.3 ± 1.0B | 9.9 ± 0.9C | 3.0 ± 0.3A | 5.9 ± 0.8B | 142.3 ± 9.6B | 40.9 ± 5.4B |
| 35S-1 | 21.7 ± 1.0C | 13.6 ± 1.1A | 1.6 ± 0.1B | 13.5 ± 1.5A | 163.0 ± 11.3A | 23.7 ± 4.2C |
| 35S-2 | 22.1 ± 1.2C | 12.9 ± 0.9B | 1.7 ± 0.1B | 13.5 ± 0.6A | 157.1 ± 10.1A | 23.9 ± 4.5C |
Numbers are presented as mean ± SE. The number of observations in each mean is 20. Means in the same column followed by the same letter are not significantly different (P < 0.05, LSD test). Date was recorded from 14-d seedlings except tiller number, which was recorded at 80-d after germination.
Phenotypes of OsPIN1 transgenic and wild-type seedlings
| Genotypes | Shoot length (cm) | Primary root length (cm) | Shoot/root ratio | Number of adventitious roots | Number of lateral roots | Number of tillers |
| WT | 30.9 ± 1.5A | 10.2 ± 1.1C | 3.1 ± 0.3A | 13.6 ± 1.1A | 146.8 ± 9.8B | 24.0 ± 3.8C |
| RNAi-1 | 29.5 ± 1.1B | 10.1 ± 0.6C | 2.9 ± 0.2A | 5.7 ± 0.8B | 138.7 ± 10.4B | 46.2 ± 4.8A |
| RNAi-2 | 29.3 ± 1.0B | 9.9 ± 0.9C | 3.0 ± 0.3A | 5.9 ± 0.8B | 142.3 ± 9.6B | 40.9 ± 5.4B |
| 35S-1 | 21.7 ± 1.0C | 13.6 ± 1.1A | 1.6 ± 0.1B | 13.5 ± 1.5A | 163.0 ± 11.3A | 23.7 ± 4.2C |
| 35S-2 | 22.1 ± 1.2C | 12.9 ± 0.9B | 1.7 ± 0.1B | 13.5 ± 0.6A | 157.1 ± 10.1A | 23.9 ± 4.5C |
| Genotypes | Shoot length (cm) | Primary root length (cm) | Shoot/root ratio | Number of adventitious roots | Number of lateral roots | Number of tillers |
| WT | 30.9 ± 1.5A | 10.2 ± 1.1C | 3.1 ± 0.3A | 13.6 ± 1.1A | 146.8 ± 9.8B | 24.0 ± 3.8C |
| RNAi-1 | 29.5 ± 1.1B | 10.1 ± 0.6C | 2.9 ± 0.2A | 5.7 ± 0.8B | 138.7 ± 10.4B | 46.2 ± 4.8A |
| RNAi-2 | 29.3 ± 1.0B | 9.9 ± 0.9C | 3.0 ± 0.3A | 5.9 ± 0.8B | 142.3 ± 9.6B | 40.9 ± 5.4B |
| 35S-1 | 21.7 ± 1.0C | 13.6 ± 1.1A | 1.6 ± 0.1B | 13.5 ± 1.5A | 163.0 ± 11.3A | 23.7 ± 4.2C |
| 35S-2 | 22.1 ± 1.2C | 12.9 ± 0.9B | 1.7 ± 0.1B | 13.5 ± 0.6A | 157.1 ± 10.1A | 23.9 ± 4.5C |
Numbers are presented as mean ± SE. The number of observations in each mean is 20. Means in the same column followed by the same letter are not significantly different (P < 0.05, LSD test). Date was recorded from 14-d seedlings except tiller number, which was recorded at 80-d after germination.
Roots number of wild-type and OsPIN1 transgenic seedlings with NAA or NPA treatment
| Treatments | Genotypes | Number of adventitious roots a | ||
| 7-day-old seedlings | 14-day-old seedlings | 21-day-old seedlings | ||
| CK | WT | 5.2 ± 0.8BCD | 12.1 ± 1.1B | 22.4 ± 1.0B |
| RNAi-1 | 4.7 ± 0.8CD | 5.4 ± 0.8EF | 10.7 ± 1.5F | |
| RNAi-2 | 4.8 ± 0.8D | 5.5 ± 0.8EF | 11.1 ± 1.4F | |
| 35S-1 | 5.1 ± 0.6BCD | 12.0 ± 1.5B | 22.1 ± 1.8BC | |
| 35S-2 | 5.1 ± 1.0BCD | 11.5 ± 1.7BC | 22.2 ± 1.7BC | |
| NAA b | WT | 6.9 ± 1.1A | 14.3 ± 1.1A | 24.6 ± 1.1A |
| RNAi-1 | 5.1 ± 1.1BCD | 11.8 ± 1.1BC | 21.4 ± 1.1CD | |
| RNAi-2 | 4.9 ± 1.0CD | 11.1 ± 1.1C | 20.9 ± 0.9D | |
| 35S-1 | 7.2 ± 0.9A | 14.3 ± 1.5A | 24.7 ± 1.5A | |
| 35S-2 | 7.0 ± 0.8A | 14.1 ± 0.8A | 24.2 ± 1.4A | |
| NPA c | WT | 5.3 ± 0.6BC | 5.9 ± 1.0E | 8.7 ± 1.6G |
| RNAi-1 | 4.7 ± 0.9D | 4.7 ± 0.9G | NA d | |
| RNAi-2 | 4.8 ± 0.7CD | 4.8 ± 0.7FG | NA | |
| 35S-1 | 5.2 ± 1.1BCD | 7.9 ± 1.4D | 12.9 ± 1.6E | |
| 35S-2 | 5.5 ± 0.8B | 7.3 ± 1.1D | 12.2 ± 1.7E |
| Treatments | Genotypes | Number of adventitious roots a | ||
| 7-day-old seedlings | 14-day-old seedlings | 21-day-old seedlings | ||
| CK | WT | 5.2 ± 0.8BCD | 12.1 ± 1.1B | 22.4 ± 1.0B |
| RNAi-1 | 4.7 ± 0.8CD | 5.4 ± 0.8EF | 10.7 ± 1.5F | |
| RNAi-2 | 4.8 ± 0.8D | 5.5 ± 0.8EF | 11.1 ± 1.4F | |
| 35S-1 | 5.1 ± 0.6BCD | 12.0 ± 1.5B | 22.1 ± 1.8BC | |
| 35S-2 | 5.1 ± 1.0BCD | 11.5 ± 1.7BC | 22.2 ± 1.7BC | |
| NAA b | WT | 6.9 ± 1.1A | 14.3 ± 1.1A | 24.6 ± 1.1A |
| RNAi-1 | 5.1 ± 1.1BCD | 11.8 ± 1.1BC | 21.4 ± 1.1CD | |
| RNAi-2 | 4.9 ± 1.0CD | 11.1 ± 1.1C | 20.9 ± 0.9D | |
| 35S-1 | 7.2 ± 0.9A | 14.3 ± 1.5A | 24.7 ± 1.5A | |
| 35S-2 | 7.0 ± 0.8A | 14.1 ± 0.8A | 24.2 ± 1.4A | |
| NPA c | WT | 5.3 ± 0.6BC | 5.9 ± 1.0E | 8.7 ± 1.6G |
| RNAi-1 | 4.7 ± 0.9D | 4.7 ± 0.9G | NA d | |
| RNAi-2 | 4.8 ± 0.7CD | 4.8 ± 0.7FG | NA | |
| 35S-1 | 5.2 ± 1.1BCD | 7.9 ± 1.4D | 12.9 ± 1.6E | |
| 35S-2 | 5.5 ± 0.8B | 7.3 ± 1.1D | 12.2 ± 1.7E |
Means in the same column followed by the same letter arenot significantly different (P < 0.05, LSD test).
a Numbers are presented as mean ± SE. The number of observations in each mean is 20.
b The seedlings were grown under 0.1 µM NAA treatment after germination.
c The seedlings were grown under 0.5 µM NPA treatment after 7 d normal cultivation.
d NA, not available. Seedlings died after 21 d treatment.
Roots number of wild-type and OsPIN1 transgenic seedlings with NAA or NPA treatment
| Treatments | Genotypes | Number of adventitious roots a | ||
| 7-day-old seedlings | 14-day-old seedlings | 21-day-old seedlings | ||
| CK | WT | 5.2 ± 0.8BCD | 12.1 ± 1.1B | 22.4 ± 1.0B |
| RNAi-1 | 4.7 ± 0.8CD | 5.4 ± 0.8EF | 10.7 ± 1.5F | |
| RNAi-2 | 4.8 ± 0.8D | 5.5 ± 0.8EF | 11.1 ± 1.4F | |
| 35S-1 | 5.1 ± 0.6BCD | 12.0 ± 1.5B | 22.1 ± 1.8BC | |
| 35S-2 | 5.1 ± 1.0BCD | 11.5 ± 1.7BC | 22.2 ± 1.7BC | |
| NAA b | WT | 6.9 ± 1.1A | 14.3 ± 1.1A | 24.6 ± 1.1A |
| RNAi-1 | 5.1 ± 1.1BCD | 11.8 ± 1.1BC | 21.4 ± 1.1CD | |
| RNAi-2 | 4.9 ± 1.0CD | 11.1 ± 1.1C | 20.9 ± 0.9D | |
| 35S-1 | 7.2 ± 0.9A | 14.3 ± 1.5A | 24.7 ± 1.5A | |
| 35S-2 | 7.0 ± 0.8A | 14.1 ± 0.8A | 24.2 ± 1.4A | |
| NPA c | WT | 5.3 ± 0.6BC | 5.9 ± 1.0E | 8.7 ± 1.6G |
| RNAi-1 | 4.7 ± 0.9D | 4.7 ± 0.9G | NA d | |
| RNAi-2 | 4.8 ± 0.7CD | 4.8 ± 0.7FG | NA | |
| 35S-1 | 5.2 ± 1.1BCD | 7.9 ± 1.4D | 12.9 ± 1.6E | |
| 35S-2 | 5.5 ± 0.8B | 7.3 ± 1.1D | 12.2 ± 1.7E |
| Treatments | Genotypes | Number of adventitious roots a | ||
| 7-day-old seedlings | 14-day-old seedlings | 21-day-old seedlings | ||
| CK | WT | 5.2 ± 0.8BCD | 12.1 ± 1.1B | 22.4 ± 1.0B |
| RNAi-1 | 4.7 ± 0.8CD | 5.4 ± 0.8EF | 10.7 ± 1.5F | |
| RNAi-2 | 4.8 ± 0.8D | 5.5 ± 0.8EF | 11.1 ± 1.4F | |
| 35S-1 | 5.1 ± 0.6BCD | 12.0 ± 1.5B | 22.1 ± 1.8BC | |
| 35S-2 | 5.1 ± 1.0BCD | 11.5 ± 1.7BC | 22.2 ± 1.7BC | |
| NAA b | WT | 6.9 ± 1.1A | 14.3 ± 1.1A | 24.6 ± 1.1A |
| RNAi-1 | 5.1 ± 1.1BCD | 11.8 ± 1.1BC | 21.4 ± 1.1CD | |
| RNAi-2 | 4.9 ± 1.0CD | 11.1 ± 1.1C | 20.9 ± 0.9D | |
| 35S-1 | 7.2 ± 0.9A | 14.3 ± 1.5A | 24.7 ± 1.5A | |
| 35S-2 | 7.0 ± 0.8A | 14.1 ± 0.8A | 24.2 ± 1.4A | |
| NPA c | WT | 5.3 ± 0.6BC | 5.9 ± 1.0E | 8.7 ± 1.6G |
| RNAi-1 | 4.7 ± 0.9D | 4.7 ± 0.9G | NA d | |
| RNAi-2 | 4.8 ± 0.7CD | 4.8 ± 0.7FG | NA | |
| 35S-1 | 5.2 ± 1.1BCD | 7.9 ± 1.4D | 12.9 ± 1.6E | |
| 35S-2 | 5.5 ± 0.8B | 7.3 ± 1.1D | 12.2 ± 1.7E |
Means in the same column followed by the same letter arenot significantly different (P < 0.05, LSD test).
a Numbers are presented as mean ± SE. The number of observations in each mean is 20.
b The seedlings were grown under 0.1 µM NAA treatment after germination.
c The seedlings were grown under 0.5 µM NPA treatment after 7 d normal cultivation.
d NA, not available. Seedlings died after 21 d treatment.
Abbreviations
- ARF
auxin response factor
- FAA
formalin acetic acid-alcohol
- GUS
β-glucuronidase
- PIN
pin-formed
- ORF
open reading frame
- NAA
α-naphthylacetic acid
- NPA
N-1-naphthylphalamic acid
- RNAi
RNA interference
References
Benková, E., Michniewicz, M., Sauer, M., Teichmann, T., Seifertová, D., Jürgens, G. and Friml, J. (
Bennett, M.J., Marchant, A., Green, H.G., May, S.T., Ward, S.P., Millner, P.A., Walker, A.R., Schulz, B. and Feldmann, K.A. (
Blilou, I., Xu, J., Wildwater, M., Willemsen, V., Paponov, I., Friml, J., Heidstra, R., Aida, M., Palme, K. and Scheres, B. (
Chatfield, S.P., Stirnberg, P., Forde, B.G. and Leyser, O. (
Chen, R., Hilson, P., Sedbrook, J., Rosen, E., Caspar, T. and Masson, P.H. (
Chen, S.Y., Jin, W.Z., Wang, M.Y., Zhang, F., Zhou, J., Jia, Q.J., Wu, Y.R., Liu, F.Y. and Wu, P. (
Crozier, A., Kamiya, Y., Bishop, G. and Yokota, T. (
Friml, J. and Palme, K. (
Friml, J., Wisniewska, J., Benková, E., Mendgen, K. and Palme, K. (
Friml, J., Benková, E., Blilou, I., Wisniewska, J., Hamann, T., Ljung, K., Woody, S., Sandberg, G., Scheres, B., Jürgens, G. and Palme, K. (
Friml, J., Vieten, A., Sauer, M., Weijers, D., Schwarz, H., Hamann, T., Offringa, R. and Jürgens, G. (
Gälweiler, L., Guan, C., Müller, A., Wisman, E., Mendgen, K., Yephremov, A. and Palme. K. (
Hall, S.M. and Hillman, J.R. (
Inukai, Y., Sakamoto, T., Ueguchi-Tanaka, M., Shibata, Y., Gomi, K., Umemura, I., Hasegawa, Y., Ashikari, M., Kitano, H. and Matsuoka, M. (
Itoh, J., Nonomura, K., Ikeda, K., Yamaki, S., Inukai, Y., Yamagishi, H., Kitano, H. and Nagato, Y. (
Jefferson, R.A., Kavanagh, T.A. and Bevan, M.W. (
Laskowski, M.J., Williams, M.E., Nusbaum, C. and Sussex, I.M. (
Leyser, O. (
Liu, H.J., Wang, S.F., Yu, X.B., Yu, J., He, X.W., Zhang, S.L., Shou, H.X. and Wu, P. (
Luschnig, C., Gaxiola, R., Grisafi, P. and Fink, G. (
Morris, D.A. (
Muday, G.K. and DeLong, A. (
Müller, A., Guan, C., Gälweiler, L., Tänzler, P., Huijser, P., Marchant, A., Parry, G., Bennett, M., Wisman, E. and Palme, K. (
Ni, W.M., Chen, X.Y., Xu, Z.H. and Xue, H.W. (
Okada, K., Ueda, J., Komaki, M.K., Bell, C.J. and Shimura, Y. (
Palme, K. and Gälweiler, L. (
Paquette, A.J. and Benfey, P.N. (
Reinhardt, D., Mandel, T., and Kuhlemeier, C. (
Reinhardt, D., Pesce, E.R., Stieger, P., Mandel, T., Baltensberger, K., Bennett, M., Traas, J., Friml, J. and Kuhlemeier, C. (
Rogg, L.E., Lasswell, J. and Bartel, B. (
Romano, C.P., Hein, M.B. and Klee, H.J. (
Sambrook, J., Fritsch, E.T. and Maniatis, T. (
Schnabel, E.L. and Frugoli, J. (
Schrader, J., Baba, K., May, S.T., Palme, K., Bennett, M., Bhalerao, R.P. and Sandberg, G. (
Swarup, R., Friml, J., Marchant, A., Ljung, K., Sandberg, G., Palme, K. and Bennett, M. (
Utsuno, K., Shikanai, T., Yamada, Y. and Hashimoto, T. (
Vernoux, T., Kronenberger, J., Grandjean, O., Laufs, P. and Traas, J. (
Weijers, D. and Jürgens, G. (