-
PDF
- Split View
-
Views
-
Cite
Cite
Céline Diaz, Vera Saliba-Colombani, Olivier Loudet, Pierre Belluomo, Laurence Moreau, Françoise Daniel-Vedele, Jean-François Morot-Gaudry, Céline Masclaux-Daubresse, Leaf Yellowing and Anthocyanin Accumulation are Two Genetically Independent Strategies in Response to Nitrogen Limitation in Arabidopsis thaliana, Plant and Cell Physiology, Volume 47, Issue 1, January 2006, Pages 74–83, https://doi.org/10.1093/pcp/pci225
Close - Share Icon Share
Abstract
For the first time in Arabidopsis thaliana, this work proposes the identification of quantitative trait loci (QTLs) associated with leaf senescence and stress response symptoms such as yellowing and anthocyanin-associated redness. When Arabidopsis plants were cultivated under low nitrogen conditions, we observed that both yellowing of the old leaves of the rosette and whole rosette redness were promoted. Leaf yellowing is a senescence symptom related to chlorophyll breakdown. Redness is a symptom of anthocyanin accumulation related to whole plant ageing and nutrient limitation. In this work, Arabidopsis is used as a model system to dissect the genetic variation of these parameters by QTL mapping in the 415 recombinant inbred lines of the Bay-0×Shahdara population. Fifteen new QTLs and two epistatic interactions were described in this study. The yellowing of the rosette, estimated by visual notation and image processing, was controlled by four and five QTLs, respectively. The visual estimation of redness allowed us to detect six QTLs among which the major one explained 33% of the total variation. Two main QTLs were confirmed in near-isogenic lines (heterogenous inbred family; HIF), thus confirming the relevance of the visual notation of these traits. Co-localizations between QTLs for leaf yellowing, redness and nitrogen use efficiency described in a previous publication indicate complex interconnected pathways involved in both nitrogen management and senescence- and stress-related processes. No co-localization between QTLs for leaf yellowing and redness has been found, suggesting that the two characters are genetically independent.
Introduction
The typical leaf senescence symptom is yellowing due to chlorophyll catabolism (Matile 1992). Another senescence-related symptom, encountered in autumn leaves, is redness. Anthocyanin accumulation in tissues leads to such a typical red-purple leaf phenotype. Anthocyanin accumulation, when it occurs in senescing leaves, usually precedes chlorophyll breakdown (Feild et al. 2001). The persistence of Chl when the photosynthetic capacity is affected increases the susceptibility to light-induced oxidative damage of leaf cells. A protective role for anthocyanins as ‘sunscreen’ and as scavengers for reactive oxygen has been suggested for young expanding leaves and for senescing leaves susceptible to light damage (Baker and Hardwick 1972, Lee et al. 1987, Noodén et al. 1996). Leaf redness through anthocyanin accumulation is then commonly considered as a stress response (Chalker-Scott 1999, Chalker-Scott and Scott 2004).
Nitrogen deficiency can also cause anthocyanin synthesis (Do and Cormier 1991, Rajendran et al. 1992, Bongue-Bartelsman and Phillips 1995, Noodén et al. 1996) and red-purple leaf phenotype, that are also partially shared by other nutrient stresses such as sulfur and phosphate starvation (Stewart et al. 2001, Nikiforova et al. 2004). Nitrogen is certainly the most important nutrient that plants have to capture from the soil solution because, after water, nitrogen is a major limiting factor for plant growth (Vitousek and Howarth 1991). There is a strong relationship between nitrogen supply and the rate of CO2 assimilation as shown in many studies (Smart 1994, and references herein), and nitrogen availability is also essential for leaf lifespan. Several publications report that nitrogen deficiency can also accelerate leaf yellowing and senescence (Smart 1994, Thomas and de Villier 1996, Pourtau et al. 2004, Kato et al. 2005). Indeed, deficiency in mobile elements such as nitrogen triggers the senescence of old leaves whereas younger leaves remain healthy, presumably because of nutrient mobilization from the older leaves (Smart 1994).
The approach commonly used with the aim to enter the network of leaf senescence and stress response processes is mutant analysis and transcriptomic approaches. Several mutants altered in leaf senescence phenotypes have been detected (Lim et al. 2003, Spano et al. 2003) and are under investigation. Because senescence is a multigenic and complex trait, quantitative genetics might be a good tool to understand the genetic bases of leaf senescence. Genetic mapping of quantitative trait loci (QTLs) conferring the stay-green phenotype (late senescing) has been described in Festulolium pratensis, and the single recessive nuclear allele sid for stay green was mapped (Thomas et al. 1997). QTL mapping was also used to analyse the genetic basis of leaf senescence in rice (Jiang et al. 2004), sorghum (Xu et al. 2000), wheat (Verma et al. 2004) and maize (Beavis et al. 1994).
Most QTL mapping experiments in Arabidopsis were performed using two recombinant inbred line (RIL) populations, namely Landsberg erecta (Ler)/Colombia and Ler/Cape Verde Island populations. A population issued from the cross between the two genetically distant accessions Bay-0 and Shahdara was recently described (Loudet et al. 2002). This population has been used to date to identify several loci related to water and anion contents (Loudet, Chaillou, Krapp et al. 2003), nitrogen use efficiency (Loudet, Chaillou, Merigout et al. 2003) and flowering date (Loudet et al. 2002).
The visual observation of the Bay-0×Shahdara RIL population revealed that, depending on the genotypes, strong leaf yellowing symptoms or typical anthocyanin-related redness can appear when plants are cultivated in low nitrogen condition. The study of the physiology of five RILs, that exhibited differential leaf yellowing phenotypes under low nitrogen nutrition, allowed us to show that the quantification of the level of the leaf yellowing through visual notations and imaging was well correlated with the expression of several other well-known leaf senescence marker such as SAG12 expression for example (Diaz et al. 2005). The aim of this work was to investigate the genetic basis of the leaf senescence and stress response process in Arabidopsis when plants are grown in low nitrogen nutrition. The visual notations and the same imaging tool as described in Diaz et al. (2005) were then used to quantify the leaf yellowing symptoms on the Bay-0×Shahdara RIL population. Stress symptom responses, such as reduced plant growth and redness of the rosette, have also been quantified using a visual notation scale to evaluate the redness phenotypes and imaging to estimate rosette area.
This work allowed us to find QTLs related to yellowing, redness and leaf area rating of the rosettes, and to analyze their genetic relationship. Moreover, data were obtained on the same plants used by Loudet, Chaillou, Merigout et al. (2003) that allowed us to investigate the co-localizations of QTLs and the correlations between these traits and traits related to nitrogen use efficiency.
Results
Phenotypic variation and heritability
In this study, we observed that under low nitrogen conditions, both leaf redness and leaf yellowing symptoms can be easily observed. The anthocyanin-related leaf redness was promoted with ageing on the whole rosette surface, whereas leaf yellowing was observed on old leaves (Fig. 1).
The values obtained for (i) yellowing through visual notation (YV; Fig. 1); (ii) yellowing through image processing (YP; see Materials and Methods); (iii) rosette area (leaf area rating (LAR), see Materials and Methods); and (iv) redness visual notation (RV; Fig. 1), are presented in Fig. 2. YV showed non-normal distribution, and data transformation did not succeed in improving the normality. The frequency distribution histogram of YV (Fig. 2A) showed that 74% of the lines ranged between 0 and 1, indicating that most of the lines did not present yellowing symptoms. Among the 26% of the lines that represented yellowing visual symptoms, we observed that 3% ranged between 3 and 4, therefore exhibiting high yellowing symptoms at 35 d after sowing. Bay-0 and Shahdara varied between 0 and 1. The YP values ranged between 0 and 32% (data not shown). A logarithmic scale was used to improve the normality of YP distribution (Fig. 2B). For both yellowing traits, we observed a transgressive segregation beyond the parental values, highly significant towards higher values (Table 1).
The RV trait notation values (on a scale from 0 to 6) showed non-normal distribution and transformation did not succeed in improving the normality either (Fig. 2C). Bay-0 and Shahdara showed opposite and nearly extreme phenotypes (Table 1 and Fig. 2C). Actually, a few lines (<10%) visually exhibited more extreme phenotypes than Bay-0.
LAR values presented a normal distribution (Fig. 2D). Bay-0 and Shahdara had nearly the same LAR value as the RIL mean value (Table 1).
Fisher’s F-values associated with the ‘genotype’ variance and the corresponding heritability value were calculated for YV, YP, LAR and RV (Table 1). The genotypic effect is highly significant for all traits (P(f) < 0.001) except for LAR (P(f) > 0.05). Heritabilities of about 50% were found for YV and RV, indicating that half of the total phenotypic variation observed for these traits is due to the genetic variation. YP was mainly controlled by environmental conditions since heritability was 36%.
Correlation among traits
Correlations between YP, YV, RV and LAR values and previously published data related to nitrogen use efficiency (Loudet, Chaillou, Merigout et al. 2003) and flowering time (Loudet et al. 2002) are presented in Table 2.
Rosette dry matter (DM, as g plant–1, Loudet, Chaillou, Merigout et al. 2003) and LAR were associated by a significant positive correlation, as shown by the Pearson’s correlation coefficient presented in Table 2. Positive correlations between YP and the amino acid content (AA, as nmol (mg DM)–1, Loudet, Chaillou, Merigout et al. 2003) or the total nitrogen content (NP, as %DM, Loudet, Chaillou, Merigout et al. 2003) of the rosette were found (Table 2), whereas correlations between YP and LAR or DM were negative. The positive link between YV and YP was consistent with the fact that correlations between YP and AA or NP were conserved for YV. RV and short day (SD) flowering time (Loudet et al. 2002) were associated by a significant positive correlation.
QTL mapping
The characteristics of the QTLs related to senescence (this work) or to nitrogen use efficiency (Loudet, Chaillou, Merigout et al. 2003) are described in Table 3. Results for QTLs presented here were obtained on raw data for YV and RV and on transformed data (log10) for YP.
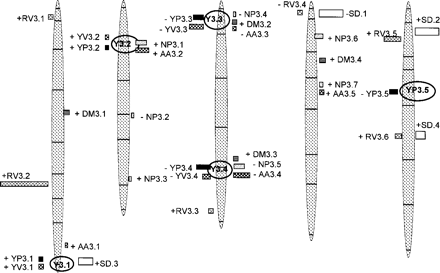
A total of nine putative QTLs and two epistatic interactions associated with leaf yellowing were detected using composite interval mapping and variance component analysis. Five QTLs showing R2 values from 2 to 12 were detected for the YP trait, while four QTLs showing R2 values from 3 to 11 were detected for the YV trait. The putative YP QTLs were located on chromosome I, II, III (two loci) and V. Interestingly, for the YV trait, the four QTLs detected co-localized with the YP QTLs found on chromosome I, II and III. No YV QTL was detected on chromosome V. For QTLs YP3.1, YP3.2, YV3.1 and YV3.2, the Bay-0 allele increased the yellowing whereas for the other QTLs YP3.3, YP3.4, YP3.5, YV3.3 and YV3.4, the Shahdara allele increased yellowing. The overlapping regions for YP3.1 and YV3.1, YP3.2 and YV3.2, YP3.3 and YV3.3, and YP3.4 and YV3.4 were named Y3.1, Y3.2, Y3.3 and Y3.4. We observed that the Y3.3 and Y3.4 loci also overlapped with some loci involved in the variation of nitrogen use efficiency (Loudet, Chaillou, Merigout et al. 2003) (Fig. 3; Table 3). These QTL regions named by Loudet, Chaillou, Merigout et al. (2003) L3 and L4 were associated with DM, NP and AA traits. We observed that Y3.2, Y3.3 and Y3.4 are always negatively associated with DM and positively associated with NP and AA. Overlapping between Y3.1 and the SD flowering time QTL, SD3 was observed (Fig. 3 and Table 3; Loudet et al. 2002).
A total of six putative QTLs associated with RV variation were detected (Table 3). The QTLs RV3.1, RV3.3 and RV3.4 localized on chromosomes I, III and IV, respectively, and RV3.5 and RV3.6 were located on chromosome V. These explained between 2 and 6% of the phenotypic variation. On chromosome I, RV3.2 explained 33% of the phenotypic variation. The Bay-0 alleles increased the visual redness, as expected from the means of the parents, for all the QTLs with one exception (RV3.4). No overlap between RV and YV or YP, and between RV and QTLs associated with nitrogen use efficiency was observed. On the other hand, RV3.4 and RV3.5, respectively, overlapped with the SD flowering time loci SD1 and SD2 described by Loudet et al. (2002) (Table 3 and Fig. 3).
Confirmation of QTLs using near-isogenic lines
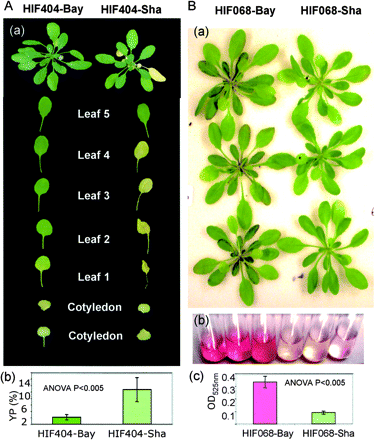
We used the HIF method to develop NILs segregating only for a QTL region (Tuinstra et al. 1997, Loudet et al. 2005). Significant trait variation between HIFs provides strong evidence supporting QTL position and effect. The QTLs YP3.4 and YV3.4 have been mapped between markers MSAT3.32 and MSAT3.21 on chromosome III. One set of NILs (HIF404-Bay/HIF404-Sha) was developed, showing the difference between the Shahdara and Bay-0 allele in the region surrounding these markers. Fig. 4A illustrates the phenotypes of these lines when grown on low nitrogen nutrition. A strong difference in yellow colour of the leaves was observed between the lines HIF404-Bay and HIF404-Sha, showing that yellowing of the old leaves of the HIF404-Bay lines was less pronounced than that of HIF404-Sha (Fig. 4A). This result was confirmed by measuring the YP value for both lines. As shown in Fig. 4A, HIF404-Bay was 5-fold less yellow than HIF404-Sha, confirming the negative effect of the YP3.4 Bay-0 allele on the expression of leaf yellowing and showing its especially strong effect in the genetic background of HIF404.
In the same way, the QTL RV3.2 for RV was confirmed. RV3.2 has been mapped between markers NGA128 and F5I14 on chromosome I. One set of NILs (HIF068-Bay/HIF068-Sha) was developed, showing the difference between the Shahdara and Bay-0 allele in the region surrounding these markers. The phenotype of these HIFs showed differences in the redness of the veins and the tips of the leaves (Fig. 4B(a)). Redness was more intense for HIF068-Bay than HIF068-Sha, and the measurement of anthocyanin contents in leaf extracts (Fig. 4B(b)) confirmed that HIF068-Bay contained significantly more anthocyanin than HIF068-Sha (Fig. 4B(c)).
Discussion
To investigate the senescence response of plants to low nitrogen nutrition, we performed QTL analysis on traits related to leaf yellowing and redness. Correlations between these traits and nitrogen use efficiency traits or flowering time were also studied, as well as QTL co-localization and candidate gene analysis.
QTL mapping
In order to dissect the genetic bases of the senescence response of leaves to low nitrogen nutrition, we decided to use the yellowing of the rosette as a Chl breakdown marker, and the redness of the rosette as an anthocyanin accumulation marker.
Using a visual notation scale for leaf yellowing quantification, the majority of the RILs were denoted 0, thus indicating that a large part of the population remained fully green at 35 d after sowing under nitrogen-limiting conditions. Quantification of yellowing by imaging permitted us to differentiate RILs presenting similar YV. Moreover, YP probably also partly measured another type of variation since a new QTL on chromosome V (YP3.5) was detected with image processing whereas it was not identified by using visual notation. Some QTLs for visual estimation had a smaller contribution (R2) than the smallest YP QTL, so we conclude that this is not a question of sensitivity but a question of normality of the distribution of data. Taking into consideration the possible co-localizations of QTL, confirmed by the sign of the respective allelic effects, we localized a total of five loci for leaf yellowing (Fig. 3). One of them, Y3.4, was confirmed using appropriate NIL comparison (HIF). We note that the percentage of phenotypic variation explained by the individual yellowing-related QTLs was no more than 12%. Plant growth traits most of the time are determined by numerous genes and very susceptible to an environmental effect. Due to this complexity, measuring growth traits often leads to the detection of relatively small effect loci, from which many might easily escape detection (Koornneef et al. 2004). The finding of numerous QTLs with a relatively small effect on yellowing emphasizes in the same manner the complexity of the senescence process and regulation (Buchanan-Wollaston et al. 2003). Two positive (Y3.1 and Y3.2) and three negative (Y3.3, Y3.4 and YP3.5) allelic effects were detected, the contribution of which explained that Bay-0 was less senescent than Shahdara.
Visual notations for the RV trait related to redness intensity allowed us to detect six QTLs. The major QTL RV3.2 found on chromosome I explained 33% of the total phenotypic variation and was confirmed when comparing redness as well as anthocyanin contents between HIF068-Bay and HIF068-Sha. The strong effect of RV3.2 on anthocyanin accumulation allows us to project fine-mapping in order to narrow down the position of the QTLs to a much smaller region in which candidate genes would be researched. The five other redness QTLs explained only 2–6% of the variability and remain to be confirmed.
One of the major issues with the QTL approach is derived from the interpretation of co-localization of QTLs related to different traits. Co-localization can theoretically be explained in two different ways (Lebreton et al. 1995), i.e. linkage (two different closely linked genes influence two different traits independently) and pleiotropy (the same genetic factor controls both traits). Then, co-localization between yellowing or redness QTLs and QTLs related to nitrogen use efficiency or flowering time described previously (Loudet et al. 2002, Loudet, Chaillou, Merigout et al. 2003) was investigated to search for evidence of the relationships between these traits.
Correlation and QTL co-localization between leaf yellowing, biomass and flowering traits
The results show a strong positive correlation between DM and LAR. In contrast to DM, the very weak heritability found for the LAR trait did not permit us to find any associated QTL. The fact that significant variation can be found for DM (Loudet, Chaillou, Merigout et al. 2003) but not for LAR with the same plants suggests that (i) image processing may be a less precise technique than measuring DM in this case; (ii) 300 plants were not enough to determine LAR variation; or (iii) other factors can interfere, such as variations in leaf shape or angle (that can interfere with light interception), variation in photosynthetic efficiency or variation in respiratory loss.
The negative links between YP and LAR or DM suggest that senescence would appear while both leaf expansion and leaf emergence have stopped. In Arabidopsis, we know that leaf emergence in the rosette stops when plants are flowering. Thus, this questions the link that could exist between leaf yellowing and flowering. In monocarpic plants, the reproductive structures often govern senescence. Arabidopsis is a monocarpic plant but there is some evidence against the control of leaf senescence by the reproductive structures (Hensel et al. 1993, Noodén et al. 1996, Noodén et al. 1997).
Since we have verified that the flowering data obtained in SD conditions, upon low (N–) and high (N+) nitrate nutrition, were highly correlated (data not shown), we explored the correlation factor existing between the SD flowering date in N+ (Loudet et al. 2002) and the YP or YV values measured in an N– environment. This allowed us to observe that no significant correlation can be detected between yellowing at 35 d and potential flowering time. This suggests that flowering did not control senescence onset.
Correlation and QTL co-localization between leaf yellowing and nitrogen use efficiency traits
Important QTLs for NP, AA and DM were detected under low nitrogen nutrition by Loudet, Chaillou, Merigout et al. (2003) and mapped especially at the three loci L2, L3 and L4. These loci co-localized with the Y3.2, Y3.3 and Y3.4 loci, respectively. A similar negative effect on NP, AA, YP and YV, and a positive influence on DM were found. Links between DM, NP, AA and YP or YV were also illustrated by the good correlation values presented in Table 2. It seems then that DM accumulation is most probably increased in plants with delayed senescence. The negative correlation between NP and DM that has been discussed by Loudet, Chaillou, Merigout et al. (2003) and attributed to a ‘nitrogen dilution’ process might also be partly controlled by leaf ontogeny (Greenwood et al. 1990, Justes et al. 1994, Plénet and Lemaire 2000).
The induction of nitrogen mobilization in leaves with ageing is well documented (Feller and Fischer 1994, Masclaux et al. 2000, Masclaux et al. 2001). Amino acids released from protein catabolism were considered as the major form of nitrogen transport and recycling in plants during senescence. Positive correlation between AA and yellowing would then suggest that the occurrence of senescence induces amino acid recycling and accumulation in the whole plant rosette. Then, some of the Y3.2, Y3.3 and Y3.4 loci could correspond to genes involved in interconnected pathways responsible for the senescence process and related to nitrogen management. The co-localizations of Y3.1 with the NIA1 gene, Y3.3 with GDH3, Y3.4 with ASN1 and YP3.5 with GS2 and GLN1;1 suggested a possible link between the glutamate dehydrogenase, glutamine synthetase, asparagine synthetase and nitrate reductase enzymes and senescence. Such links have already been proposed by physiological studies showing that nitrate reductase and chloroplastic glutamine synthetase are repressed during senescence whereas cytosolic glutamine synthetase and mitochondrial glutamate dehydrogenase are induced (Masclaux et al. 2000, Brugière et al. 2000, Guo et al. 2004). Interestingly, the glutamate dehydrogenase (NADH) activity measured in the older leaves of the rosettes positively correlated with yellowing in this genetic material, as revealed by Pearson’s and Spearmann’s tests (data not shown).
Correlation and QTL co-localization between anthocyanin-related redness and flowering traits
Whereas no co-localization between the RV loci and yellowing or nitrogen use efficiency-related QTLs was found, we observed interesting co-localizations of RV3.4 and SD1, RV3.5 and SD2, and RV3.6 and SD4 (Loudet et al. 2002). SD1 and SD2 have been hypothesized to correspond to the flowering time genes FRIGIDA and FLC, respectively. Such a finding suggests that the redness symptoms induced in low nitrogen conditions are not directly linked to nitrogen metabolism but might be linked to the flowering date and then to plant longevity. The allelic effect signs, however, would indicate that earlier flowering is associated with less anthocyaned plants. Unlike leaf yellowing, a significant correlation between redness and flowering time can be detected (Table 2). The weakness of this correlation is probably explained by the small contribution of RV3.4, RV3.5 and RV3.6 in the redness trait total variation.
The redness of the leaves is associated with anthocyanin accumulation, and a significant difference in this trait between HIF068-Bay and HIF068-Sha was shown, thus confirming the importance of the RV3.3 locus explaining 33% of redness total variation. The study of these HIFs also confirmed the positive effect of the Bay-0 allele on anthocyanin accumulation. An interesting finding is that the regulatory gene PAP1 is localized in the LOD-1 interval of RV3.2. Recently, it was shown that the two PAP1 and PAP2 anthocyanin regulatory genes are induced by nitrogen starvation (Scheible et al. 2004). This finding encourages further investigation in fine-mapping and tentative complementation of HIF068.
For the other minor effect RV loci, candidate gene analysis revealed that the CHI and F3H, CHS, DFR and AOS genes involved in anthocyanin synthesis were found in the LOD-1 intervals of RV3.3, RV3.5 and RV3.6, respectively. These QTLs, however, explained no more than 2–6% of the total variation, and their individual effects might be difficult to confirm.
Correlation and QTL co-localization between anthocyanin-related redness and leaf yellowing
The absence of coincidence between YV or YP and RV QTLs suggests that redness and yellowing are two genetically independent pathways. An alternative hypothesis could be that there is a part common to these two pathways, but it is not genetically variable in this material. The literature shows that both traits are related to plant ageing. The absence of links between the two characters studied herein could be explained by the fact that strong redness symptoms may have partially masked yellowing phenotypes, thus discriminating each trait separately. Since low nitrogen nutrition affects the photosynthetic rate by limiting the amount and/or activity of the photosynthetic apparatus, plants cultivated in low nitrogen conditions are more sensitive to the effects of light saturation associated with high incidence light levels. We propose that plants would respond to nitrogen limitation by (i) promoting Chl breakdown and inducing typical leaf senescence symptoms on their older leaves, thus inducing nitrogen remobilization for optimum nutrient allocation within the plant; or (ii) by accumulating anthocyanin that might have a protective effect from light and may influence leaf lifespan and, as a result, plant longevity. Since no co-localization between RV and YP or YV can be detected, it seems that plants would respond to nitrogen limitation promoting both process independently. We also noted many SAG (senescence-associated genes) genes localized in the LOD-1 intervals of both traits. For example, genes involved in the biosynthesis of stress- and senescence-related hormones such as jasmonic acid (OPR1-3) and ethylene (EIN4) co-localized with Y3.1, Y3.2 and Y3.3. Moreover, the gene ORE4 involved in senescence symptom expression is located within the Y3.1 region (Woo et al. 2002, Lim et al. 2003).
For the first time in Arabidopsis thaliana this work proposed the mapping of QTLs associated with leaf senescence and stress response symptoms such as yellowing and anthocyanin-associated redness. The co-localization existing between the QTLs for yellowing and nitrogen use efficiency suggests that some of the NP, AA and DM QTLs detected by Loudet, Chaillou, Merigout et al. (2003) might be a consequence of the leaf ontogeny that is also implicated in senescence and leaf yellowing symptoms. The search for QTLs explaining nitrogen mobilization or leaf senescence under high nitrogen conditions will probably cast light on this question and is in progress.
Materials and Methods
Plant material and experiments
The Bay-0×Shahdara RIL population has been fully described in a previous publication (Loudet et al. 2002) and on http://www.inra.fr/qtlat. F7 seeds obtained from the last generation of single seed descent for 415 lines were used. The production of homogeneous vegetative plant material for the 415 lines was performed in controlled conditions (growth chamber) as described previously (Loudet, Chaillou, Merigout et al. 2003). In summary, the experimental unit was a small pot (L = 60 mm, l = 65 mm, h = 60 mm) containing six plants positioned on a circle. With only one repetition per RIL (one pot, i.e. six plants) and 17 connecting controls (Bay-0 and Shahdara repetitions), the whole population studied represented 432 experimental units, organized in 18 blocks of 24 pots. The experiment was carried out in two consecutive culture cycles in the same growth chamber that represented two independent biological repeats (Loudet, Chaillou, Merigout et al. 2003). The RILs were completely and independently randomized in each cultivation repetition. Plants were cultivated under low nitrogen nutrition (3 mM nitrate) and in SDs as described previously (Loudet, Chaillou, Merigout et al. 2003).
NILs were developed as HIF (Tuinstra et al. 1997, Loudet et al. 2005). At the F6 stage, line RIL404 is homozygous everywhere on its genome, except for a small region including the 8 cM interval defined by the markers MSAT3.32 and MSAT3.21 on chromosome 3. We planted 30 F7 seeds from this line and genotyped the plants individually, selecting one plant fixed for the Bay-0 allele (named HIF404-Bay) and one other plant fixed for the Shahdara allele (named HIF404-Sha) at the two segregating markers. F8 seeds from these plants were then used for phenotyping. The same procedure was applied to RIL068, which is segregating for a small region including the 7 cM interval defined by markers NGA128 and F5I14 on chromosome I.
Phenotyping
The yellowing rate was measured visually and by image processing at 35 d after sowing, since at this time variation for senescing symptoms was high. Visual yellowing measurement was made on the whole population (415 RILs) and was designated as YV. A rating of 0 indicated a totally green rosette, while rating 4 corresponded to high yellowing symptoms. Ratings of 1, 2 and 3 indicated intermediate yellowing spreading throughout the rosette (Fig. 1A). Rosette yellowing was also determined using an image processing technique. Each pot was imaged using a Nikon digital camera (model Coolpix800; Nikon Corporation Imaging Products Division, Shinagawa-Ku, Tokyo, Japan). Digital pictures were analysed using an algorithm (Belluomo et al. 2003) running on the computer program Optimas (version 6.5; Media Cybernetics Inc, Silver Spring, MD, USA, http://www.mediacy.com). This program quantified, at first, the pixel number contained in the rosette shape. The ratio between this number and the total image pixel number reflected the rosette area. This ratio was considered as a photosynthetic area indicator and was designated as LAR. On rosette pictures, yellow and green area samples were selected and image processing permitted us to create a binary picture defining two classes containing green and yellow pixels, respectively. The percentage of yellow pixels was designated as YP. YP and LAR were measured, on two independent biological repeats, on a subset of the original population (300 RILs), because it was a time-consuming process.
Redness was considered as indicative of low nitrate susceptibility and anthocyanin accumulation. The visual redness of rosettes, designated as RV, was also measured on the whole population. A rating of 0 indicated no redness, while rating 6 corresponded to a complete red rosette. Scores from 1 to 5 measured intermediate symptoms (Fig. 1B).
Data for NP, DM and AA, used for comparison, were measured on the same plants and were published previously by Loudet, Chaillou, Merigout et al. (2003). Flowering dates were presented in Loudet et al. (2002). All published phenotypes can be found at http://www.inra.fr/qtlat/.
Statistical analysis and QTL mapping
Data were collected from the two independent replications, and mean values of each trait measurement were used for statistical analysis and QTL detection. RV and YV frequency distributions were measured on the whole F7 population (415 RILs), YP and LAR were measured on 300 RILs. The mean value of the YP trait was transformed (log10) to improve the normality of the distribution. The complete set of data was included in an analysis of variance (ANOVA) model to determine the specific effect of ‘genotype’ (i.e. the RIL). This ANOVA allowed the quantification of the broad-sense heritability (genetic variance/total phenotypic variance). Standard statistical procedures, such as ANOVA, frequency distributions and Pearson’s correlation coefficients were performed using the software XLSTAT-PRO version 6.1.8. The original set of markers (38 microsatellite markers) and the genetic map obtained with MAPMAKER 3.0, as previously described (Loudet et al. 2002; http://www.inra.fr/qtlat), were used in this study. All QTL analyses were performed using the Unix version of QTL Cartographer 1.14 (Basten et al. 1994, Basten et al. 2000). We used standard methods as previously described (Loudet, Chaillou, Merigout et al. 2003), and composite interval mapping (CIM) will be shown. LOD significance thresholds were estimated for all traits from permutation test analyses (1,000 permutations, overall error level: 5%), as suggested by Churchill and Doerge (1994). The additive effects of detected QTLs were estimated from CIM results and represented the mean effect of the replacement of both Shahdara alleles by Bay-0 alleles at the studied locus. Thus, a QTL with a positive effect means that the Bay-0 allele increases the trait value. The contribution of each identified QTL to the total phenotypic variance (R2) was estimated by variance component analysis.
QTL analysis of DM, NP, AA and SD flowering time has been published previously for this population (Loudet et al. 2002, Loudet, Chaillou, Merigout et al. 2003).
Candidate genes for senescence-associated yellowing, anthocyanin accumulation and nitrogen nutrition
Candidate genes at the QTLs for yellowing and redness were screened in silico by comparing the physical locations of the QTLs (inferred from their estimated genetic position and the physical position of the adjacent markers) with the position of genes that are known to be involved in leaf senescence, Chl degradation or synthesis, anthocyanin synthesis or nitrate management. Moreover, the putative co-localization of the senescence-enhanced genes previously described (Guo et al. 2004) and our QTLs was analysed. Candidate genes were identified when their position coincided with the QTL interval.
Anthocyanin extraction and OD measurement on HIF
To estimate anthocyanin concentration, an entire rosette was ground in liquid nitrogen before the powder was mixed with 1.5 ml of 80% methanol in water (v/v) containing 0.01 M HCl and incubated for 24 h at 4°C in darkness. After centrifugation (13,000×g, 10 min), the remaining Chl was eliminated from the supernatant by addition of 100 µl of chloroform. Anthocyanin content was then estimated by measuring the absorbance of the methanolic phase at 525 nm using a spectrophotometer.
Acknowledgments
The authors thank Joël Talbotec for technical assistance, Dr. Alain Charcosset (INRA, Moulon, France) for his help with QTL mapping, and Dr. Guy Fouillou (INRA Versailles, France) for his help with other statistical analysis. We thank the CETIOM (Centre Technique Interprofessionnel des Oléagineux Metropolitains; http://www.cetiom.fr) and INRA for providing financial support for the thesis of C.D. V.S.-C. was supported by NATURAL (European Community Project # QLRT-2000-01097).
Fig. 1 Pictures of the five classes representing the gradation of visual yellowing symptoms (A) and of the six classes for redness visual symptoms (B; grades 3 and 5 are not represented) as observed in the Bay-0×Shahdara RIL population. In the YV and RV scales, 0 is less yellow/red and higher values are more yellow/red.
Fig. 2 Frequency distribution of traits in the Bay-0×Shahdara RIL population. Senescence traits: rosette yellowing estimated using visual notation (A) or image processing (log10(YP) presented) (B). Stress-related trait: redness visual estimation (C). Growth trait measured as leaf area rating (D). ‘B’ and ‘S’ arrow positions indicate the average values obtained for parental accessions Bay-0 and Shahdara, respectively.
Fig. 3 The Bay-0×Shahdara linkage map shows the locations of QTLs associated with yellowing degree estimated by visual notation (YV, grey bars); yellowing degree estimated by image processing (YP, black bars); redness visual estimation (RV, white diamond bars); dry matter (DM, horizontal ray bars); total amino acid content (AA, draught board bars); total nitrogen content (NP, dotted bars) and short day flowering time (SD, white bars). Each QTL is represented by a bar, located at its most probable position. The QTLs on the left of the chromosomes are those detected in this study; QTLs on the right of the chromosome are those described in previous publications (Loudet et al. 2002, Loudet, Chaillou, Merigout et al. 2003). Bar length is proportional to the QTL contribution (R2). The sign of the allelic effect is indicated for each QTL. The framework genetic map (indicating markers position) is from Loudet et al. (2002).
Fig. 4 Confirmation of QTL YV3.4, YP3.4 and RV3.2 in near-isogenic lines. (A) Heterogeneous inbred family HIF404 segregates only for a region that overlaps with Y3.4 and is homozygous elsewhere. Leaves were dissected and are presented according to their emergence number, to facilitate the comparison of the leaf yellowing phenotypes (A.a). YP values were quantified (A.b) for each genotypic class. (B) Heterogeneous inbred family HIF068 segregates only for a region that overlaps with RV3.2 and is homozygous elsewhere. Three rosettes from HIF068-Bay and HIF068-Sha are presented to show the redness symptoms that are more visible on midribs (B.a). The redness of leaf methanolic extracts can be observed (B.b) and anthocyanin contents in leaf extracts were quantified (B.c). Values are the mean of six repeats ± SD, and ANOVA results are shown.
Phenotypic variation for yellowing and redness traits
| Name | Trait | Units | Bay-0 mean | Shahdara mean | RIL mean | RIL range (min–max) | Fisher’s F | Heritability (%) |
| YP | Yellowing imaging | % (log) | 1.6 (0.52) | 4.5 (0.77) | 5.03 (0.69) | 0–32 (0–1.5) | 2.14*** | 36 |
| YV | Yellowing notation | 0–4 | 0 | 0.44 | 0.5 | 0–4 | 2.89*** | 49 |
| RV | Redness notation | 0–6 | 3.75 | 0.5 | 1.77 | 0–6 | 3.28*** | 53 |
| LAR | Leaf area rating | % | 0.43 | 0.39 | 0.41 | 0.18–0.58 | 1.09 NS | ND |
| Name | Trait | Units | Bay-0 mean | Shahdara mean | RIL mean | RIL range (min–max) | Fisher’s F | Heritability (%) |
| YP | Yellowing imaging | % (log) | 1.6 (0.52) | 4.5 (0.77) | 5.03 (0.69) | 0–32 (0–1.5) | 2.14*** | 36 |
| YV | Yellowing notation | 0–4 | 0 | 0.44 | 0.5 | 0–4 | 2.89*** | 49 |
| RV | Redness notation | 0–6 | 3.75 | 0.5 | 1.77 | 0–6 | 3.28*** | 53 |
| LAR | Leaf area rating | % | 0.43 | 0.39 | 0.41 | 0.18–0.58 | 1.09 NS | ND |
The mean values of the parents are shown. RILs are described by their mean, range of variation, Fisher’s value for the genotype effect, its significance and heritability. Values in parentheses are transformed data for YP.
***Significant at the 0.1% level; NS, not significant; ND, not determined
Phenotypic variation for yellowing and redness traits
| Name | Trait | Units | Bay-0 mean | Shahdara mean | RIL mean | RIL range (min–max) | Fisher’s F | Heritability (%) |
| YP | Yellowing imaging | % (log) | 1.6 (0.52) | 4.5 (0.77) | 5.03 (0.69) | 0–32 (0–1.5) | 2.14*** | 36 |
| YV | Yellowing notation | 0–4 | 0 | 0.44 | 0.5 | 0–4 | 2.89*** | 49 |
| RV | Redness notation | 0–6 | 3.75 | 0.5 | 1.77 | 0–6 | 3.28*** | 53 |
| LAR | Leaf area rating | % | 0.43 | 0.39 | 0.41 | 0.18–0.58 | 1.09 NS | ND |
| Name | Trait | Units | Bay-0 mean | Shahdara mean | RIL mean | RIL range (min–max) | Fisher’s F | Heritability (%) |
| YP | Yellowing imaging | % (log) | 1.6 (0.52) | 4.5 (0.77) | 5.03 (0.69) | 0–32 (0–1.5) | 2.14*** | 36 |
| YV | Yellowing notation | 0–4 | 0 | 0.44 | 0.5 | 0–4 | 2.89*** | 49 |
| RV | Redness notation | 0–6 | 3.75 | 0.5 | 1.77 | 0–6 | 3.28*** | 53 |
| LAR | Leaf area rating | % | 0.43 | 0.39 | 0.41 | 0.18–0.58 | 1.09 NS | ND |
The mean values of the parents are shown. RILs are described by their mean, range of variation, Fisher’s value for the genotype effect, its significance and heritability. Values in parentheses are transformed data for YP.
***Significant at the 0.1% level; NS, not significant; ND, not determined
Correlation between traits among RILs
| SD | DM | NP | AA | YP | YV | RV | LAR | |
| SD (flowering time) a | 1 | |||||||
| DM (g plant–1) b | 0.13** | 1 | ||||||
| NP (% DM) b | –0.14** | –0.27*** | 1 | |||||
| AA [nmol (mg DM)–1] b | NS | –0.33*** | 0.84*** | 1 | ||||
| YP (%) | NS | –0.38*** | 0.55*** | 0.63*** | 1 | |||
| YV (notation) | NS | N0.29*** | 0.39*** | 0.47 *** | 0.55*** | 1 | ||
| RV (notation) | 0.38*** | NS | –0.13** | NS | NS | NS | 1 | |
| LAR (%) | 0.16** | 0.67** | NS | NS | –0.26*** | –0.23** | NS | 1 |
| SD | DM | NP | AA | YP | YV | RV | LAR | |
| SD (flowering time) a | 1 | |||||||
| DM (g plant–1) b | 0.13** | 1 | ||||||
| NP (% DM) b | –0.14** | –0.27*** | 1 | |||||
| AA [nmol (mg DM)–1] b | NS | –0.33*** | 0.84*** | 1 | ||||
| YP (%) | NS | –0.38*** | 0.55*** | 0.63*** | 1 | |||
| YV (notation) | NS | N0.29*** | 0.39*** | 0.47 *** | 0.55*** | 1 | ||
| RV (notation) | 0.38*** | NS | –0.13** | NS | NS | NS | 1 | |
| LAR (%) | 0.16** | 0.67** | NS | NS | –0.26*** | –0.23** | NS | 1 |
Pearson’s coefficients are shown.
aLoudet et al. (2002), bLoudet, Chaillou, Merigout et al. (2003).
NS not significant; *P < 0.05; **P < 0.01; ***P < 0.001.
Correlation between traits among RILs
| SD | DM | NP | AA | YP | YV | RV | LAR | |
| SD (flowering time) a | 1 | |||||||
| DM (g plant–1) b | 0.13** | 1 | ||||||
| NP (% DM) b | –0.14** | –0.27*** | 1 | |||||
| AA [nmol (mg DM)–1] b | NS | –0.33*** | 0.84*** | 1 | ||||
| YP (%) | NS | –0.38*** | 0.55*** | 0.63*** | 1 | |||
| YV (notation) | NS | N0.29*** | 0.39*** | 0.47 *** | 0.55*** | 1 | ||
| RV (notation) | 0.38*** | NS | –0.13** | NS | NS | NS | 1 | |
| LAR (%) | 0.16** | 0.67** | NS | NS | –0.26*** | –0.23** | NS | 1 |
| SD | DM | NP | AA | YP | YV | RV | LAR | |
| SD (flowering time) a | 1 | |||||||
| DM (g plant–1) b | 0.13** | 1 | ||||||
| NP (% DM) b | –0.14** | –0.27*** | 1 | |||||
| AA [nmol (mg DM)–1] b | NS | –0.33*** | 0.84*** | 1 | ||||
| YP (%) | NS | –0.38*** | 0.55*** | 0.63*** | 1 | |||
| YV (notation) | NS | N0.29*** | 0.39*** | 0.47 *** | 0.55*** | 1 | ||
| RV (notation) | 0.38*** | NS | –0.13** | NS | NS | NS | 1 | |
| LAR (%) | 0.16** | 0.67** | NS | NS | –0.26*** | –0.23** | NS | 1 |
Pearson’s coefficients are shown.
aLoudet et al. (2002), bLoudet, Chaillou, Merigout et al. (2003).
NS not significant; *P < 0.05; **P < 0.01; ***P < 0.001.
QTLs associated with yellowing value detected by image processing (YP) or by visual notation (YV) and visual redness (RV) in the Bay-0×Shahdara RIL population
| Trait QTL | Chromosome marker a | Map position b | LOD score | R2c | Allelic effect 2a d |
| Yellowing -image processing (log) | |||||
| YP3.1 | Chrom I-MSAT1.5 | 80.6 | 2.3 | 2 | +0.1 |
| YP3.2 | Chrom II-MSAT2.38 | 15.1 | 4.7 | 7 | +0.14 |
| YP3.3 | Chrom III- ATHCHIB2 | 1.4 | 6.9 | 6 | –0.16 |
| YP3.4 | Chrom III-MSAT3.21 | 54.7 | 9.6 | 12 | –0.22 |
| YP3.5 | Chrom III-MSAT5.22 | 36.7 | 5.9 | 5 | –0.15 |
| YP complete model | 32 | ||||
| Yellowing visual notation | |||||
| YV3.1 | Chrom I-MSAT1.5 | 83.9 | 3.5 | 3 | +0.14 |
| YV3.2 | Chrom II-MSAT2.38 | 15.5 | 7.6 | 7 | +0.23 |
| YV3.3 | Chrom III-ATHCHIB2 | 5.4 | 11.9 | 11 | –0.28 |
| YV3.4 | Chrom III-MSAT3.21 | 52.3 | 7.4 | 7 | –0.23 |
| YV3.2 x YV3.3 | 2 | ||||
| YV3.3 x YV3.4 | 2 | ||||
| YV complete model | 32 | ||||
| Redness visual notation | |||||
| RV3.1 | Chrom I-T1G11 | 0.0 | 3.7 | 2 | +0.18 |
| RV3.2 | Chrom I-NGA128 | 59.4 | 53.6 | 33 | +0.79 |
| RV3.3 | Chrom III-MSAT3.18 | 57.7 | 3.9 | 3 | +0.22 |
| RV3.4 | Chrom IV-MSAT4.8 | 2.0 | 9.1 | 3 | –0.29 |
| RV3.5 | Chrom V-NGA249 | 11.9 | 19.5 | 6 | +0.49 |
| RV3.6 | Chrom V-MSAT5.9 | 42.3 | 6.2 | 2 | +0.26 |
| RV complete model | 49 |
| Trait QTL | Chromosome marker a | Map position b | LOD score | R2c | Allelic effect 2a d |
| Yellowing -image processing (log) | |||||
| YP3.1 | Chrom I-MSAT1.5 | 80.6 | 2.3 | 2 | +0.1 |
| YP3.2 | Chrom II-MSAT2.38 | 15.1 | 4.7 | 7 | +0.14 |
| YP3.3 | Chrom III- ATHCHIB2 | 1.4 | 6.9 | 6 | –0.16 |
| YP3.4 | Chrom III-MSAT3.21 | 54.7 | 9.6 | 12 | –0.22 |
| YP3.5 | Chrom III-MSAT5.22 | 36.7 | 5.9 | 5 | –0.15 |
| YP complete model | 32 | ||||
| Yellowing visual notation | |||||
| YV3.1 | Chrom I-MSAT1.5 | 83.9 | 3.5 | 3 | +0.14 |
| YV3.2 | Chrom II-MSAT2.38 | 15.5 | 7.6 | 7 | +0.23 |
| YV3.3 | Chrom III-ATHCHIB2 | 5.4 | 11.9 | 11 | –0.28 |
| YV3.4 | Chrom III-MSAT3.21 | 52.3 | 7.4 | 7 | –0.23 |
| YV3.2 x YV3.3 | 2 | ||||
| YV3.3 x YV3.4 | 2 | ||||
| YV complete model | 32 | ||||
| Redness visual notation | |||||
| RV3.1 | Chrom I-T1G11 | 0.0 | 3.7 | 2 | +0.18 |
| RV3.2 | Chrom I-NGA128 | 59.4 | 53.6 | 33 | +0.79 |
| RV3.3 | Chrom III-MSAT3.18 | 57.7 | 3.9 | 3 | +0.22 |
| RV3.4 | Chrom IV-MSAT4.8 | 2.0 | 9.1 | 3 | –0.29 |
| RV3.5 | Chrom V-NGA249 | 11.9 | 19.5 | 6 | +0.49 |
| RV3.6 | Chrom V-MSAT5.9 | 42.3 | 6.2 | 2 | +0.26 |
| RV complete model | 49 |
The name of the QTL is the name of the trait, 3 for 3 mM of nitrate and the number of the QTL.
a The corresponding marker is the cofactor used in CIM model 6.
b The position of the QTL is expressed in cM from the first marker of the chromosome.
c Percentage of the variation explained by the QTL, calculated by variance component analysis.
d The mean effect of the replacement of Shahdara alleles by Bay-0 alleles at the QTL.
QTLs associated with yellowing value detected by image processing (YP) or by visual notation (YV) and visual redness (RV) in the Bay-0×Shahdara RIL population
| Trait QTL | Chromosome marker a | Map position b | LOD score | R2c | Allelic effect 2a d |
| Yellowing -image processing (log) | |||||
| YP3.1 | Chrom I-MSAT1.5 | 80.6 | 2.3 | 2 | +0.1 |
| YP3.2 | Chrom II-MSAT2.38 | 15.1 | 4.7 | 7 | +0.14 |
| YP3.3 | Chrom III- ATHCHIB2 | 1.4 | 6.9 | 6 | –0.16 |
| YP3.4 | Chrom III-MSAT3.21 | 54.7 | 9.6 | 12 | –0.22 |
| YP3.5 | Chrom III-MSAT5.22 | 36.7 | 5.9 | 5 | –0.15 |
| YP complete model | 32 | ||||
| Yellowing visual notation | |||||
| YV3.1 | Chrom I-MSAT1.5 | 83.9 | 3.5 | 3 | +0.14 |
| YV3.2 | Chrom II-MSAT2.38 | 15.5 | 7.6 | 7 | +0.23 |
| YV3.3 | Chrom III-ATHCHIB2 | 5.4 | 11.9 | 11 | –0.28 |
| YV3.4 | Chrom III-MSAT3.21 | 52.3 | 7.4 | 7 | –0.23 |
| YV3.2 x YV3.3 | 2 | ||||
| YV3.3 x YV3.4 | 2 | ||||
| YV complete model | 32 | ||||
| Redness visual notation | |||||
| RV3.1 | Chrom I-T1G11 | 0.0 | 3.7 | 2 | +0.18 |
| RV3.2 | Chrom I-NGA128 | 59.4 | 53.6 | 33 | +0.79 |
| RV3.3 | Chrom III-MSAT3.18 | 57.7 | 3.9 | 3 | +0.22 |
| RV3.4 | Chrom IV-MSAT4.8 | 2.0 | 9.1 | 3 | –0.29 |
| RV3.5 | Chrom V-NGA249 | 11.9 | 19.5 | 6 | +0.49 |
| RV3.6 | Chrom V-MSAT5.9 | 42.3 | 6.2 | 2 | +0.26 |
| RV complete model | 49 |
| Trait QTL | Chromosome marker a | Map position b | LOD score | R2c | Allelic effect 2a d |
| Yellowing -image processing (log) | |||||
| YP3.1 | Chrom I-MSAT1.5 | 80.6 | 2.3 | 2 | +0.1 |
| YP3.2 | Chrom II-MSAT2.38 | 15.1 | 4.7 | 7 | +0.14 |
| YP3.3 | Chrom III- ATHCHIB2 | 1.4 | 6.9 | 6 | –0.16 |
| YP3.4 | Chrom III-MSAT3.21 | 54.7 | 9.6 | 12 | –0.22 |
| YP3.5 | Chrom III-MSAT5.22 | 36.7 | 5.9 | 5 | –0.15 |
| YP complete model | 32 | ||||
| Yellowing visual notation | |||||
| YV3.1 | Chrom I-MSAT1.5 | 83.9 | 3.5 | 3 | +0.14 |
| YV3.2 | Chrom II-MSAT2.38 | 15.5 | 7.6 | 7 | +0.23 |
| YV3.3 | Chrom III-ATHCHIB2 | 5.4 | 11.9 | 11 | –0.28 |
| YV3.4 | Chrom III-MSAT3.21 | 52.3 | 7.4 | 7 | –0.23 |
| YV3.2 x YV3.3 | 2 | ||||
| YV3.3 x YV3.4 | 2 | ||||
| YV complete model | 32 | ||||
| Redness visual notation | |||||
| RV3.1 | Chrom I-T1G11 | 0.0 | 3.7 | 2 | +0.18 |
| RV3.2 | Chrom I-NGA128 | 59.4 | 53.6 | 33 | +0.79 |
| RV3.3 | Chrom III-MSAT3.18 | 57.7 | 3.9 | 3 | +0.22 |
| RV3.4 | Chrom IV-MSAT4.8 | 2.0 | 9.1 | 3 | –0.29 |
| RV3.5 | Chrom V-NGA249 | 11.9 | 19.5 | 6 | +0.49 |
| RV3.6 | Chrom V-MSAT5.9 | 42.3 | 6.2 | 2 | +0.26 |
| RV complete model | 49 |
The name of the QTL is the name of the trait, 3 for 3 mM of nitrate and the number of the QTL.
a The corresponding marker is the cofactor used in CIM model 6.
b The position of the QTL is expressed in cM from the first marker of the chromosome.
c Percentage of the variation explained by the QTL, calculated by variance component analysis.
d The mean effect of the replacement of Shahdara alleles by Bay-0 alleles at the QTL.
Abbreviations
- AA
amino acid content
- CIM
composite interval mapping
- DM
dry matter
- HIF
heterogenous inbred family
- LAR
leaf area rating
- NIL
near-isogenic line
- NP
nitrogen content
- QTL
quantitative trait locus
- RIL
recombinant inbred line
- RV
red visual notation
- SD
short day
- YP
yellowing by image processing
- YV
yellowing by visual notation
References
Baker, N.R. and Hardwick. (
Basten, C., Weir, B. and Zeng, Z.-B. (
Basten, C., Weir, B. and Zeng, Z.-B. (
Beavis, W.D., Smith, O.S., Grant, D. and Fincher, R. (
Belluomo, P., Robert, C. and Bancal, M.O. (
Bongue-Bartelsman, M. and Phillips, D.A. (
Brugière, N., Dubois, F., Masclaux, C., Sangwan, R.S. and Hirel, B. (
Buchanan-Wollaston, V., Earl, S., Harrison, E., Mathas, E., Navabpour, S., Page, T. and Pink, D. (
Chalker-Scott, L. (
Chalker-Scott, L. and Scott, J.D. (
Churchill, G.A. and Doerge, R.W. (
Diaz, C., Purdy, S., Christ, A., Morot-Gaudry, J.-F., Wingler, A. and Masclaux-Daubresse, C. (
Do, C.B. and Cormier, F. (
Feild, T.S., Lee, D.W. and Holbrook, N.M. (
Feller, U. and Fischer, A. (
Greenwood, D., Lemaire, G., Gosse, G., Cruz, P., Draycott, A. and Neeteson, J. (
Guo, Y., Cai, Z. and Gan, S. (
Hensel, L.L., Grbic, V., Baumgarten, D.A. and Bleecker, A.B. (
Jiang, G.H., He, Y.Q., Xu, C.G., Li, X.H. and Zhang, Q. (
Justes, E., Mary, B., Meynard, J.-M., Machet, J.-M. and Thelier-Huche, L. (
Kato, Y., Yamamoto, Y., Murakami, S. and Sato, F. (
Koornneef, M., Alonso-Blanco, C. and Vreugdenhil, D. (
Lebreton, C., Lazic-Jancic, V., Steed, A., Pekic, S. and Quarrie, S. (
Lee, D.W., Bremmeier, S. and Smith, A.P. (
Lim, P.O., Woo, H.R. and Nam, H.G. (
Loudet, O., Chaillou, S., Camilleri, C., Bouchez, D. and Daniel-Vedele, F. (
Loudet, O., Chaillou, S., Krapp, A. and Daniel-Vedele, F. (
Loudet, O., Chaillou, S., Merigout, P., Talbotec, J. and Daniel-Vedele, F. (
Loudet, O., Gaudon, V., Trubuil, A and Daniel-Vedele, F. (
Masclaux, C., Quilleré, I., Gallais, A. and Hirel, B. (
Masclaux, C., Valadier, M., Brugière, N., Morot-Gaudry, J. and Hirel, B. (
Matile, P. (
Nikiforova, V.J., Gakiere, B., Kempa, S., Adamik, M., Willmitzer, L., Hesse, H. and Hoefgen, R. (
Noodén, L.D., Hillsberg, J.W. and Schneider, M.J. (
Plénet, D. and Lemaire, G. (
Pourtau, N., Mares, M., Purdy, S., Quentin, N., Ruel, A. and Wingler, A. (
Rajendran, L., Ravishankar, G.A., Venkataraman, L.V. and Prathiba, K.R. (
Scheible, W.R., Morcuende, R., Czechowski, T., Fritz, C., Osuna, D., Palacios-Rojas, N., Schindelasch, D., Thimm, O., Udvardi, M.K. and Stitt, M. (
Spano, G., Di Fonzo, N., Perrota, C., Platini, C., Ronga, G., Lawlor, D.W., Napier, J.A. and Shewery, P.R. (
Stewart, A.J., Chapman, W., Jenkins, G.I., Graham, I., Martin, T. and Crozier, A. (
Thomas, H. and de Villier, L. (
Thomas, H., Evans, C., Humphreys, M., Morgan, G., Hauck, B. and Donnison, I. (
Tuinstra, M., Ejeta, G. and Goldsbrough, P. (
Verma, V., Foulkes, M.J., Worland, A.J., Sylvester-Bradley, R., Caligari, P.D.S. and Snape, J.W. (
Vitousek, P.M. and Howarth, R.V. (
Woo, H.R., Goh, C.H., Park, J.H., Teyssendier de la Serve, B., Kim, J.H., Park, Y.I. and Nam, H.G. (