-
PDF
- Split View
-
Views
-
Cite
Cite
Yuko Inoue, Takao Suzuki, Masaki Hattori, Kohki Yoshimoto, Yoshinori Ohsumi, Yuji Moriyasu, AtATG Genes, Homologs of Yeast Autophagy Genes, are Involved in Constitutive Autophagy in Arabidopsis Root Tip Cells , Plant and Cell Physiology, Volume 47, Issue 12, December 2006, Pages 1641–1652, https://doi.org/10.1093/pcp/pcl031
Close - Share Icon Share
Abstract
In Arabidopsis root tips cultured in medium containing sufficient nutrients and the membrane-permeable protease inhibitor E-64d, parts of the cytoplasm accumulated in the vacuoles of the cells from the meristematic zone to the elongation zone. Also in barley root tips treated with E-64, parts of the cytoplasm accumulated in autolysosomes and pre-existing central vacuoles. These results suggest that vacuolar and/or lysosomal autophagy occurs constitutively in these regions of cells. 3-Methyladenine, an inhibitor of autophagy, inhibited the accumulation of such inclusions in Arabidopsis root tip cells. Such inclusions were also not observed in root tips prepared from Arabidopsis T-DNA mutants in which AtATG2 or AtATG5 , an Arabidopsis homolog of yeast ATG genes essential for autophagy, is disrupted. In contrast, an atatg9 mutant, in which another homolog of ATG is disrupted, accumulated a significant number of vacuolar inclusions in the presence of E-64d. These results suggest that both AtAtg2 and AtAtg5 proteins are essential for autophagy whereas AtAtg9 protein contributes to, but is not essential for, autophagy in Arabidopsis root tip cells. Autophagy that is sensitive to 3-methyladenine and dependent on Atg proteins constitutively occurs in the root tip cells of Arabidopsis .
Introduction
Autophagy is a process in which components of cells, including cytosol and cell organelles, are degraded in the lytic organelles, lysosomes and/or vacuoles. Cellular components are enclosed by membrane sacs to make double membrane-bounded organelles called autophagosomes, and transported into lysosomes/vacuoles, where they are eventually degraded (Dunn 1994 , Klionsky 2004 ). Autophagy is known to occur under nutrient-deficient conditions not only in mammalian and yeast cells but also in plant cells (Chen et al. 1994 , Aubert et al. 1996 , Moriyasu and Ohsumi 1996 ), and thus is supposed to contribute to intracellular nutrient supply when nutrient uptake from the environment is limiting. In plant cells, it has also been observed in various developmental processes (Moriyasu and Hillmer 2000 ); membranes derived from endoplasmic reticulum (ER) or the trans -Golgi network have been observed to enclose a part of the cytoplasm and become a vacuole through autophagy in root tip cells, suggesting that autophagy is involved in vacuole genesis (Marty 1978 , Amelunxen and Heinze 1984 , Hilling and Amelunxen 1985 , Marty 1999 ). Recently, it has been shown that autophagy is also involved in the regulation of the programmed cell death that occurs in pathogen-infected plant cells (Liu et al. 2005 , Seay and Dinesh-Kumar 2005 ).
Protease inhibitors are useful tools for analyzing autophagy. The treatment of mammalian cells undergoing autophagy with cysteine protease inhibitors such as leupeptin causes the accumulation of lysosomes containing cytoplasmic components, the autolysosomes (Kominami et al. 1983 ). Similarly, treatment of yeast cells with the serine protease inhibitor phenylmethylsulfonyl fluoride results in the accumulation of membrane-bounded parts of the cytoplasm, the autophagic bodies, in the vacuole (Takeshige et al. 1992 ). In cultured tobacco cells, autophagy, which accompanies the net degradation of cellular proteins, is induced by transfer to culture medium lacking sucrose (Moriyasu and Ohsumi 1996 ). The addition of a cysteine protease inhibitor such as E-64c, leupeptin or antipain to the culture medium inhibits net protein degradation, and particles of cytoplasm accumulate in membrane-bounded structures. They were designated as plant autolysosomes by analogy with mammalian cells (Moriyasu and Ohsumi 1996 ). In the absence of inhibitors, the particles of cytoplasm segregated in autolysosomes are thought to be degraded by the action of proteases there. Of various cysteine protease inhibitors, E-64d is a more membrane-permeable analog of E-64 (Tamai et al. 1986 , Moriyasu 1995 ). After treatment with E-64d, autolysosomes similar to those in cultured tobacco cells accumulated in root cells prepared from barley seedlings (Moriyasu et al. 2003 ). These results strongly suggest that also in plant cells, the application of a cysteine protease inhibitor causes the accumulation of intermediate structures formed by autophagy.
3-Methyladenine (3-MA) was found to be a potent inhibitor in mammalian autophagy by screening of various kinds of chemicals (Gordon and Seglen 1982 , Seglen and Gordon 1982 ). It has been thought to block the formation of autophagosomes. Recently, 3-MA has been shown to block autophagy almost completely in tobacco cells cultured under nutrient-deficient conditions (Takatsuka et al. 2004 , Inoue and Moriyasu 2006 ).
Molecular genetic analysis of autophagy in yeast cells cultured under starvation conditions revealed more than a dozen genes essential for autophagy (Tsukada and Ohsumi 1993 , Thumm et al. 1994 , for a review, see also Klionsky and Ohsumi 1999 ). These genes were designated as ATG genes (Klionsky et al. 2003 ). The Arabidopsis genome has 25 ATG genes that are homologous to 12 of the yeast ATG genes (Hanaoka et al. 2002 ), and T-DNA mutant plants in which ATG4, ATG5, ATG7 and ATG9 were disrupted have been isolated and characterized (Doelling et al. 2002 , Hanaoka et al. 2002 , Yoshimoto et al. 2004 , Thompson et al. 2005 ). These mutants entered senescence slightly earlier than wild-type plants. Moreover, they clearly displayed earlier senescence syndromes under nutrient-deficient conditions, which include earlier degradation of Chl, chloroplast proteins or cytoplasmic rRNA.
In yeast cells, a product of the ATG8 gene is known to be localized to autophagosomes and thus can be used as a marker of autophagosomes formation (Kirisako et al. 1999 ). The fusion protein of Atg8 and green fluorescent protein (GFP) expressed in Arabidopsis cells was localized on punctate structures, which were presumed to be autophagosomes by analogy with yeast cells (Yoshimoto et al. 2004 , Sláviková et al. 2005 , Thompson et al. 2005 ). When the plants expressing the fusion protein were treated with the vacuolar H + -ATPase inhibitor concanamycin, fluorescent autophagic bodies, structures deriving from autophagosomes, accumulate in the central vacuoles (Yoshimoto et al. 2004 , Sláviková et al. 2005 , Thompson et al. 2005 ). In contrast, such accumulation did not occur in the Arabidopsis atg mutants (Yoshimoto et al. 2004 , Thompson et al. 2005 ). These results were interpreted as showing that autophagy does not occur in the atg mutants. In two of these studies, autophagy was examined mainly under starvation conditions or in the dark where photosynthetic energy supply is limited (Yoshimoto et al. 2004 , Thompson et al. 2005 ), whereas in the other study, it has been reported that autophagosome-like structures are observed in the cytoplasm and accumulate in the central vacuoles even under favorable growth conditions (Sláviková et al. 2005 ). In the present study, using E-64d, a reliable autophagy inhibitor other than concanamycin, we examined autophagy in the cells of root tips excised from Arabidopsis and barley seedlings. We found that in root cells, autophagy occurs constitutively, although it is further activated under nutrient-limiting conditions. We examined the effect of 3-MA on this autophagy and whether it occurs in the Arabidopsis atg mutant plants in which an ATG2, ATG5 or ATG9 gene is disrupted.
Results
Morphology of root tips immediately after excision
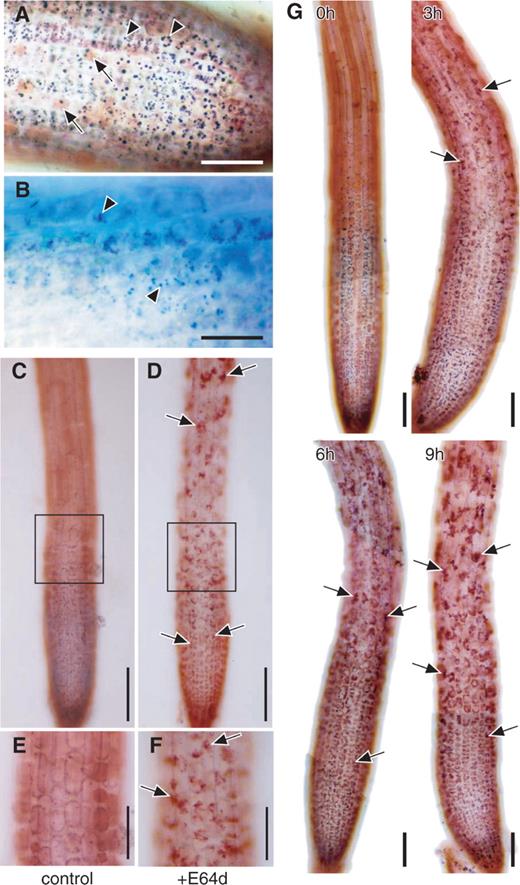
We first stained root tips immediately after excision from Arabidopsis seedlings with neutral red, which stains the acidic compartments of cells ( Fig. 1 A). As expected, the central vacuoles were stained in the cells of root tips ( Fig. 1 A, arrows). In addition to the central vacuoles, spherical structures of 0.5–3 μm in diameter were stained red in a region from the tips of the roots to approximately 200 µm away from the tips ( Fig. 1 A, arrowheads). Almost all of these spherical structures were found in the cytoplasm, but some were moving in a Brownian manner and thus were thought to exist in the central vacuoles. With the exception of these structures, almost all vacuoles of the cells in this region appeared to be empty. In this study, we did not analyze these structures in detail. However, when root tips were stained with toluidine blue, similar spherical structures were stained blue ( Fig. 1 B, arrowheads). Since toluidine blue mainly stains protein and/or RNA, some of these spherical structures could be granules of storage proteins located in protein storage vacuoles. In addition, large central vacuoles in more mature cells above this region also appeared empty (image not shown).
Effect of the membrane-permeable cysteine protease inhibitor E-64d on the morphology of Arabidopsis root tips. Root tips were excised from the seedlings of Arabidopsis (Landsberg in A and B, and Columbia in C–G) grown under nutrient-sufficient conditions. (A and B) Immediately after excision, root tips were stained with neutral red (A) or with toluidine blue (B). Arrows in A, central vacuole stained with neutral red; arrowheads in A, spherical structures stained with neutral red; arrows in B, spherical structures stained with toluidine blue. Bar in A = 50 μm; bar in B = 25 μm. (C–F). Root tips were incubated in the culture medium containing 100 μM E-64d (D and F) or 1% (v/v) methanol as a solvent control of E-64d addition (C and E) for 1 d. They were stained with neutral red and observed by light microcopy. E and F are magnified images of parts of the images C and D, respectively. Arrows in D and F point to vacuolar inclusions. Bars in C and D = 100 μm; bars in E and F = 50 μm. (G) Root tips were incubated in the culture medium containing 100 μM E-64d for various times indicated in each image, and stained with neutral red. Arrows point to vacuolar inclusions. Bar = 50 μm.
Parts of the cytoplasm accumulate in autolysosomes and/or pre-existing vacuoles of root cells after treatment with E-64d
Marked morphological changes did not occur in cells of root tips incubated in culture medium for 1 d ( Fig. 1 C, E). However, particulate structures were found to accumulate in the vacuoles of root tip cells 3 h after the addition of E-64d to the culture medium ( Fig. 1 G, 3 h; arrows). The number of these structures gradually increased for several hours ( Fig. 1 G, 6 and 9 h; arrows), and the accumulation of many particulate structures was clearly seen after 1 d ( Fig. 1 D, F; arrows). These structures were stained more intensely with neutral red than the lumen of the central vacuole, probably because of the stronger absorption of the dye. Such particulate structures were observed in all 20 root tips examined in four independent experiments.
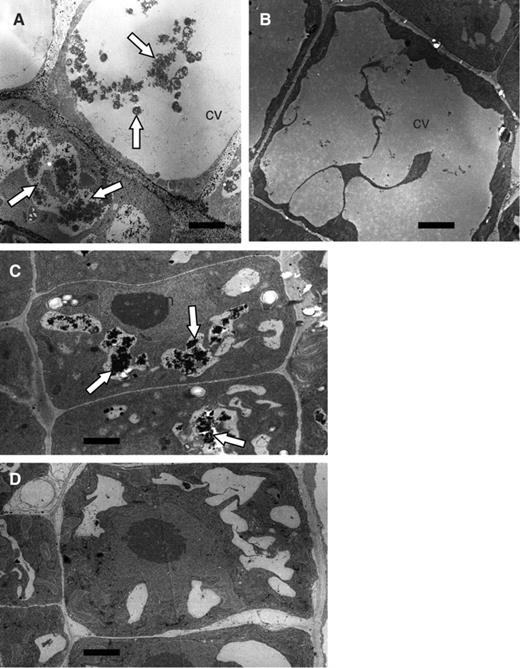
To analyze these particulate structures further, we observed the sections of Arabidopsis root tips by electron microscopy ( Fig. 2 ). In both the relatively young and mature cells of root tips treated with E-64d for 1 d, many degradation products of cytoplasmic organelles were observed in the vacuoles ( Fig. 2 A, C; arrows). In contrast, the vacuole appeared to be almost empty in the cells of root tips treated only with 1% (w/v) methanol used as a solvent control ( Fig. 2 B, D). These results suggest that cytoplasmic particles accumulate in autolysosomes that are newly formed or in large pre-existing vacuoles by the treatment with E-64d.
Electron micrographs of the cells of Arabidopsis root tips cultured in the presence of E-64d. Root tips were excised from the seedlings of Arabidopsis (Landsberg) grown under nutrient-sufficient conditions and incubated in the culture medium containing sucrose in the presence (A and C) or absence (B and D) of E-64d for 1 d. They were fixed and embedded in plastic blocks. Sections made from these blocks were stained with uranyl acetate and lead nitrate, and observed by electron microscopy. Images of relatively young root tip cells (C and D) and mature vacuolated cells (A and B) are shown. Arrows point to vacuolar inclusions. CV, the central vacuoles; n, nucleus. Bar = 2 μm.
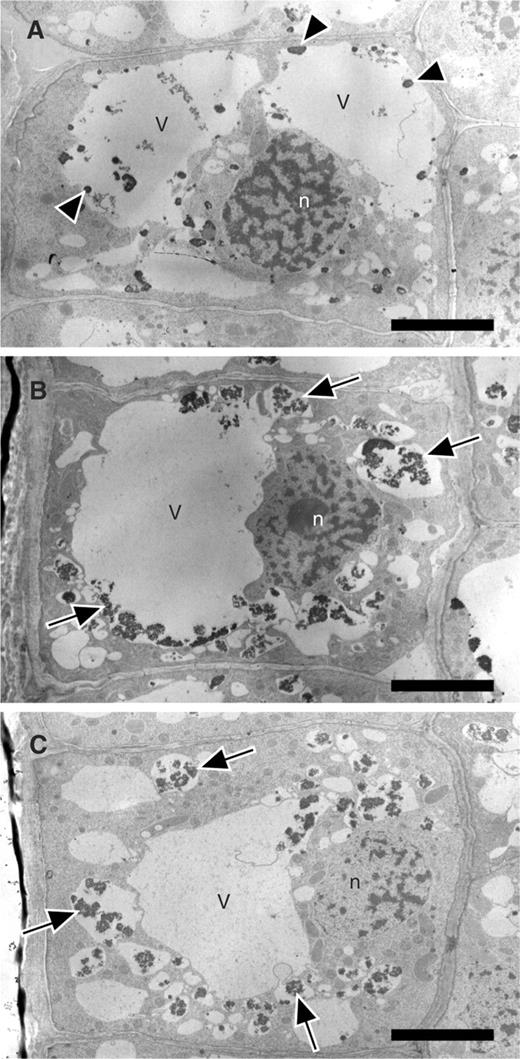
In a previous study, we reported that when immature root tip cells of barley, which lack large vacuoles, are treated with E-64d, parts of the cytoplasm mainly accumulate in newly synthesized autolysosomes (Moriyasu et al. 2003 ). To examine the consistency of the results from Arabidopsis root tips with those from barley root tips, we observed relatively mature cells of barley root tips treated with the inhibitor ( Fig. 3 ). In these cells, parts of the cytoplasm accumulated not only in small vacuoles but also in pre-existing large vacuoles ( Fig. 3 , arrows); these small vacuoles appeared to be newly formed autolysosomes, and the large vacuoles appeared to be pre-existing central vacuoles. Even after 1 d, we occasionally observed small autolysosomes outside of the central vacuole (data not shown). These results suggest that autolysosomes formed in the cytoplasm fuse with the central vacuole and, as a result, particles in autolysosomes are transferred into the central vacuole.
Electron micrograph of the relatively mature cells of barley root tips cultured in the presence of E-64d. Barley root tips were kept in the culture medium containing 3% (w/v) sucrose in the presence of 100 μM E-64d for 4 h (B) or 8 h (C), or in the presence of only methanol for 8 h (A). They were then fixed with glutaraldehyde and embedded in the blocks of Spurr's resin. Sections made from these blocks were stained with KMnO 4 and uranyl acetate, and observed by electron microscopy. Arrows point to vacuolar inclusions. Some central vacuoles of the control cells (A) also contain several electron-dense particles (A, arrowheads). The surfaces of these particles were smooth and were different from those in autolysosomes (B and C, arrows). V, vacuole; n, nucleus. Bar = 5 μm.
Autophagy in root tip cells is inhibited by 3-MA
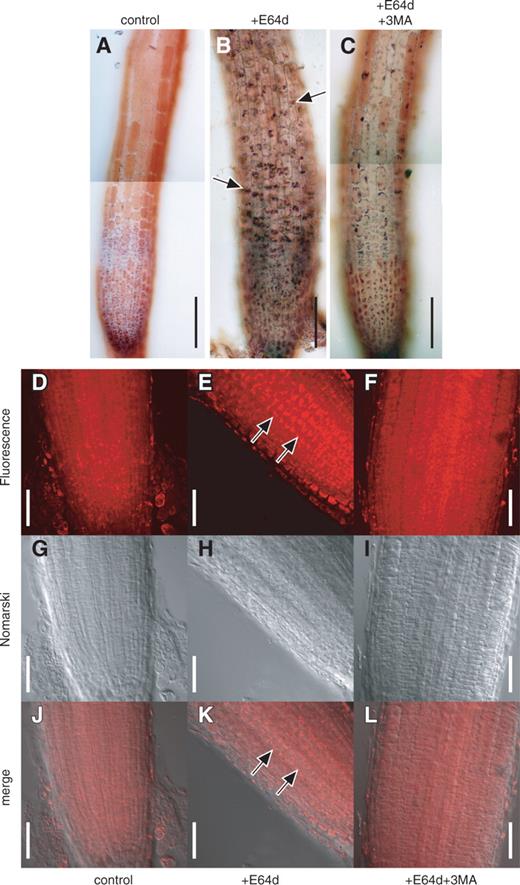
To investigate whether or not 3-MA inhibits autophagy in the root tip cells, we cultured Arabidopsis root tips in the presence of E-64d and/or 3-MA for 1 d ( Fig. 4 ). After incubation, root tips were stained with neutral red and observed by light microscopy ( Fig. 4 A–C). This result was consistent with our previous observations; empty vacuoles were mostly observed in control root tips, which were treated with 1% (v/v) methanol as a solvent control of E-64d addition and 5% (v/v) water as a solvent control of 3-MA addition, whereas particulate structures accumulated in central vacuoles in root tips that were cultured in the presence of E-64d and water ( Fig. 4 B vs. A). In contrast, in root tip cells cultured in the presence of both E-64d and 3-MA, the number of particulate structures decreased ( Fig. 4 C vs. B). Thirteen out of 17 root tips showed a clear decrease (a representative image is presented in Fig. 4 C), and the other four root tips showed a slight decrease.
Effect of 3-MA on the accumulation of vacuolar inclusions in the cells of Arabidopsis root tips cultured in the presence of E-64d. (A–C) Root tips were excised from the seedlings of Arabidopsis (Columbia), and cultured in the culture medium containing 1% (v/v) methanol and 5% (v/v) water (A), 100 μM E-64d and 5% (v/v) water (B) or 100 μM E-64d and 5 mM 3-MA (C) for 1 d. Root tips were stained with neutral red and their bright field images were obtained by light microscopy. Arrows in B point to the accumulation of vacuolar inclusions. Bar = 100 μm. (D–L) Root tips cultured in the presence of 1% (v/v) methanol and 5% (v/v) water (D, G and J), 100 μM E-64d and 5% (v/v) water (E, H and K) or 100 μM E-64d and 5 mM 3-MA (F, I and L) were stained with LysoTracker Red and fixed with formaldehyde. They were then observed with a confocal laser microscope, and the fluorescence images of LysoTracker Red (D, E and F) and Nomarski images (G, H and I) were obtained. Images J, K and L are merges of the images D, E and F, and the images G, H and I, respectively. Arrows in E and K point to the accumulation of vacuolar inclusions. Bar = 50 μm.
3-MA blocks autophagy in tobacco cells cultured under sucrose starvation conditions almost completely (Takatsuka et al. 2004 ). In contrast, the inhibition of autophagy by 3-MA did not appear to be complete in the root tips cells of Arabidopsis , and we detected some accumulation of inclusion bodies in the vacuole even in the presence of 3-MA ( Fig. 4 , compare A and C). Thus it is possible that a sufficient concentration of 3-MA is unable to diffuse into intact root tips, and thus the concentration of 3-MA in root tip cells could be suboptimal.
Inclusions accumulating in autolysosomes in tobacco culture cells and barley root tip cells can be stained with LysoTracker Red (Moriyasu et al. 2003 ). We stained Arabidopsis root tips using this fluorescent dye ( Fig. 4 D–L) and confirmed by this staining that parts of the cytoplasm accumulate in vacuoles or lysosomes in the presence of E-64d ( Fig. 4 , compare D and E, or J and K). Treatment with 3-MA decreased the quantity of these inclusions ( Fig. 4 ; compare E and F, or K and L). We interpret these data as demonstrating that 3-MA blocks a step upstream from the step of proteolysis that is sensitive to E-64d and, as a result, autophagic inclusions did not accumulate even in the presence of E-64d.
Putting the results obtained thus far in this study and the results from a previous study (Moriyasu et al. 2003 ) together, we conclude that autophagy occurs in root tip cells of barley and Arabidopsis and that we can observe autophagy by light microscopy after treatment with E-64d.
Isolation of atg2, atg5 and atg9 mutant plants
ATG genes are essential for autophagy to occur in yeast cells under nutrient starvation conditions (Klionsky and Ohsumi 1999 ). To examine whether Arabidopsis genes homologous to these yeast ATG genes ( AtATGs ) are involved in autophagy in root tip cells, we used mutant plants in which AtATG genes were disrupted. We chose AtATG2 (At3g19190), AtATG5 (At5g17290) and AtATG9 (At2g31260) because the BLASTP search for the homologs of yeast ATG2, ATG5 and ATG9 genes revealed only one homolog of each gene in the Arabidopsis genome. The amino acid sequences of AtAtg5 and AtAtg9 proteins have already been aligned with their counterparts from other organisms (Thompson et al. 2005 , Hanaoka et al. 2002 , respectively). Here we present the alignment of AtAtg2 with its homologs from other organisms (Supplementary Fig. 1). AtAtg2 protein has 29 and 33% amino acid similarity with its counterparts of yeast (Atg2 protein) and human (KIAA0404 protein), respectively. AtAtg2 protein is presumed to have 1,862 amino acids and a mol. wt of 205,471.
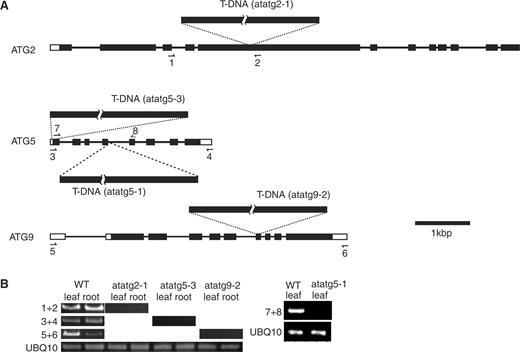
Fig. 5 shows the structures of these AtATG genes. The AtATG5 gene is composed of eight exons ( Fig. 5 A), which was already shown in a previous study (Thompson et al. 2005 ). In one of the two mutant alleles of ATG5 used in this study (designated as atatg5-3 considering the numbering of atatg5 mutants in Thompson et al. 2005 ), T-DNA was inserted into the first exon. In the other ATG5 mutant allele ( atatg5-1 ), which is the same as atatg5-1 reported in Thompson et al. ( 2005 ), T-DNA was inserted into the fourth intron. The AtATG9 gene consists of nine exons (Hanaoka et al. 2002 ), and T-DNA had been introduced into the sixth exons in the atatg9 mutant [designated as atatg9-2 considering the numbering of atatg9 mutants in Hanaoka et al. ( 2002 )] used in this study ( Fig. 5 A). In the atatg2 mutant (designated as atatg2-1 ), T-DNA was introduced into the fifth long exon of AtATG2 composed of 12 exons ( Fig. 5 A). DNA sequencing around the inserted T-DNA in the atatg2-1 mutant clarified that seven bases in the fifth exon (+3,661 to +3,667) are deleted in place of the insertion of T-DNA.
Expression of AtATG genes in atatg mutants used in this study. (A) Diagrams of AtATG2, AtATG5 and AtATG9 genes. Exons and introns in each gene are shown by boxes and lines, respectively. The sites into which T-DNA is inserted in AtATG2, AtATG5 and AtATG9 genes are shown in atatg2-1, atatg5-3, atatg5-1 and atatg9-2 mutants. Arrows show the priming sites in RT–PCR analysis. (B) RT–PCR analysis of atatg2-1, atatg5-3, atatg9-2 and atatg5-1 mutants. Total RNAs were extracted from the leaves and roots of the wild type (WT), atatg2-1, atatg5-3 and atatg9-2 plants and were subjected to RT–PCR using one of the UBQ10 primer pairs (see Materials and Methods) and either the primers 1 and 2 (1 + 2), the primers 3 and 4 (3 + 4) or the primers 5 and 6 (5 + 6). The total RNAs of the atatg5-1 and WT plants were extracted from leaves in separate experiments. They were subjected to RT–PCR using the other UBQ10 primer pairs (see Materials and Methods) and the primers 7 and 8 (7 + 8). The products of PCR were separated on agarose gels and stained with ethidium bromide.
We confirmed that normal mRNAs for the AtATG2, AtATG5 and AtATG9 genes were not present in either the leaf or root of atatg2-1, atatg5-3 and atatg9-2 mutants, respectively, by reverse tanscription–PCR (RT–PCR) ( Fig. 5 B) using the pair of primers shown in Fig. 5 A. Normal mRNA for the AtATG5 gene was not present in the leaves of atatg5-1 plants either ( Fig. 5 B).
Autophagy in root tip cells is dependent on ATG genes
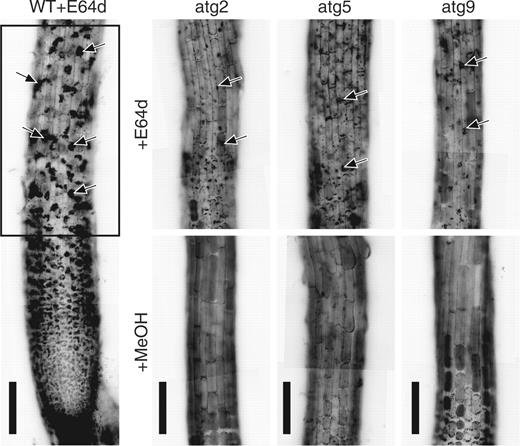
The gross morphology of the root tips in the mutants atatg2-1, atatg5-3, atatg5-1 and atatg9-2 was similar to that in wild-type plants. We examined whether autophagy occurs or not in these mutants by the use of E-64d ( Fig. 6 ). In the atatg2 mutant, only a very slight amount of accumulation of cytoplasmic particles was detected ( Fig. 6 , arrowheads; compare atg2 + E64d with WT + E64d). This result was confirmed in all 10 root tips examined in three independent experiments. Furthermore, this phenotype was also found in the descendants (designated as atatg2-1a and atatg2-1b in this study) that were obtained by two successive backcrosses of the atatg2-1 mutant to the wild-type Columbia ecotype. These show that the autophagy in Arabidopsis root tip cells is dependent on the AtATG2 gene. Also in the atatg5-3 and atatg5-1 mutants, few cytoplasmic particles accumulated after incubation with E-64d ( Fig. 6 , atg5 + E64d), and the result was confirmed in all 10 root tips in atatg5-3 mutants and all six root tips in atatg5-1 mutants examined. This shows that the autophagy in Arabidopsis root tip cells is also dependent on the AtATG5 gene. In contrast, more cytoplasmic particles accumulated in the atatg9 mutants than in the atatg2 and atatg5 mutants ( Fig. 6 , arrowheads; compare atg9 with atg2 and atg5 in +E64d) although the amount was still less than that in the wild-type plant ( Fig. 6 , compare atg9 + E64d with WT + E64d). This result suggests that the AtATG9 gene is not essential for autophagy although it contributes significantly to autophagy in Arabidopsis root tip cells.
Involvement of AtATG genes in autophagy in Arabidopsis root tips. Root tips excised from the wild-type Columbia ecotype and atatg2-1, atatg5-3 and atatg9-2 mutant seedlings were incubated in medium containing 100 μM E-64d (+E64d) or 1% (v/v) methanol (+MeOH) for 1 d. They were stained with neutral red and observed by light microscopy. The same parts as enclosed by a rectangle in the root tip of the wild-type seedling (WT+E64d) are shown in atatg2-1 (atg2), atatg5-3 (atg5) and atatg9-2 (atg9) mutants. Arrows point to vacuolar inclusions. Bar = 100 μm.
Depriving the incubation medium of sucrose activates autophagy
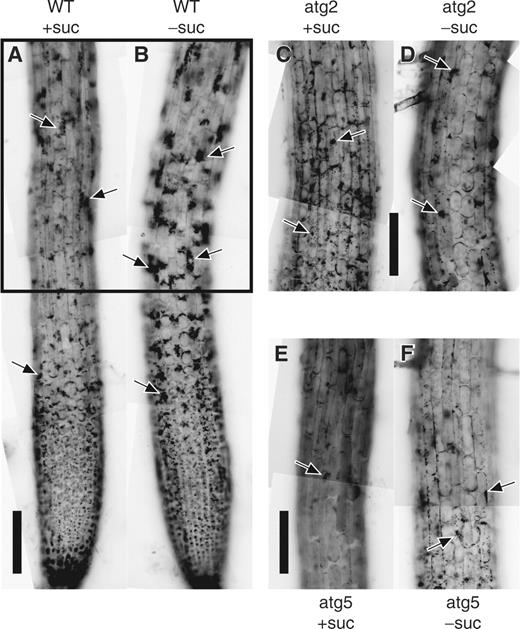
The amount of cytoplasmic particles accumulating in vacuoles significantly increased in root tips of wild-type seedlings incubated in sucrose-free medium in the presence of E-64d for 1 d ( Fig. 7 , compare +suc and −suc in WT). This result suggests that the transport of parts of the cytoplasm to vacuoles by autophagy is enhanced in sucrose-free culture medium. In contrast, no further accumulation of cytoplasmic particles was found in root tips of atatg2-1, atatg5-3 or atatg5-1 mutants treated in the same way ( Fig. 7 , compare +suc and −suc in atg2 and atg5), showing that the enhanced portion of autophagic transport of cytoplasmic components into vacuoles is also dependent on ATG2 and ATG5 gene products.
Activation of autophagy in Arabidopsis root tips by sucrose deprivation. Root tips from the wild-type of the Columbia ecotype (WT) and atatg2-1 (atg2) and atatg5-3 (atg5) mutant seedlings of Arabidopsis were incubated in culture medium containing (+suc) or lacking (−suc) sucrose in the presence of E-64d for 1 d. They were stained with neutral red and observed. The parts corresponding to that enclosed by a rectangle in the WT are shown in the mutants. Arrows point to vacuolar inclusions. Bar = 100 μm.
Quantification of autophagy in the root cells of Arabidopsis
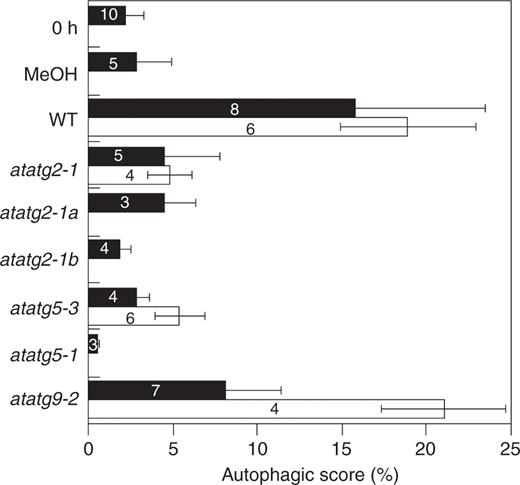
By analyzing images of root tips, we tried to quantify the accumulation of cytoplasmic materials due to the inhibition of autophagy by E-64d, and defined the autophagic score as described in Materials and Methods ( Fig. 8 ). The score reflected our interpretation of the photographs well with respect to the accumulation of cytoplasmic materials in vacuoles. It was <5 in root tips used in this study ( Fig. 8 ; 0 h, filled bar), and did not alter during 24 h of incubation of root tips in culture medium (MeOH, filled bar). However, it increased up to approximately 16 after 24 h of incubation with E-64d (WT, filled bar), showing that the score can reasonably estimate the accumulation of cytoplasmic materials due to the inhibition of protein degradation. In contrast, increases of the score were not observed in atatg2 mutants ( atatg2-1, atatg2-1a and atatg2-1b ; filled bar), or atatg5 mutants ( atatg5-3 and atatg5-1 , filled bar), which is consistent with our observation that only a very small amount of cytoplasmic particles accumulated in the vacuoles of these mutants. Also consistent with our observation, the autophagic score of atatg9-2 mutants was a little higher than that of atatg2 and atatg5 mutants ( atatg9-2 , filled bar). Furthermore, it increased up to approximately the same value as that of wild-type plants under sucrose starvation conditions ( atatg9-2 , open bar), supporting the notion that the AtAtg9 protein is not essential for autophagy in Arabidopsis root cells. In contrast, the autophagic score remained <7 under sucrose starvation conditions in atatg2 and atatg5 mutants ( atatg2-1 and atatg5-3 , open bar). The score seemed to be saturated around 20, and the activation of autophagy in wild-type plants under sucrose starvation conditions was not reflected enough in the score ( Fig. 8 , WT, open bar vs. filled bar).
Image analysis on the accumulation of cytoplasmic materials in the vacuoles of Arabidopsis root cells. Root tips from the wild-type seedlings of the Columbia ecotype (WT) and atatg2, atatg5 and atatg9 mutant seedlings of Arabidopsis were incubated in culture medium containing (filled bar) or lacking (open bar) sucrose in the presence of E-64d for 1 d. They were stained with neutral red and photographed. As control experiments, root tips of the wild type were stained with neutral red immediately after excision (0 h, filled bar), or after incubation for 1 d in culture medium containing sucrose in the absence of E-64d (MeOH, filled bar). Autophagic scores were obtained as described in Materials and Methods. Numbers in the bars show the numbers of root tips examined in each treatment.
Discussion
Treatment of barley and Arabidopsis root tips with E-64d led to the accumulation of undegraded cytoplasmic components in newly formed autolysosomes and/or in the pre-existing central vacuole of root tip cells (Moriyasu et al. 2003 , Figs. 1–3 in this study). In contrast, in cultured tobacco cells treated with E-64c, parts of the cytoplasm accumulate in autolysosomes and little accumulation is observed in the central vacuoles (Moriyasu and Ohsumi 1996 ). It remains unclear why the sites of accumulation of cytoplasmic components are different in these cell types. However, Arabidopsis cells cultured in the absence of sucrose in the presence of E-64c accumulate parts of the cytoplasm in the central vacuole and the accumulation of autolysosomes is not observed (Y. Murayama, H. Kubo and Y. Moriyasu, unpublished results). These observations support the notion that the lytic compartments of autophagy in plant cells are autolysosomes and/or central vacuoles, and that the contribution of these two compartments is regulated differently depending on the cell type.
In contrast to such a complex situation in plant cells, where the two organelles, the central vacuoles and autolysosomes, are used as lytic compartments for autophagy, only one organelle appears to work as a lytic compartment in mammalian and yeast cells. Parts of the cytoplasm accumulate in autolysosomes in mammalian cells that are subjected to nutrient starvation in the presence of a cysteine protease inhibitor (Kominami et al. 1983 ), whereas they accumulate in the vacuole of yeast cells under starvation conditions in the presence of a serine protease inhibitor (Takeshige et al. 1992 ). In the present study, we examined the cells for the presence or absence of autophagy using a light microscope without differentiating autolysosomes from central vacuoles. How the contribution of these two organelles to autophagy is regulated will be a future issue in autophagy of plant cells.
In the atatg2 and atatg5 mutants, cytoplasmic components were accumulated only slightly even in the presence of E-64d ( Fig. 6 ). This suggests that AtATG2 and AtATG5 gene products are essential for autophagy in root tip cells as in yeast cells. In yeast cells, Atg5 protein binds to other Atg proteins, Atg12 and Atg16, and makes a large protein complex. This complex is localized to the possible precursory structure of an autophagosome called the pre-autophagosomal structure (PAS) (Suzuki et al. 2001 ), which is thought to work at the initiation step of an autophagosome formation. Atg2 is a peripheral membrane-associated protein, which is also localized to the PAS. Although whether plant cells possess a PAS is not yet known, it is reasonable to speculate that a PAS or a structure like a PAS is also formed in autophagy of plant root cells and that AtAtg2 and AtAtg5 proteins play essential roles in the PAS in the process of autophagy.
The atatg9 mutant showed less accumulation of cytoplasmic components in the presence of E-64d than the wild-type plant, but more than the atatg2 and atatg5 mutants. Thus, AtAtg9 protein may not contribute to autophagy in Arabidopsis root tip cells as much as the AtAtg2 and AtAtg5 proteins. Yeast Atg9 protein is an integral membrane protein with several membrane-spanning regions (Noda et al. 2000 ). It is localized to various structures existing in the peripheral region of the vacuole, including the PAS and mitochondria (Reggiori et al. 2004 , Reggiori et al. 2005 ). Although the significance of its localization to these structures is unclear, Atg9 is essential for the formation of autophagosomes, i.e. autophagy does not occur in yeast mutants lacking the ATG9 gene. Taken together, although root tip cells in Arabidopsis seem to use machinery for autophagy similar to that used in yeast cells, it is likely that the essentiality of some part in the machinery has been diverted.
Even in the atatg2 and atatg5 mutants, a very small amount of cytoplasmic components was recognized after incubation with E-64d ( Fig. 6 ). This suggests the presence of the transport of cytoplasmic components into vacuoles that are independent of ATG genes. Recently, an ER-resident membrane protein was reported to be constitutively transported to vacuoles by a mechanism different from autophagy, and degraded there (Tamura et al. 2004 ). Such transport may contribute to this significant accumulation of cytoplasmic materials in the presence of E-64d. Alternatively, the presence of ATG -independent autophagic pathways may contribute to such an accumulation.
Autophagy in Arabidopsis root cells appears to be enhanced under starvation conditions ( Fig. 7 in this study, Yoshimoto et al. 2004 ). This suggests that as in other organisms, autophagy in root cells contributes to provide substrates for respiration and molecules for the synthesis of new cellular components under starvation conditions. However, autophagy was found to occur even under nutrient-sufficient conditions in root cells, whereas it does not appear to occur under nutrient-sufficient conditions in tobacco suspension-cultured cells (Inoue and Moriyasu 2006 ). Since the genesis and enlargement of vacuoles occur constitutively in root tip cells, the result showing the constitutive occurrence of autophagy in root tip cells supports the notion that autophagy plays a role in these events in Euphorbia and Linus root tip cells (Amelunxen and Heinze 1984 , Marty 1978 ; for a review, see also Marty 1999 , Moriyasu and Hillmer 2000 ). Taken together, it is likely that plant root cells use autophagy for providing membranes for the enlargement of the central vacuole.
If autophagy contributes to the formation of vacuoles in root tip cells, some defects will be expected in vacuolar morphology in atg mutant plants. However, there were no obvious defects found in the morphology of the central vacuoles in the atatg7 and atatg9 mutants grown under nutrient-sufficient conditions (Doelling et al. 2002 , Hanaoka et al. 2002 ), whereas the central vacuole seemed smaller (Yoshimoto et al. 2004 ) and the growth of roots is slower (Doelling et al. 2002 , Yoshimoto et al. 2004 ) in the atatg4 and atatg7 mutants grown under nutrient-deprived conditions. Such phenotypes of the mutants were explained by the notion that autophagy contributes to the supply of nitrogen and carbon for continued growth when nutrient uptake is limiting (Doelling et al. 2002 ). However, we suppose that the slower growth of the root may also be caused by slower cell growth, which in turn originates from the defect of vacuole genesis and enlargement. A more detailed examination may clarify some defect in the vacuolar morphology of the mutant even under nutrient-sufficient conditions. To test this possibility, we are now closely examining the vacuolar morphology in root tip cells of atatg2 and atatg5 mutants.
Materials and Methods
Preparation and incubation of root tips
Arabidopsis thaliana (ecotype Columbia or Landsberg) was used. The seeds were sterilized for about 5 min with 5% (v/v) commercially available bleach. The seeds were rinsed with sterile water, and then sown on dishes containing 0.3% (w/v) Gelrite containing half-strength Murashige and Skoog (MS) salt mixture and 3% (w/v) sucrose. The dishes were kept at 4°C for 2–3 d, and then at 22 ± 2°C for 6–10 d under continuous light from fluorescent lamps. Root tips (4–6 in number, 8–12 mm in length) excised from the seedlings were incubated in 2 ml of liquid culture medium consisting of half-strength MS salt mixture and 3% (w/v) sucrose at 22 ± 2°C in the dark under agitation at 100 r.p.m. In sucrose-free culture medium, sucrose was replaced with mannitol to keep the osmotic value constant.
Barley (cv. Doriru) seedlings were prepared as described previously (Moriyasu et al. 2003 ). Root tips (about 10 mm long) were incubated in medium consisting of 1× MS salt mixture and 3% (w/v) sucrose, at 25 ± 2°C with rotation of 100 r.p.m. as described previously (Moriyasu et al. 2003 ).
Neutral red and LysoTracker Red staining of Arabidopsis root tips
Whole root tips were stained with 0.01% (w/v) neutral red in 50 mM Mes-Tris (pH 6.5) for about 2 min and their bright field images were observed under a light microscope (OptiPhoto, Nikon). Negative color film (Fujicolor, ISO400, Fuji) photographs were taken using a photomicrographic camera (Microflex UFX-II, Nikon) through 40× or 20× objective lenses and 2.5× or 5× projection lenses. Alternatively, whole root tips were stained with 1 μM LysoTracker Red (Molecular Probes) as described (Moriyasu et al. 2003 ), and their fluorescence and Nomarski images were observed using a confocal microscope (LSM510, Zeiss).
Sectioning of root tips
Root tips of Arabidopsis were fixed with 2% (w/v) glutaraldehyde and 1% (w/v) formaldehyde in 100 mM sodium cacodylate-HCl buffer (pH 6.9) at room temperature for 1 h, and then at 4°C overnight, and post-fixed with 1% (w/v) osmium tetroxide for 2 h at room temperature. They were then dehydrated using a series of ethanol and propylene oxide, and embedded in Spurr's resin. At the 50% ethanol stage of dehydration, they were en bloc stained with 1% (w/v) uranyl acetate in 50% ethanol for 1 h at room temperature. For light microscopy, sections of 1 μm thickness were made from the blocks of Spurr's resin, and stained with toluidine blue; for electron microscopy (H-7000, Hitachi, Japan), thin sections of 60–80 nm were made and stained with uranyl acetate and lead citrate.
Root tips from barley were fixed with 2.5% (w/v) glutaraldehyde in Millonig's phosphate buffer consisting of 100 mM Pi-Na (pH 7.4) and 86 mM NaCl, and post-fixed with 1% (w/v) osmium tetroxide. They were embedded in Spurr's resin in the same way as Arabidopsis root tips, but the sections were stained with a mixture (2 : 1) of 4% (w/v) uranyl acetate and 1% (w/v) KMnO 4 (Moriyasu et al. 2003 ).
Isolation of atg mutant plants
The seeds of atatg2, atatg5 and atatg9 mutants were obtained from the Arabidopsis Biological Resource Center (Columbus, OH, USA). Genomic DNA was isolated from the leaves of 6- to 10-day-old seedlings using a DNA isolation kit (Isoplant II, Nippon Gene, Toyama, Japan). The homozygous plants with respect to atatg2-1, atatg5-3 and atatg9-2 mutation were selected by PCR according to the Protocol for SIGnAL T-DNA Verification Primer Design from Salk Institute Genomic Analysis Laboratory (Alonso et al. 2003 ). The primers used are 5′-GCGTGGACCGCTTGCTGCAACT-3′ designated as LBb1, and 5′-TCCTACGGCCGAGATCGTACA-3′ and 5′-GCACTTTCCATCAGCTACTCGC-3′ for verifying the atatg2-1 mutant, 5′-GAGCATGAGATTCATCTCCACTT-3′ and 5′-TGCGTGCCAAACAAATAGCAA-3′ for the atatg5-3 mutant, or 5′-GGTGATTTGCATTGTGGATGC-3′ and 5′-TTTCCGCATAAGGCGCATAAC-3′ for the atatg9-2 mutants. The homozygous plants with respect to the atatg5-1 mutation were selected by PCR according to Sessions et al. ( 2002 ) using the primers 5′-TAGCATCTGAATTTCATAACCAATCTCGATACAC-3′ designated as LB3, and 5′-ATTCACTTCCTCCTGGTGAAG-3′ and 5′-TTGTGCCTGCAGGATAAGCG-3′.
Isolation of RNA and RT–PCR
RNA was isolated from the leaves and roots of 10- to 14-day-old Arabidopsis seedlings using an RNA isolation kit (AquaPure RNA Isolation Kit, Bio-Rad, CA, USA, or RNeasy Plant Mini Kit, Qiagen, Hilden, Germany). The first-strand cDNA was synthesized by the Omniscript reverse transcriptase (Qiagen, Hilden, Germany) or SuperScript First-Strand Synthesis System for RT–PCR (Invitrogen, CA, USA) using the oligo(dT) 15 primer. PCR was performed using these cDNAs as templates. The primers used were 5′-CATCTAGCCTTGCTTCGGGGT-3′ (primer 1 in Fig. 5 ) and 5′-GCTGCCATACGCGATAGCCAA-3′ (primer 2) for the amplification of AtATG2 , 5′-ATGGCGAAGGAAGCGGTCAA-3′ (primer 3) and 5′-TCACCTTTGAGGAGCTTTCACAAG-3′ (primer 4) or 5′-GGAAGGAGCAATTCCTCTGC-3′ (primer 7) and 5′-GAGGTCCATAGATCCTCTTG-3′ (primer 8) for AtATG5 , 5′-GGGTCGACCATGAGCAGTGGGCATAAG-3′ (primer 5) and 5′-GGAGAACATGTACCGTAATGTGGTGCTTG-3′ (primer 6) for AtATG9 , and 5′-CAAAGAGCTCTTCTTCTTCAC-3′ and 5′-ACCACCACGGAGCCTGA-3′ or 5′-TTCACTTGGTCCTGCGTCTTCGTGGTGGTTTC-3′ and 5′-CGAAGCGATGATAAAGAAGAAGTTCGACTTG-3′ for UBQ10 .
Quantification of autophagy in the root tip cells of Arabidospsis
Color photographs of parts of roots, 800–1,200 μm distant from the tips, were digitalized using a scanner (GT-9300UF, Epson, Tokyo, Japan). From each image, a rectangular region (150 μm in length × 50 μm in width) was cropped and converted to a 8-bit grayscale image using the computer software Photoshop (version 7.0, Adobe). The resultant image was then converted to binary using Photoshop by defining a grayscale cut-off point, which was set as the lowest grayscale value of the cytoplasm in the rectangle. In this conversion, grayscale values below the cut-off became pure black and those above became pure white. The ratio of the number of pure black pixels to all the number of pixels in the rectangle was calculated using the software Image J (version 3.11, NIH, Bethesda, MD, USA), and defined as the autophagic score.
Supplementary material
Supplementary material mentioned in the article is available to online subscribers at the journal website www.pcp.oxfordjournals.org .
Acknowledgments
We thank Dr. Tetsuro Mimura for providing barley seeds. We also thank Dr. John Rogers for critical reading of the manuscript and helpful comments. This work was supported in part by ISAS Grant for Basic Biology Study oriented to utilization of Space station, and ‘Ground-based Research Announcement for Space Utilization’ promoted by Japan Space Forum to Y. M.
References
Abbreviations:
- ER
endoplasmic reticulum
- GFP
green fluorescent protein
- 3-MA
3-methyladenine
- MS
Murashige and Skoog
- PAS
pre-autophagosomal structure
- RT–PCR
reverse transcription–PCR.
Author notes
These authors contributed equally to this work.